A Protein Asteroid with PIN Domain in Silkworm Bombyx mori Is Involved in Anti-BmNPV Infection
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Insect Rearing and Microorganism
2.2. Sequence Analysis of BmAst Gene
2.3. Quantitative Real-Time Polymerase Chain Reaction
2.4. Tissue Distribution and Developmental Profiles of BmAst Gene
2.5. Different Microbial Infections
2.6. dsRNA Synthesis
2.7. Expressions of BmAst Gene after dsEGFP Injection
2.8. RNAi Assays
2.9. Statistic Analysis
3. Results
3.1. Sequence Analysis of BmAst Gene and Its Predicted Protein
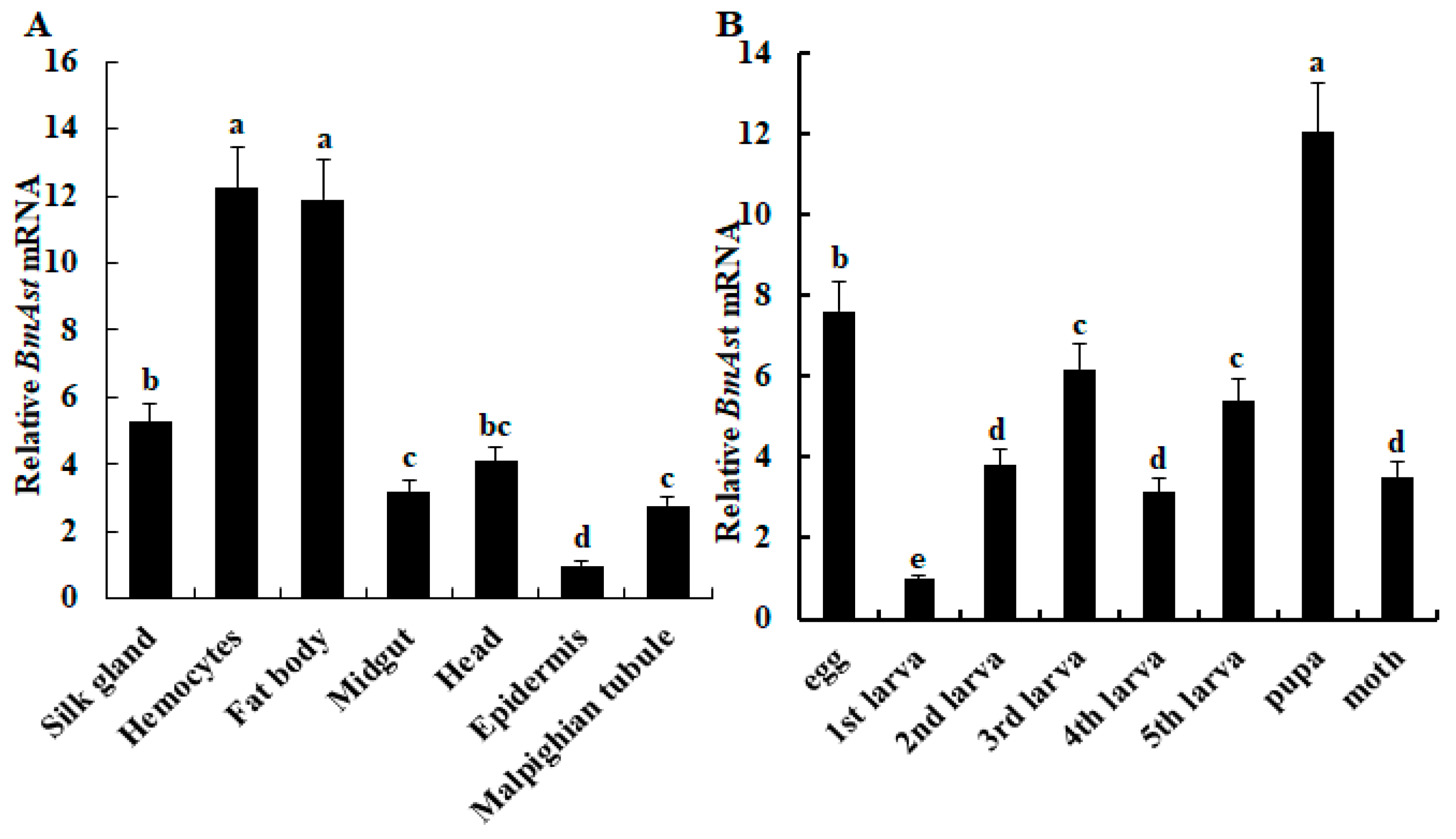
3.2. Tissues Distribution and Developmental Expression of BmAst
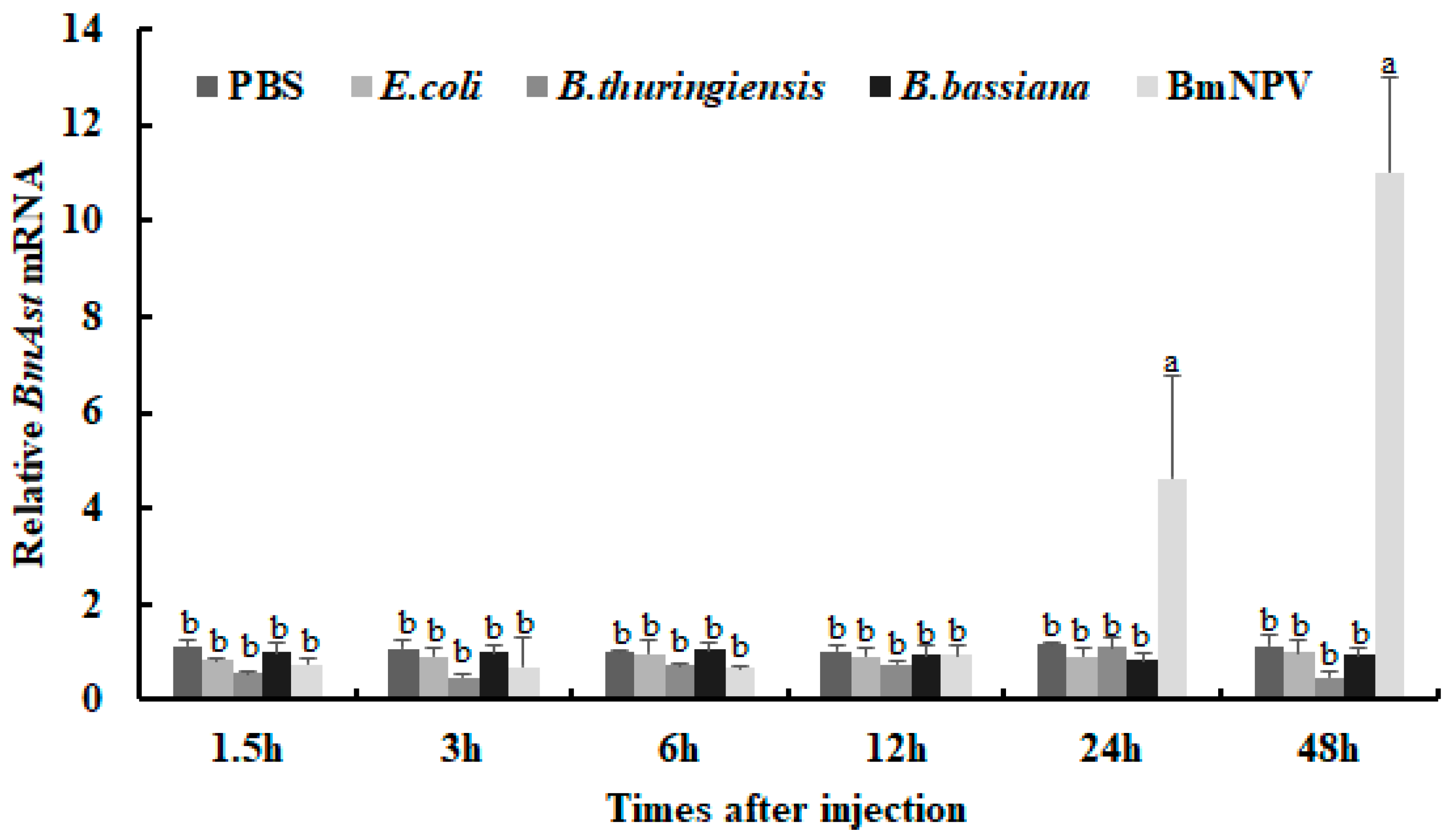
3.3. BmAst Genes Respond to Different Pathogenic Microorganisms
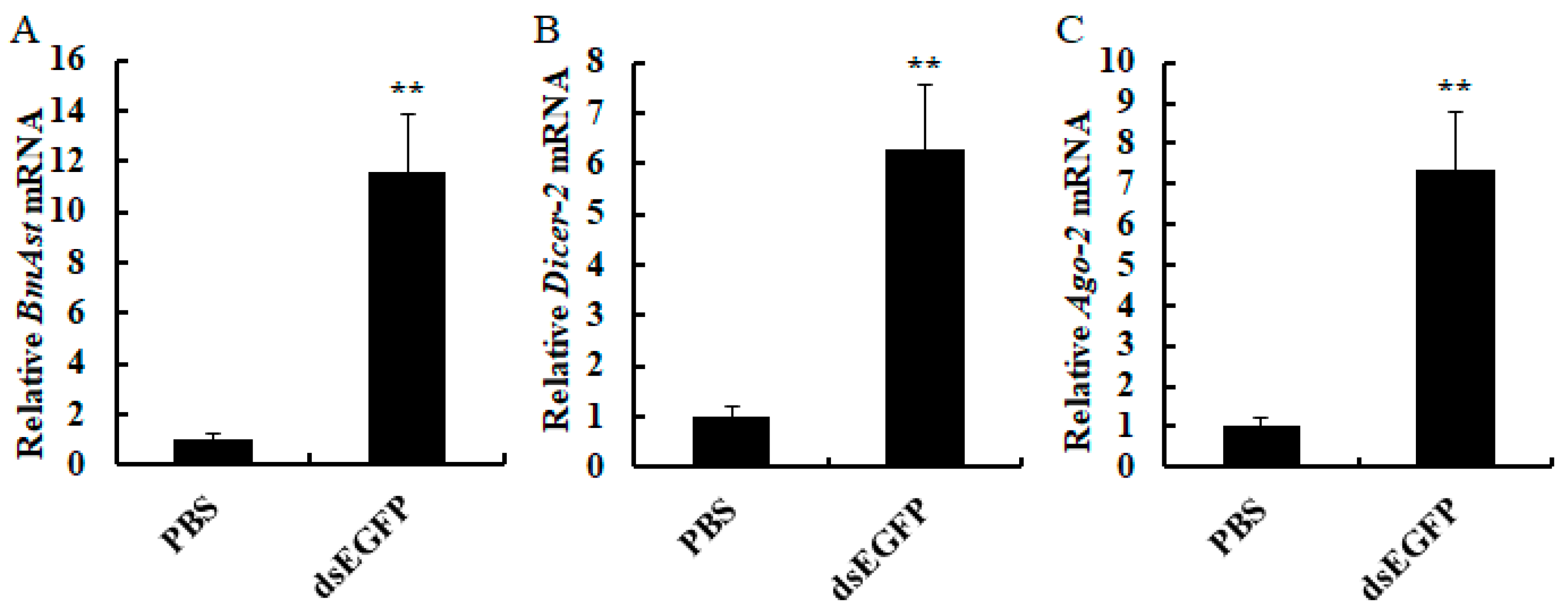
3.4. The Effect of dsRNA on BmAst Expression
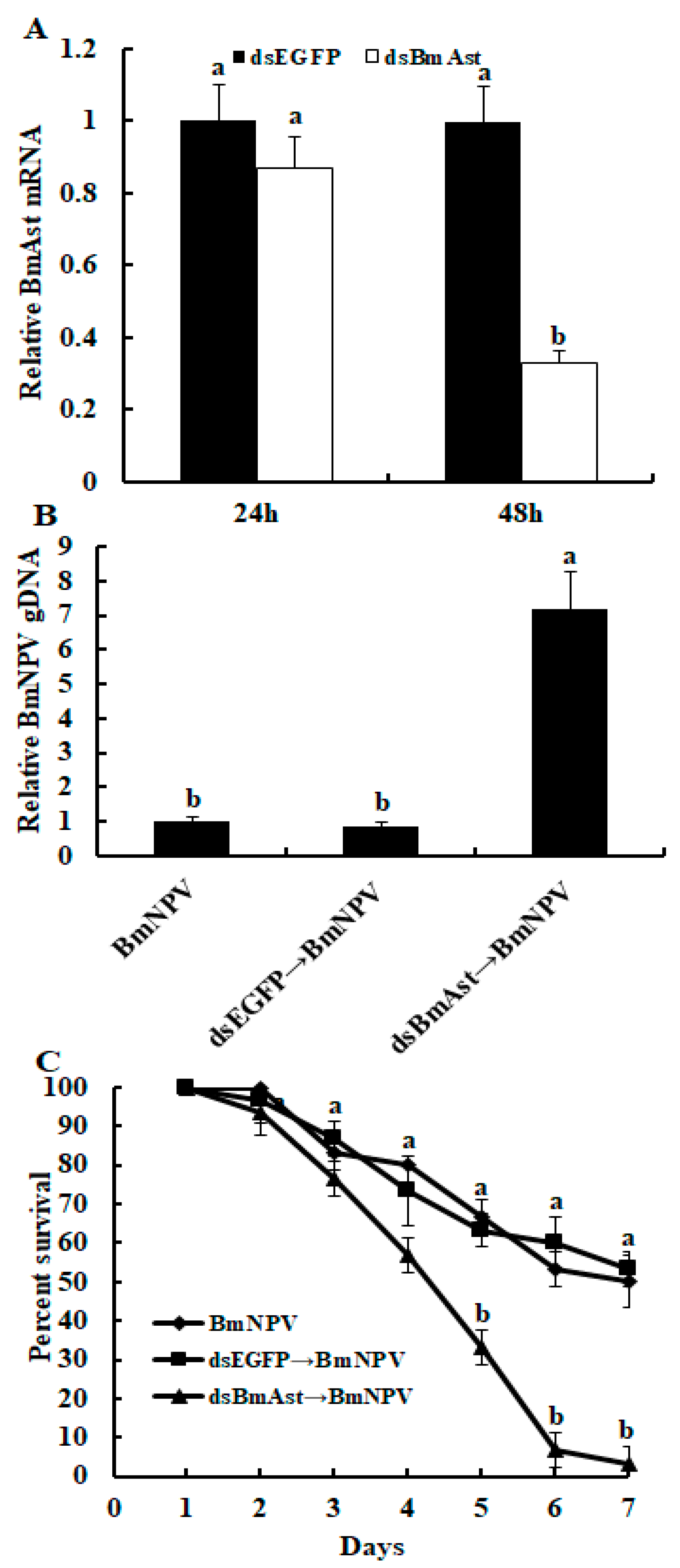
3.5. Knocking down of BmAst Gene Enhances BmNPV Replication in Silkworm
4. Discussion
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Matelska, D.; Steczkiewicz, K.; Ginalski, K. Comprehensive classification of the PIN domain-like superfamily. Nucleic Acids Res. 2017, 45, 6995–7020. [Google Scholar] [CrossRef]
- Siwecka, M.A. Purification and some properties of a novel dsRNA degrading nuclease bound to rye germ ribosomes. Acta Biochim. Pol. 1997, 44, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Arimatsu, Y.; Kotani, E.; Sugimura, Y.; Furusawa, T. Molecular characterization of a cDNA encoding extracellular dsRNase and its expression in the silkworm, Bombyx mori. Insect Biochem. Mol. 2007, 37, 176–183. [Google Scholar] [CrossRef] [PubMed]
- Wynant, N.; Santos, D.; Verdonck, R.; Spit, J.; Van Wielendaele, P.; Vanden Broeck, J. Identification, functional characterization and phylogenetic analysis of double stranded RNA degrading enzymes present in the gut of the desert locust, Schistocerca gregaria. Insect Biochem. Mol. 2014, 46, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Spit, J.; Philips, A.; Wynant, N.; Santos, D.; Plaetinck, G.; Broeck, J.V. Knockdown of nuclease activity in the gut enhances RNAi efficiency in the Colorado potato beetle, Leptinotarsa decemlineata, but not in the desert locust, Schistocerca gregaria. Insect Biochem. Mol. 2017, 81, 103–116. [Google Scholar]
- Fan, Y.H.; Song, H.F.; Abbas, M.; Wang, Y.L.; Li, T.; Ma, E.B.; Cooper, A.M.W.; Silver, K.; Zhu, K.Y.; Zhang, J.Z. A dsRNA-degrading nuclease (dsRNase2) limits RNAi efficiency in the Asian corn borer (Ostrinia furnacalis). Insect Sci. 2020, 28, 1677–1689. [Google Scholar] [CrossRef]
- Cooper, A.M.W.; Song, H.; Shi, X.; Yu, Z.; Lorenzen, M.; Silver, K.; Zhang, J.; Zhu, K.Y. Molecular Characterizations of Double-Stranded RNA Degrading Nuclease Genes from Ostrinia nubilalis. Insects 2020, 11, 652. [Google Scholar] [CrossRef]
- Prentice, K.; Smagghe, G.; Gheysen, G.; Christians, O. Nuclease activity decreases the RNAi response in the sweetpotato weevil Cylas puncticollis. Insect Biochem. Mol. 2019, 110, 80–89. [Google Scholar]
- Peng, Y.; Zhu, G.-H.; Wang, K.; Chen, J.; Liu, X.; Wu, M.; Zhao, C.; Xiao, H.; Palli, S.R.; Han, Z. Knockout of SldsRNase1 and SldsRNase2 revealed their function in dsRNA degradation and contribution to RNAi efficiency in the tobacco cutworm, Spodoptera litura. J. Pest Sci. 2021, 94, 1449–1460. [Google Scholar]
- Glavan, F.; Behm-Ansmant, I.; Izaurralde, E.; Conti, E. Structures of the PIN domains of SMG6 and SMG5 reveal a nuclease within the mRNA surveillance complex. EMBO J. 2006, 25, 5117–5125. [Google Scholar]
- Arcus, V.L.; Backbro, K.; Roos, A.; Daniel, E.L.; Baker, E.N. Distant structural homology leads to the functional characterization of an archaeal PIN domain as an exonuclease. J. Biol. Chem. 2004, 279, 16471–16478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arcus, V.L.; McKenzie, J.L.; Robson, J.; Cook, G.M. The PIN-domain ribonucleases and the prokaryotic VapBC toxin-antitoxin array. Protein Eng. Des. Sel. 2011, 24, 33–40. [Google Scholar] [CrossRef] [Green Version]
- Guan, R.B.; Li, H.C.; Fan, Y.J.; Hu, S.R.; Christiaens, O.; Smagghe, G.; Miao, X.X. A nuclease specific to lepidopteran insects suppresses RNAi. J. Biol. Chem. 2018, 293, 6011–6021. [Google Scholar]
- Meng, X.; Zhu, F.F.; Chen, K.P. Silkworm: A promising model organism in life science. J. Insect Sci. 2017, 7, 97. [Google Scholar]
- Yang, F.Y.; Zhang, Z.J.; Hu, B.; Yu, Y.; Tan, A.J. A CCCH zinc finger gene regulates doublesex alternative splicing and male development in Bombyx mori. Insect Sci. 2021, 28, 1253–1261. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.Q.; Li, Z.Q.; Xu, J.; Zeng, B.S.; Ling, L.; You, L.; Chen, Y.Z.; Huang, Y.P.; Tan, A.J. The CRISPR/Cas System mediates efficient genome engineering in Bombyx mori. Cell Res. 2013, 23, 1414–1416. [Google Scholar] [CrossRef] [Green Version]
- Toufeeq, S.; Wang, J.; Zhang, S.Z.; Li, B.; Hu, P.; Zhu, L.B.; You, L.L.; Xu, J.P. Bmserpin2 is involved in BmNPV infection by suppressing melanization in Bombyx mori. Insects 2019, 10, 399. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Zhang, Y.; Fei, S.; Awais, M.M.; Zheng, H.; Feng, M.; Sun, J. Heat Shock Protein 75 (TRAP1) facilitate the proliferation of the Bombyx mori nucleopolyhedrovirus. Int. J. Biol. Macromol. 2021, 175, 372–378. [Google Scholar] [CrossRef]
- Jiang, L.; Xie, E.Y.; Guo, H.Z.; Sun, Q.; Liuli, H.Y.; Wang, Y.M.; Li, Q.; Xia, Q.Y. Heat shock protein 19.9 (Hsp19.9) from Bombyx mori is involved in host protection against viral infection. Dev. Comp. Immunol. 2021, 114, 103790. [Google Scholar]
- Jiang, L.; Cheng, T.C.; Zhao, P.; Yang, Q.; Wang, G.H.; Jin, S.K.; Lin, P.; Xiao, Y.; Xia, Q.Y. Resistance to BmNPV via Overexpression of an Exogenous Gene Controlled by an Inducible Promoter and Enhancer in Transgenic Silkworm, Bombyx mori. PLoS ONE 2012, 7, e41838. [Google Scholar] [CrossRef] [Green Version]
- Guo, H.Z.; Xu, G.W.; Wang, B.B.; Xia, F.; Sun, Q.; Wang, Y.M.; Xie, E.Y.; Lu, Z.Y.; Jiang, L.; Xia, Q.Y. Phosphoenolpyruvate carboxykinase is involved in antiviral immunity against Bombyx mori nucleopolyhedrovirus. Dev. Comp. Immunol. 2019, 92, 193–198. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Wang, G.H.; Cheng, T.C.; Yang, Q.; Jin, S.K.; Lu, G.; Wu, F.Q.; Xiao, Y.; Xu, H.F.; Xia, Q.Y. Resistance to Bombyx mori nucleopolyhedrovirus via overexpression of an endogenous antiviral gene in transgenic silkworms. Arch. Virol. 2012, 157, 1323–1328. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.; Hu, Z.; Qin, Q.; Dong, F.; Huang, L.; Long, J.; Chen, P.; Lu, C.; Pan, M. CRISPR/Cas9-mediated disruption of the immediate early-0 and 2 as a therapeutic approach to Bombyx mori nucleopolyhedrovirus in transgenic silkworm. Insect Mol. Biol. 2019, 28, 112–122. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.J.; Zhang, X.Q.; Chen, D.B.; Yang, D.H.; Zhu, C.X.; Tang, L.M.; Yang, X.; Wang, Y.H.; Luo, X.Y.; Wang, M.L.; et al. CRISPR/Cas9-Mediated Disruption of the lef8 and lef9 to Inhibit Nucleopolyhedrovirus Replication in Silkworms. Viruses 2022, 14, 1119. [Google Scholar] [CrossRef]
- Xiao, Q.; Wang, L.; Zhou, X.L.; Zhu, Y.; Dong, Z.Q.; Chen, P.; Lu, C.; Pan, M.H. BmAtg13 promotes the replication and proliferation of Bombyx mori nucleopolyhedrovirus. Pesticide Biochem. Phys. 2019, 157, 143–151. [Google Scholar] [CrossRef]
- Liu, Y.X.; Zhu, L.B.; Huang, Z.H.; Cao, H.H.; Yu, H.Z.; Liu, S.H.; Xu, J.P. Bombyx mori ferritin heavy-chain homolog facilitates BmNPV proliferation by inhibiting reactive oxygen species-mediated apoptosis. Int. J. Biol. Macromol. 2022, 217, 842–852. [Google Scholar] [CrossRef]
- Jiang, L.; Xia, Q.Y. The progress and future of enhancing antiviral capacity by transgenic technology in the silkworm Bombyx mori. Insect Biochem. Mol. 2014, 48, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Guo, H.Z.; Sun, Q.; Wang, B.B.; Wang, Y.M.; Xie, E.Y.; Xia, Q.Y.; Jiang, L. Spry is downregulated by multiple viruses to elevate ERK signaling and ensure viral reproduction in silkworm. Dev. Comp. Immunol. 2019, 98, 1–5. [Google Scholar]
- Bonning, B.C.; Saleh, M.C. The interplay between viruses and RNAi pathways in insects. Annu. Rev. Entomol. 2021, 66, 61–79. [Google Scholar]
- Cooper, A.M.; Silver, K.; Zhang, J.Z.; Park, Y.; Zhu, K.Y. Molecular mechanisms influencing efficiency of RNA interference in insects. Pest Manag. Sci. 2019, 75, 18–28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, T.H.; Xia, Y.C.; Xu, X.Y.; Wei, G.Q.; Wang, L. Functional analysis of Dicer-2 gene in Bombyx mori resistance to BmNPV virus. Arch. Insect Biochem. Physiol. 2020, 105, e21724. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Bao, Y.Y.; Tang, X.D.; Lv, Z.Y.; Wang, X.Y.; Tian, C.H.; Xu, Y.P.; Zhang, C.X. Gene expression profiling of resistant and susceptible Bombyx mori strains reveals nucleopolyhedrovirus-associated variations in host gene transcript levels. Genomics 2009, 94, 138–145. [Google Scholar] [PubMed] [Green Version]
- Bao, Y.Y.; Lv, Z.Y.; Liu, Z.B.; Xue, J.; Xu, Y.P.; Zhang, C.X. Comparative analysis of Bombyx mori nucleopolyhedrovirus responsive genes in fat body and haemocyte of B. mori resistant and susceptible strains. Insect Mol. Biol. 2010, 19, 347–358. [Google Scholar] [CrossRef]
- Tang, Q.Y.; Zhang, C.X. Data Processing System (DPS) software with experimental design, statistical analysis and data mining developed for use in entomological research. Insect Sci. 2013, 20, 254–260. [Google Scholar] [CrossRef]
- Domeier, M.E.; Morse, D.P.; Knight, S.W.; Portereiko, M.; Bass, B.L.; Mango, S.E. A link between RNA interference and nonsense-mediated decay in Caenorhabditis elegans. Science 2000, 289, 1928–1930. [Google Scholar] [CrossRef]
- Tayler, A.; Heschuk, D.; Giesbrecht, D.; Park, J.Y.; Whyard, S. Efficiency of RNA interference is improved by knockdown of dsRNA nucleases in tephritid fruit flies. Open Biol. 2019, 9, 190198. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Swevers, L.; Iatrou, K.; Huvenne, H.; Smagghe, G. Bombyx mori DNA/RNA non-specific nuclease: Expression of isoforms in insect culture cells, subcellular localization and functional assays. J. Insect Physiol. 2012, 58, 1166–1176. [Google Scholar] [CrossRef]
- Gammon, D.B.; Mello, C.C. RNA interference-mediated antiviral defense in insects. Curr. Opin. Insect Sci. 2015, 8, 111–120. [Google Scholar]
- Li, H.W.; Li, W.X.; Ding, S.W. Induction and suppression of RNA silencing by an animal virus. Science 2002, 296, 1319–1321. [Google Scholar] [PubMed] [Green Version]
- Bronkhorst, A.W.; van Cleef, K.W.R.; Vodovar, N.; Ince, I.A.; Blanc, H.; Vlak, J.M.; Saleh, M.C.; van Rij, R.P. The DNA virus Invertebrate iridescent virus 6 is a target of the Drosophila RNAi machinery. Proc. Natl. Acad. Sci. USA 2012, 109, E3604–E3613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jayachandran, B.; Hussain, M.; Asgari, S. RNA Interference as a cellular defense mechanism against the DNA virus baculovirus. J. Virol. 2012, 86, 13729–13734. [Google Scholar] [CrossRef] [Green Version]
- Mehrabadi, M.; Hussain, M.; Matindoost, L.; Asgari, S. The baculovirus antiapoptotic p35 protein functions as an inhibitor of the host RNA interference antiviral response. J. Virol. 2015, 89, 8182–8192. [Google Scholar] [CrossRef] [Green Version]
- van Rij, R.P.; Saleh, M.C.; Berry, B.; Foo, C.; Houk, A.; Antoniewski, C.; Andino, R. The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster. Genes. Dev. 2006, 20, 2985–2995. [Google Scholar] [CrossRef] [Green Version]
- de Faria, I.J.S.; Aguiar, E.; Olmo, R.P.; da Silva, J.A.; Daeffler, L.; Carthew, R.W.; Imler, J.L.; Marques, J.T. Invading viral DNA triggers dsRNA synthesis by RNA polymerase II to activate antiviral RNA interference in Drosophila. Cell Rep. 2022, 39, 110976. [Google Scholar] [PubMed]
- Zhao, S.D.; Kong, X.S.; Wu, X.F. RNAi-based immunity in insects against baculoviruses and the strategies of baculoviruses involved in siRNA and miRNA pathways to weaken the defense. Dev. Comp. Immunol. 2021, 122, 104116. [Google Scholar] [CrossRef]

| Primers | Sequence (5′–3′) | Purpose |
|---|---|---|
| BmAst-F | CGGGATCCATGGGAGTACGAGGACTAACTACTT | Protein expression |
| BmAst-R | CCGCTCGAGTTATATAACAACTTCAGATTCAAAACC | |
| qBmAst-F | GCGTTGGTACAATTCGAGGT | qRT-PCR |
| qBmAst-R | TGTAAAAGCATTTTCGCTGCT | |
| qBmDicer2-F | GTCGATTGTCAAGTCGCTGA | |
| qBmDicer2-R | AAACCGGACAAGCGTGTATC | |
| qBmAgo2-F | GCTCCTAAAACCGAGGCTCT | |
| qBmAgo2-R | TAGGTTTCCTGCTGCGAGTT | |
| qBmActin3-F | ATCACCATCGGAAACGAAAG | |
| qBmActin3-R | GGTGTTGGCGTACAAGTCCT | |
| qBmGAPDH-F | CATTCCGCGTCCCTGTTGCTAAT | |
| qBmGAPDH-R | GCTGCCTCCTTGACCTTTTGC | |
| Vp39-F | CAACTTTTTGCGAAACGACTT | |
| Vp39-R | GGCTACACCTCCACTTGCTT | |
| dsBmAst-F | GGATCCTAATACGACTCACTATAGGGGTATCCCTGATTGGTTT | dsRNA synthesis |
| dsBmAst-R | GGATCCTAATACGACTCACTATAGGCTCCAGCATAATGTAGCA | |
| dsEGFP-F | GGATCCTAATACGACTCACTATAGGCAGTGCTTCAGCCGCTACCC | |
| dsEGFP-R | GGATCCTAATACGACTCACTATAGGACTCCAGCAGGACCATGTGAT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xia, Y.; Jiang, M.; Hu, X.; Wang, Q.; Qian, C.; Zhu, B.; Wei, G.; Wang, L. A Protein Asteroid with PIN Domain in Silkworm Bombyx mori Is Involved in Anti-BmNPV Infection. Insects 2023, 14, 550. https://doi.org/10.3390/insects14060550
Xia Y, Jiang M, Hu X, Wang Q, Qian C, Zhu B, Wei G, Wang L. A Protein Asteroid with PIN Domain in Silkworm Bombyx mori Is Involved in Anti-BmNPV Infection. Insects. 2023; 14(6):550. https://doi.org/10.3390/insects14060550
Chicago/Turabian StyleXia, Yuchen, Mouzhen Jiang, Xiaoxuan Hu, Qing Wang, Cen Qian, Baojian Zhu, Guoqing Wei, and Lei Wang. 2023. "A Protein Asteroid with PIN Domain in Silkworm Bombyx mori Is Involved in Anti-BmNPV Infection" Insects 14, no. 6: 550. https://doi.org/10.3390/insects14060550
APA StyleXia, Y., Jiang, M., Hu, X., Wang, Q., Qian, C., Zhu, B., Wei, G., & Wang, L. (2023). A Protein Asteroid with PIN Domain in Silkworm Bombyx mori Is Involved in Anti-BmNPV Infection. Insects, 14(6), 550. https://doi.org/10.3390/insects14060550






