Molecular Identification of Culicoides Species and Host Preference Blood Meal in the African Horse Sickness Outbreak-Affected Area in Hua Hin District, Prachuap Khiri Khan Province, Thailand
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
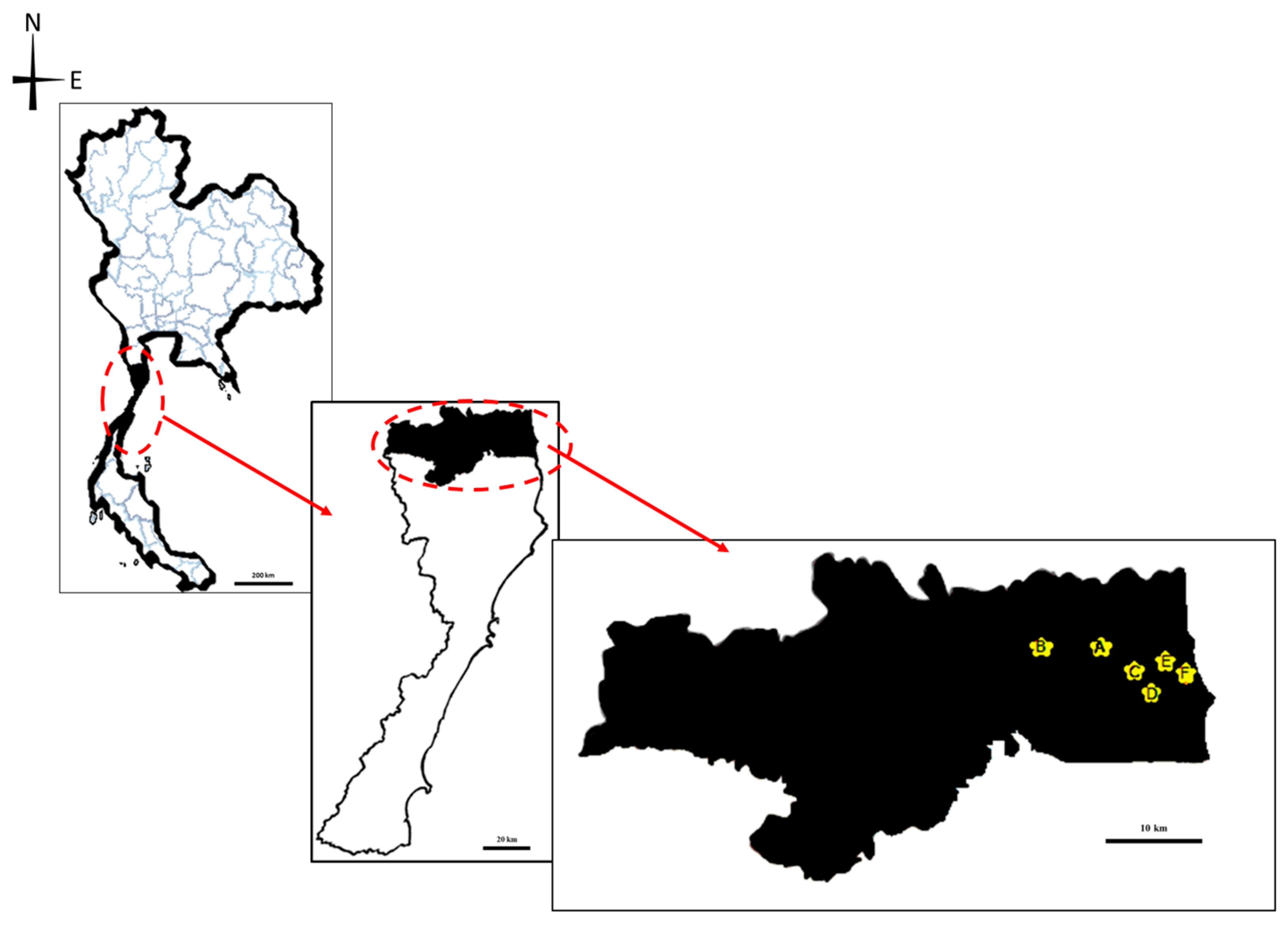
2.1. Study Design and Data Collection
2.2. Sample Collection and Identification
2.3. Blood Meal Identification
2.4. Sequencing and Bioinformatics Analysis
3. Results
3.1. Culicoides Identified in Horse Farms
3.2. Blood Meal Identification in Fully Fed Culicoides in Hua Hin District, Prachuab Khiri Khan Province, Thailand
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- van Rijn, P.A. African Horse Sickness Virus. In Reference Module in Life Science; Elsevier: Amsterdam, The Netherlands, 2019; pp. 1–5. [Google Scholar] [CrossRef]
- Mullen, G.R.; Murphree, C.S. Chapter 13 Biting Midges (Ceratopogonidae). In Medical and Veterinary Entomology; Academic Press: Cambridge, MA, USA, 2016; pp. 213–235. [Google Scholar]
- Coxon, C.; Bowen, J.; Gauntlett, F.; Stephan, L. Updated Situation Assessment: African Horse Sickness in Thailand#3. May 07, 2020. Department for Environment, Food and Rural Affairs, Animal and Plant Health Agency, Advice Services-International Diseases Monitoring. 4 Pages. 2020. Available online: https://www.gov.uk/government/collections/animal-diseases-internationalmonitoring (accessed on 20 December 2022).
- King, S.; Rajko-Nenow, P.; Ashby, M.; Frost, L.; Carpenter, S.; Batten, C. Outbreak Alerts: Outbreak of African horse sickness in Thailand, 2020. Transbound. Emerg. Dis. 2020, 67, 1764–1767. [Google Scholar] [CrossRef]
- Bunpapong, N.; Charoenkul, K.; Nasamran, C.; Chamsai, E.; Udom, K.; Boonyapisitsopa, S.; Tantilertcharoen, R.; Kesdangsakonwut, S.; Techakriengkrai, N.; Suradhat, S.; et al. African Horse Sickness Virus Serotype 1 on Horse Farm, Thailand, 2020. Emerg. Infect. Dis. 2021, 27, 2208–2211. [Google Scholar] [CrossRef]
- Vintzel, L.W. Interview: Coordinating a Regional Response to African Horse Sickness OIE. 2021. Available online: https://www.report2020oie.fr/en/a-regional-response-to-african-horse-sickness/ (accessed on 20 December 2022).
- Sunantaraporn, S.; Hortiwakul, T.; Kraivichian, K.; Siriyasatien, P.; Brownell, N. Molecular Identification of Host Blood Meals and Detection of Blood Parasites in Culicoides Latreille (Diptera: Ceratopogonidae) Collected from Phatthalung Province, Southern Thailand. Insects 2022, 13, 912. [Google Scholar] [CrossRef]
- Garros, C.; Gardès, L.; Allène, X.; Rakotoarivony, I.; Viennet, E.; Rossi, S.; Balenghien, T. Adaptation of species-specific multiplex PCR assay for the identification of blood meal source in Culicoides (Ceratopogonidae: Diptera): Applications on Palaearctic biting midge species, vector of Orbiviruses. Infect. Genet. Evol. 2012, 11, 1103–1110. [Google Scholar] [CrossRef]
- Martinez-de la Puente, J.; Figuerola, J.; Soriguer, R. Fur or feather? Feeding preferences of species of Culicoides biting midges in Europe. Trends Parasitol. 2015, 31, 16–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ninio, C.; Augot, D.; Delecolle, J.C.; Dufour, B.; Depaquit, J. Contribution to the knowledge of Culicoides (Diptera: Ceratopogonidae) host preferences in France. Parasitol. Res. 2011, 108, 657–663. [Google Scholar] [CrossRef]
- Lassen, S.B.; Nielsen, S.A.; Kristensen, M. Identity and diversity of blood meal hosts of biting midges (Diptera: Ceratopogonidae: Culicoides Latreille) in Denmark. Parasites Vectors 2012, 5, 143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thepparat, A.; Tauruishi, T.; Ketawal, C. Species Diversity and Abundance of Culicoides (Diptera: Ceratopogonidae) in Sakaew Province. Ramkhamhaeng Res. J. Sci. Technol. 2012, 15, 65–80. [Google Scholar]
- Thepparat, A.; Bellis, G.; Ketavan, C.; Ruangsitichai, J.; Sumruayphol, S.; Apiwathnasorn, C. Ten species of Culicoides Latreille (Diptera: Ceratopogonidae) newly recorded from Thailand. Zootaxa 2015, 4033, 48–56. [Google Scholar] [CrossRef] [Green Version]
- Satheesha, S.P.; Udupa, K.G.; Appannavar, M.M.; Labuschagne, K. A study on Culicoides midges associated with buffaloes. Buffalo Bull. 2014, 33, 400–406. [Google Scholar]
- Jomkumsing, P.; Surapinit, A.; Saengpara, T.; Pramual, P. Genetic variation, DNA barcoding and blood meal identification of Culicoides Latreille biting midges (Diptera: Ceratopogonidae) in Thailand. Acta Trop. 2021, 217, 105866. [Google Scholar] [CrossRef] [PubMed]
- Pramual, P.; Jomkumsing, P.; Jumpato, W.; Bunauea, S. Molecular detection of avain haemosporidian parasites in biting midges (Diptera: Ceratopogonidae) from Thailand. Acta Trop. 2021, 224, 106118. [Google Scholar] [CrossRef] [PubMed]
- Sunantaraporn, S.; Thepparat, A.; Phumee, A.; Sor-Suwan, S.; Boonserm, R.; Bellis, G.; Siriyasatien, P. Culicoides Latreille (Diptera: Ceratopogonidae) as potential vectors for Leishmania martiniquensis and Trypanosoma sp. in northern Thailand. PLoS Negl. Trop. Dis. 2021, 15, e0010014. [Google Scholar] [CrossRef] [PubMed]
- Choocherd, S.; Pattanatanang, K.; Chimnoi, W.; Kamyingkird, K.; Tongyoo, P.; Phasuk, J. Preliminary study on comparative efficacy of four light sources for trapping Culicoides spp. (Diptera: Ceratopogonidae) in Prachuab Khiri Khan Province, Thailand. J. Econ. Entomol. 2022, 115, 1719–1723. [Google Scholar] [CrossRef] [PubMed]
- Pramual, P.; Jomkumsing, P.; Wongpakam, K.; Vaisusuk, K.; Chatan, W.; Gomontean, B. Population genetic structure and population history of the biting midge Culicoides mahasarakhamense (Diptera: Ceratopogonidae). Insects 2022, 13, 724. [Google Scholar] [CrossRef] [PubMed]
- Burkett, D.A.; Butler, J.F.; Kline, D.L. Field evaluation of colored light-emitting diodes as attractants for woodland mosquitoes and other diptera in north central Florida. J. Am. Mosq. Control Assoc. 1998, 14, 186–195. [Google Scholar]
- Wirth, W.W.; Hubert, A.A. The Culicoides of Southeast Asia (Diptera: Ceratopogonidae). Mem. Am. Entomol. Soc. 1989, 44, 1–508. [Google Scholar]
- Dyce, A.L.; Bellis, G.A.; Muller, M.J. Pictorial Atlas of Australasian Culicoides Wings (Diptera: Ceratopogonidae); Australian Biological Resources Study: Canberra, Australia, 2007. [Google Scholar]
- Bellis, G.A.; Halling, L.; Anderson, S.J. Pictorial key to adult female Culicoides Latreille, 1809 (Diptera: Ceratopogonidae) from the Northern Territory, Western Australia and South Australia. Austral Entomol. 2015, 54, 28–59. [Google Scholar] [CrossRef]
- Hakima, B.; Hwang, H.S.; Lee, K.Y. Molecular identification of Culicoides (Diptera: Ceratopogonidae) species in Algeria. Acta Trop. 2020, 202, 105261. [Google Scholar] [CrossRef]
- Okonechnikov, K.; Golosova, O.; Fursov, M.; The UGENE Team. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef] [Green Version]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA 11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Thepparat, A.; Boonkerd, S. Ecological studies of Ceratopogonid in Chonburi Province. Ramkhamhaeng Res. J. Sci. Technol. 2009, 12, 11–21. [Google Scholar]
- Fall, M.; Fall, A.G.; Seck, M.T.; Bouyer, J.; Diarra, M.; Balenghien, T.; Garros, C.; Bakhoum, M.T.; Faye, O.; Baldet, T.; et al. Circadian activity of Culicoides oxystoma (Diptera: Ceratopogonidae), potential vector of bluetongue and African horse sickness viruses in the Niayes area, Senegal. Parasitol. Res. 2015, 114, 3151–3158. [Google Scholar] [CrossRef] [Green Version]
- Bakhoum, M.T.; Fall, M.; Seck, M.T.; Gardes, K.; Fall, A.G.; Dio, M.; Mall, I.; Balenghien, T.; Baldet, T.; Gimonneau, G.; et al. Foraging range of arthropods with veterinary interest: New insights for Afrotropical Culicoides biting midges (Diptera: Ceratopogonidae) using the ring method. Acta Trop. 2016, 157, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Hadj-Henni, L.; De Meulemeester, T.; Depaquit, J.; Noël, P.; Germain, A.; Helder, R.; Augot, D. Comparison of vertebrate cytochrome b and prepronociceptin for blood meal analyses in Culicoides. Front. Vet. Sci. 2015, 2, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martínez-de la Puente, J.; Martínez, J.; Ferraguti, M.; Morales-de la Nuez, A.; Castro, N.; Figuerola, J. Genetic characterization and molecular identification of the bloodmeal sources of the potential bluetongue vector Culicoides obsoletus in the Canary Islands, Spain. Parasites Vectors 2012, 5, 147. [Google Scholar] [CrossRef] [Green Version]
- Slama, D.; Haouas, N.; Mezhoud, H.; Babba, H.; Chaker, E. Blood Meal Analysis of Culicoides (Diptera: Ceratopogonidae) in Central Tunisia. PLoS ONE 2015, 10, e0120528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tomazatos, A.; Jost, H.; Schulze, J.; Spinu, M.; Schmidt-Chanasit, J.; Cadar, D.; Lukken, R. Blood-meal analysis of Culicoides (Diptera: Ceratopogonidae) reveals a broad host range and new species records for Romania. Parasites Vectors 2020, 13, 79. [Google Scholar] [CrossRef] [PubMed]
- Vasic, A.; Zdravkovic, N.; Anita, D.; Bojkovski, J.; Marinov, M.; Mathias, A.; Niculaua, M.; Oslobanu, E.L.; Pavlovic, I.; Petric, D.; et al. Species diversity, host preference and arbovirus detection of Culicoides (DipteraL Ceratopogonidae) in south-eastern Serbia. Parasites Vectors 2019, 12, 61. [Google Scholar] [CrossRef] [Green Version]
- Baum, M.; de Castro, E.A.; Pinto, M.C.; Goulart, T.M.; Baura, W.; Klisiowicz Ddo, R.; Vieira da Costa-Ribeiro, M.C. Molecular detection of the blood meal source of sand flies (Diptera: Psychodidae) in a transmission area of American cutaneous leishmaniasis, Paraná State, Brazil. Acta Trop. 2015, 143, 8–12. [Google Scholar] [CrossRef]
- Bartsch, S.; Bauer, B.; Wiemann, A.; Clausen, P.H.; Steuber, S. Feeding patterns of biting midges of the Culicoides obsoletus and Culicoides pulicaris groups on selected farms in Brandenburg, Germany. Parasitol. Res. 2009, 105, 373–380. [Google Scholar] [CrossRef] [PubMed]




| Horse Farm | Number of Collected Female Culicoides | ||
|---|---|---|---|
| Position A (Near the Horse <2 m) (%) | Position B (Far from the Horse >5 m) (%) | Total (%) | |
| A | 289 (85.5%) | 49 (56%) | 338 (33.5%) |
| B | 125 (92.6%) | 10 (4.7%) | 135 (13.4%) |
| C | 3 (75.0%) | 1 (25.0%) | 4 (0.40%) |
| D | 227 (52.7%) | 204 (47.3%) | 431 (42.8%) |
| E | 48 (82.8%) | 10 (17.2%) | 58 (5.8%) |
| F | 16 (38.1%) | 26 (61.9%) | 42 (4.2%) |
| Total | 708 (70.2%) | 300 (29.8%) | 1008 (100%) |
| Culicoides Species | Horse Farm A * | Horse Farm B * | Horse Farm C * | Horse Farm D * | Horse Farm E * | Horse Farm F | Total | (%) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Position A | Position B | Position A | Position B | Position A | Position B | Position A | Position B | Position A | Position B | Position A | Position B | |||
| C. oxystoma | 264 | 40 | 30 | 2 | 1 | 1 | 204 | 91 | 48 | 9 | 15 | 20 | 725 | (71.92%) |
| C. imicola | 12 | 3 | 77 | 5 | 1 | 0 | 14 | 92 | 0 | 0 | 1 | 1 | 206 | (20.44%) |
| C. actoni | 5 | 2 | 4 | 0 | 0 | 0 | 5 | 3 | 0 | 0 | 0 | 4 | 23 | (2.28%) |
| C. flavipunctatus | 6 | 1 | 1 | 0 | 0 | 0 | 1 | 11 | 0 | 0 | 0 | 0 | 20 | (1.98%) |
| C. asiana | 0 | 1 | 8 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 10 | (0.99%) |
| C. peregrinus | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 6 | (0.60%) |
| C. huffi | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 2 | 0 | 1 | 0 | 0 | 6 | (0.60%) |
| C. brevatarsis | 0 | 0 | 0 | 2 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 4 | (0.40%) |
| C. innoxius | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | (0.30%) |
| C. histrio | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 3 | (0.30%) |
| C. minimus | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | (0.10%) |
| C. geminus | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | (0.10%) |
| Total | 338 | 135 | 4 | 431 | 58 | 42 | 1008 | (100.00%) | ||||||
| Culicoides Species/Host Species |  Equus caballus Position A/B (%) |  Canis lupus familiaris Position A/B (%) |  Sus scrofa Position A/B (%) |  Homo sapiens Position A/B (%) | ||||
|---|---|---|---|---|---|---|---|---|
| C. oxystoma | 15/10 | (36.23%) | 2/1 | (60.00%) | 2/ND | (66.67%) | 2/ND | (66.67%) |
| C. peregrinus | 2/1 | (4.35%) | ND | ND | ND | |||
| C. actoni | 7/7 | (20.29%) | ND | ND | ND | |||
| C. flavipunctatus | 4/5 | (13.04%) | ND | ND | ND | |||
| C. imicola | 7/3 | (14.49%) | 1/ND | (20.00%) | ND/1 | (33.33%) | ND/1 | (33.33%) |
| C. brevatarsis | 1/ND | (1.45%) | ND/1 | (20.00%) | ND | ND | ||
| C. innoxius | 3/ND | (4.35%) | ND | ND | ND | |||
| C. histrio | ND/2 | (2.90%) | ND | ND | ND | |||
| C. arakawae | ND/1 | (1.45%) | ND | ND | ND | |||
| C. geminus | 1/ND | (1.45%) | ND | ND | ND | |||
| Total (80 females Culicoides) | 40/29 | (86.25%) | 3/2 | (6.25%) | 2/1 | (3.75%) | 2/1 | (3.75%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kamyingkird, K.; Choocherd, S.; Chimnoi, W.; Klinkaew, N.; Kengradomkij, C.; Phoosangwalthong, P.; Thammasonthijarern, N.; Pattanatanang, K.; Inpankaew, T.; Phasuk, J.; et al. Molecular Identification of Culicoides Species and Host Preference Blood Meal in the African Horse Sickness Outbreak-Affected Area in Hua Hin District, Prachuap Khiri Khan Province, Thailand. Insects 2023, 14, 369. https://doi.org/10.3390/insects14040369
Kamyingkird K, Choocherd S, Chimnoi W, Klinkaew N, Kengradomkij C, Phoosangwalthong P, Thammasonthijarern N, Pattanatanang K, Inpankaew T, Phasuk J, et al. Molecular Identification of Culicoides Species and Host Preference Blood Meal in the African Horse Sickness Outbreak-Affected Area in Hua Hin District, Prachuap Khiri Khan Province, Thailand. Insects. 2023; 14(4):369. https://doi.org/10.3390/insects14040369
Chicago/Turabian StyleKamyingkird, Ketsarin, Suchada Choocherd, Wissanuwat Chimnoi, Nutsuda Klinkaew, Chanya Kengradomkij, Pornkamol Phoosangwalthong, Nipa Thammasonthijarern, Khampee Pattanatanang, Tawin Inpankaew, Jumnongjit Phasuk, and et al. 2023. "Molecular Identification of Culicoides Species and Host Preference Blood Meal in the African Horse Sickness Outbreak-Affected Area in Hua Hin District, Prachuap Khiri Khan Province, Thailand" Insects 14, no. 4: 369. https://doi.org/10.3390/insects14040369







