Simple Summary
The phylogenetic status of the family Nitidulidae and its sister group relationship remain controversial. Also, the phylogenetic status of the subfamily Meligethinae and its phylogenetic relationships with other subfamilies of Nitidulidae are not fully understood. Mitochondrial genome sequences can be used to study species identification, phylogeny, and population genetic structure, and to provide valuable molecular markers for further genetic studies. In this paper, two complete mitochondrial genomes of Meligethinae were provided for the first time, and the phylogenetic status of the family Nitidulidae and subfamily Meligethinae were explored based on the complete mitochondrial genomes. A comparative analysis of the general characteristics and non-coding region patterns of the complete mitochondrial genomes of Meligethinus tschungseni and Brassicogethes affinis revealed that the base composition and mitochondrial genome structure of these two species are markedly different. Given the results of the phylogenetic analysis based on 20 mitochondrial genomes, the status of Nitidulidae and its sister group relationship is discussed. We also attempted to analyze the taxonomic status of Meligethinae and its sister group relationship. This study will provide a basis for further studies on the higher phylogeny of Nitidulidae.
Abstract
The phylogenetic status of the family Nitidulidae and its sister group relationship remain controversial. Also, the status of the subfamily Meligethinae is not fully understood, and previous studies have been mainly based on morphology, molecular fragments, and biological habits, rather than the analysis of the complete mitochondrial genome. Up to now, there has been no complete mitochondrial genome report of Meligethinae. In this study, the complete mitochondrial genomes of Meligethinus tschungseni and Brassicogethes affinis (both from China) were provided, and they were compared with the existing complete mitochondrial genomes of Nitidulidae. The phylogenetic analysis among 20 species of Coleoptera was reconstructed via PhyloBayes analysis and Maximum likelihood (ML) analysis, respectively. The results showed that the full lengths of Meligethinus tschungseni and Brassicogethes affinis were 15,783 bp and 16,622 bp, and the AT contents were 77% and 76.7%, respectively. Each complete mitochondrial genome contains 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), 2 ribosomal RNA genes (rRNAs), and a control region (A + T-rich region). All the PCGs begin with the standard start codon ATN (ATA, ATT, ATG, ATC). All the PCGs terminate with a complete terminal codon, TAA or TAG, except cox1, cox2, nad4, and nad5, which terminate with a single T. Furthermore, all the tRNAs have a typical clover-leaf secondary structure except trnS1, whose DHU arm is missing in both species. The two newly sequenced species have different numbers and lengths of tandem repeat regions in their control regions. Based on the genetic distance and Ka/Ks analysis, nad6 showed a higher variability and faster evolutionary rate. Based on the available complete mitochondrial genomes, the results showed that the four subfamilies (Nitidulinae, Meligethinae, Carpophilinae, Epuraeinae) of Nitidulidae formed a monophyletic group and further supported the sister group relationship of Nitidulidae + Kateretidae. In addition, the taxonomic status of Meligethinae and the sister group relationship between Meligethinae and Nitidulinae (the latter as currently circumscribed) were also preliminarily explored.
1. Introduction
Nitidulidae includes 11 subfamilies with approximately 350 genera and nearly 4500 species worldwide [1,2,3]. Meligethinae (Coleoptera: Nitidulidae) is the second largest subfamily of Nitidulidae, also known as “pollen beetles”, with 46 genera and approximately 700 species worldwide and 12 genera and approximately 130 species in China [4,5,6,7,8,9,10,11,12,13,14,15]. Meligethinae is widely distributed in the Nearctic, Afrotropical, Oriental, and Palaearctic realms, and (very marginally) in Australia, except the Neotropical realm [4,16]. It is worth noting that Meligethinae is the only subfamily among Nitidulidae that independently and entirely became strictly anthophagous, with all members of this lineage using pollen as the main food resource [17,18,19,20,21,22]. Meligethinae represent an important group to reveal the different and regular interactions between morphological structure, biological habit, and ecological adaptability among the various subfamilies of the family Nitidulidae [1,4,5,7,11,12,14,15,16]. The first two complete mitochondrial genomes of the subfamily Meligethinae analyzed here are Meligethinus tschungseni Kirejtshuk, 1987 and Brassicogethes affinis Jelínek, 1982, collected from palm flowers and rape flowers in China, respectively [23,24].
There are many studies on the status of Nitidulidae: Bocak et al. (2014) [25] supported Passandridae nested within Nitidulidae, and Tang et al. (2019) [26] supported Nitidulidae nested within Erotylidae based on the mitochondrial genome. Other studies supported that Nitidulidae is monophyletic based on morphological and molecular data analysis [1,3,27,28,29,30]. However, previous analyses based on the mitochondrial genome had a small sample size, without a complete mitochondrial genome of Meligethinae. Regarding the sister group relationship of Nitidulidae, most studies supported the sister group relationship of Nitidulidae + Kateretidae based on morphological characters [7,31,32]. The following studies supported the sister group relationship of Nitidulidae + Kateretidae: Cline et al. (2014) [28], based on seven molecular fragments (12S, 16S, 18S, 28S, COI, COII, and H3); Bocak et al. (2014) [25], based on four molecular fragments (18S, 28S, rrnL, and COI); Robertson et al. (2015) [2], based on eight molecular fragments (18S, 28S, H3, CAD, 12S, 16S, COI, and COII); and Cai et al. (2022) [3], based on single-copy nuclear protein-coding (NPC) genes and fossil data. Only the phylogenetic trees constructed by Chen et al. (2020) [29] based on the complete mitochondrial genomes of 17 species (seven species of Nitidulidae and ten species of other Coleoptera) supported that the sister group of Nitidulidae could be Monotomidae, but this clade had low bootstrap support values in ML trees. In fact, this potentially spurious sister group relationship of (Nitidulidae + Monotomidae) was probably due to the mismatch between the dataset and the selected nucleotide substitution model [33].
Meligethinae, as the second largest subfamily in Nitidulidae, has always attracted much attention. Many scholars have used morphological characteristics and a small amount of molecular data to explore the taxonomic status of Meligethinae. Kirejtshuk et al. (1982, 1986, 1995, 2008) [34,35,36,37] supported Meligethinae as monophyletic based on a few morphological characters of the adults and biological habits such as larval host plants. Trizzino et al. (2009) [38] and Audisio et al. (2009) [39] also supported that Meligethinae is monophyletic based on morphological characters, molecular fragments, and larval host plants, respectively. Cline et al. (2014) [28] reconstructed the phylogenetic relationships among Nitidulidae based on seven molecular fragments (12S, 16S, 18S, 28S, COI, COII, H3) and showed that Meligethinae nested in Nitidulinae, but Cline et al. (2014) only selected one species as a representative of Meligethinae. Lee et al. (2020) [1] reconstructed the phylogenetic relationships of Nitidulidae based on five molecular fragments (COI, 28S, CAD, H3, Wingless), and proposed that Meligethinae (represented by three genera and seven species) is monophyletic, but also showed that Meligethinae nested in Nitidulinae. The phylogenetic trees constructed by Cline et al. and Lee et al. based on molecular fragments were insufficient to resolve the status of Meligethinae and its phylogenetic relationship with Nitidulinae (which, as presently circumscribed, very likely represent a polyphyletic lineage). Therefore, there is an urgent need to supplement new representative genera and species as well as molecular data (such as complete mitochondrial genomes) to continue studying Meligethinae.
In recent years, the complete mitochondrial genome has been widely used to study the phylogenetic relationships among insects [40,41,42], phylogeography [43], and molecular evolution [40,44]. The mitochondrial genome of insects has unique features, such as maternal inheritance, rapid evolution rate, stable gene composition, and high independence and integrity, making it a powerful genetic marker for studying the evolution of insects [45,46]. Previously, there were only seven complete mitochondrial genomes among three subfamilies of Nitidulidae in GenBank [29,47,48,49]. Meligethinae, as the second largest subfamily of Nitidulidae, does not yet have a complete mitochondrial genome. Therefore, for the first time, this study provided the complete mitochondrial genomes of two species (Meligethinus tschungseni and Brassicogethes affinis) of Meligethinae, with a detailed annotation and analysis of their sequences. There is also another complete mitochondrial genome sequence of Meligethinae (Teucriogethes sp.) that has been uploaded to GenBank by authors but not yet published. These four subfamilies (Nitidulinae, Carpophilinae, Epuraeinae, Meligethinae) for which mitochondrial genome data are available so far are the most species-rich groups in Nitidulidae, accounting for approximately 3/4 of the total species in Nitidulidae [1,4,7]. Therefore, it is possible to further analyze the status of Nitidulidae and the sister group relationship of Nitidulidae. Moreover, it is necessary to preliminarily explore the status and the sister group relationship of Meligethinae based on the complete mitochondrial genomes for the first time. In this study, we reconstructed the phylogenetic relationships of 20 species (including 17 ingroups and 3 outgroups) under the site-heterogeneous mixture CAT + GTR substitution model (BI trees) and the best model (ML trees), respectively.
2. Materials and Methods
2.1. Sample Preparation and DNA Extraction
Adults of M. tschungseni in this study were collected from palm flowers and B. affinis from rape flowers in April and May 2022 at the West Campus of Yangtze University, Jingzhou, Hubei, China. All the specimens were immediately preserved in absolute ethanol. The total genomic DNA was extracted using the Ezup Column Animal Genomic DNA Purification Kit (Sangon Biotech, Shanghai, China).
2.2. Sequence Analysis
The mitochondrial genomes of M. tschungseni and B. affinis were sequenced using next-generation sequencing (NGS; Illumina NovaSeq6000; Berry Genomics, Beijing, China). The raw paired reads were trimmed and assembled using Geneious 8.1.3 (Biomatters, Auckland, New Zealand) with default parameters [50]. The complete mitochondrial genome of Carpophilus pilosellus Motschulsky, 1858 (Nitidulidae: Carpophilinae; NC_046035) [47] was selected as the reference sequence.
The positions of 13 protein-coding genes (PCGs) were determined by finding ORFs based on the invertebrate mitochondrial genetic codon and comparing with reference sequences. The positions of 22 tRNAs were determined according to the prediction results of the MITOS Web Server (http://mitos.bioinf.uni-leipzig.de/index.py (accessed on 13 May 2022)) [51]. The secondary structures of 22 tRNAs were predicted according to MITOS and the tRNAscan-SE Online Search Server [52] and then drawn using Adobe Illustrator CS5. The positions of rRNAs (rrnL and rrnS) and the control region (A + T-rich region) were determined based on the positions of the tRNAs and comparison with other homologous sequences. Tandem repeats in the control region were determined using the Tandem Repeats Finder Online server (http://tandem.bu.edu/trf/trf.html (accessed on 23 May 2022)) [53]. Circular maps of the mitochondrial genome were drawn using Organellar Genome DRAW (OGDRAW) (https://chlorobox.mpimp-golm.mpg.de/OGDraw.html (accessed on 16 May 2022)) [54]. The base composition, AT and GC skew, and relative synonymous codon usage (RSCU) of 10 species of Nitidulidae were calculated using PhyloSuite v1.2.2 [55]. The nucleotide diversity (Pi) of 13 PCGs, 22 tRNAs, and 2 rRNAs of 10 species of Nitidulidae were calculated using DnaSP v6.0 [56] with a step size of 20 bp and a sliding window of 200 bp. The non-synonymous (Ka)/synonymous (Ks) mutation rate ratios for 13 PCGs of 10 species of Nitidulidae were also calculated using DnaSP v6.0 [56]. The genetic distances between the mitochondrial genomes of 10 species of Nitidulidae were calculated using MEGA-X [57] based on the Kimura-2-parameter (K2P) model.
2.3. Phylogenetic Analysis
In this study, we analyzed the phylogenetic relationships among 20 species of Coleoptera based on the complete mitochondrial genomes. The information regarding these 20 species is shown in Table 1. Two newly sequenced mitochondrial genomes (M. tschungseni and B. affinis) were provided and analyzed in this study, one mitochondrial genome (Teucriogethes sp. from China) was sequenced and uploaded to GenBank by authors but has not yet been published, and the remaining mitochondrial genomes were downloaded from GenBank. Firstly, 13 PCGs and two rRNAs of these 20 species were aligned using Mafft v7.313 (PCG alignment strategy: G-INS-i; RNA alignment strategy: Q-INS-i). Secondly, poorly aligned and highly scattered regions were removed using Gblocks v0.91b. Then, the aligned and modified sequences were concatenated using PhyloSuite. Phylogenetic trees were constructed based on four datasets: (1) the first and second codon positions of 13 PCGs (PCG12); (2) all three codon positions of 13 PCGs (PCG123); (3) the first and second codon positions of 13 PCGs and two rRNAs (PCG12R); (4) all three codon positions of 13 PCGs and two rRNAs (PCG123R).

Table 1.
Summary of mitochondrial genome information used in this study.
BI trees were established under the site-heterogeneous mixture CAT + GTR substitution model using PhyloBayes MPI v1.5a, running four Markov Chain Monte Carlo (MCMC) independently. When the sampled tree had stabilized and the four runs had reached satisfactory convergence (maxdiff < 0.3), the first 25% of the samples were discarded as “burn-in”. The ML trees were constructed using IQ-TREE v1.6.8 [61]. ModelFinder was used to select the substitution models (Table S2) for the ML analysis. A “greedy” algorithm and BIC (Bayesian information criterion) [55] were used to obtain the best model and optimal partitioning strategy for each partition. The ML analysis was performed using ultrafast bootstrap parameters of 1000 repetitions. The phylogenetic trees were visualized and edited using iTOL [62].
3. Results and Discussion
3.1. Genome Structure and Base Composition
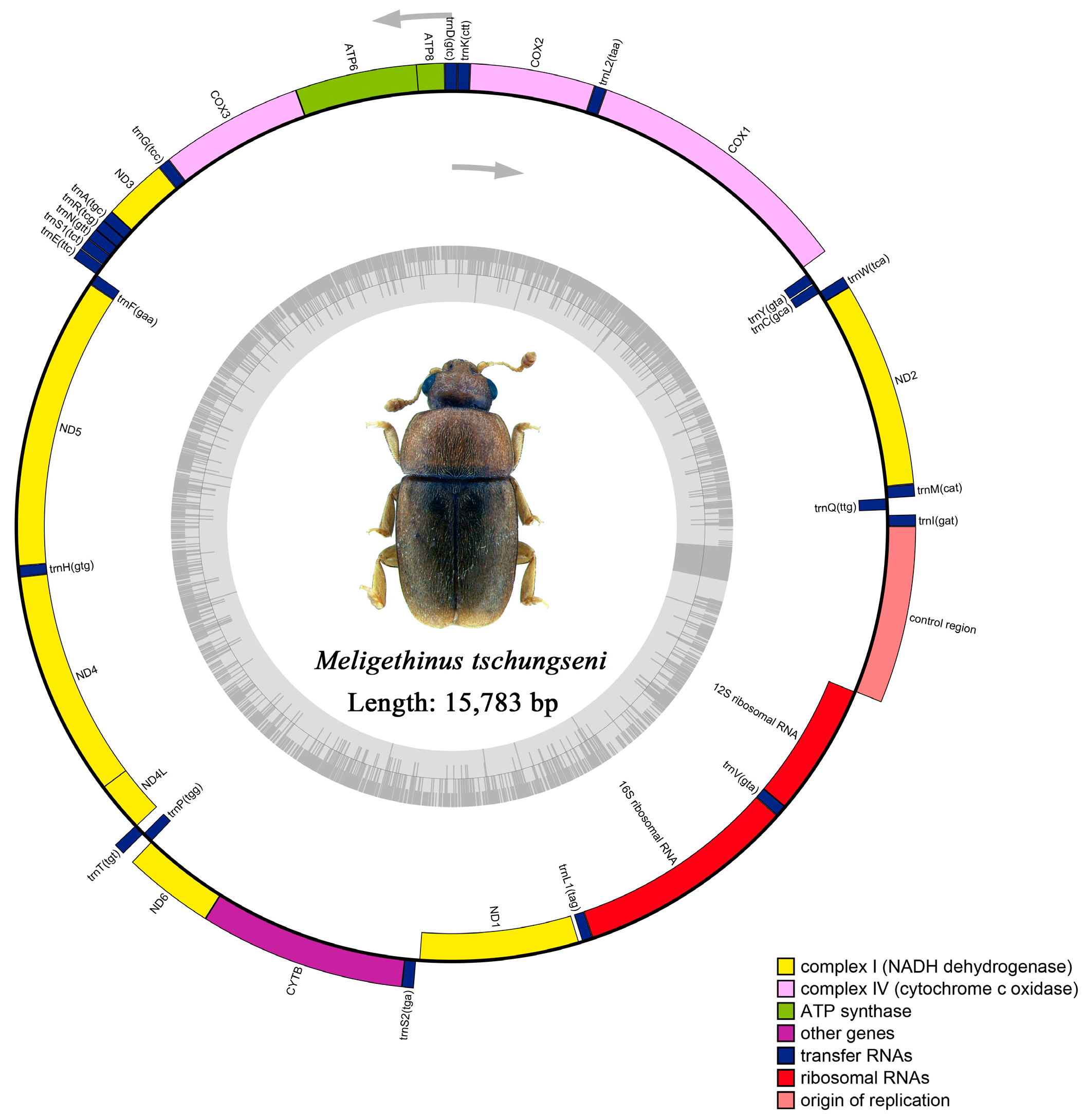
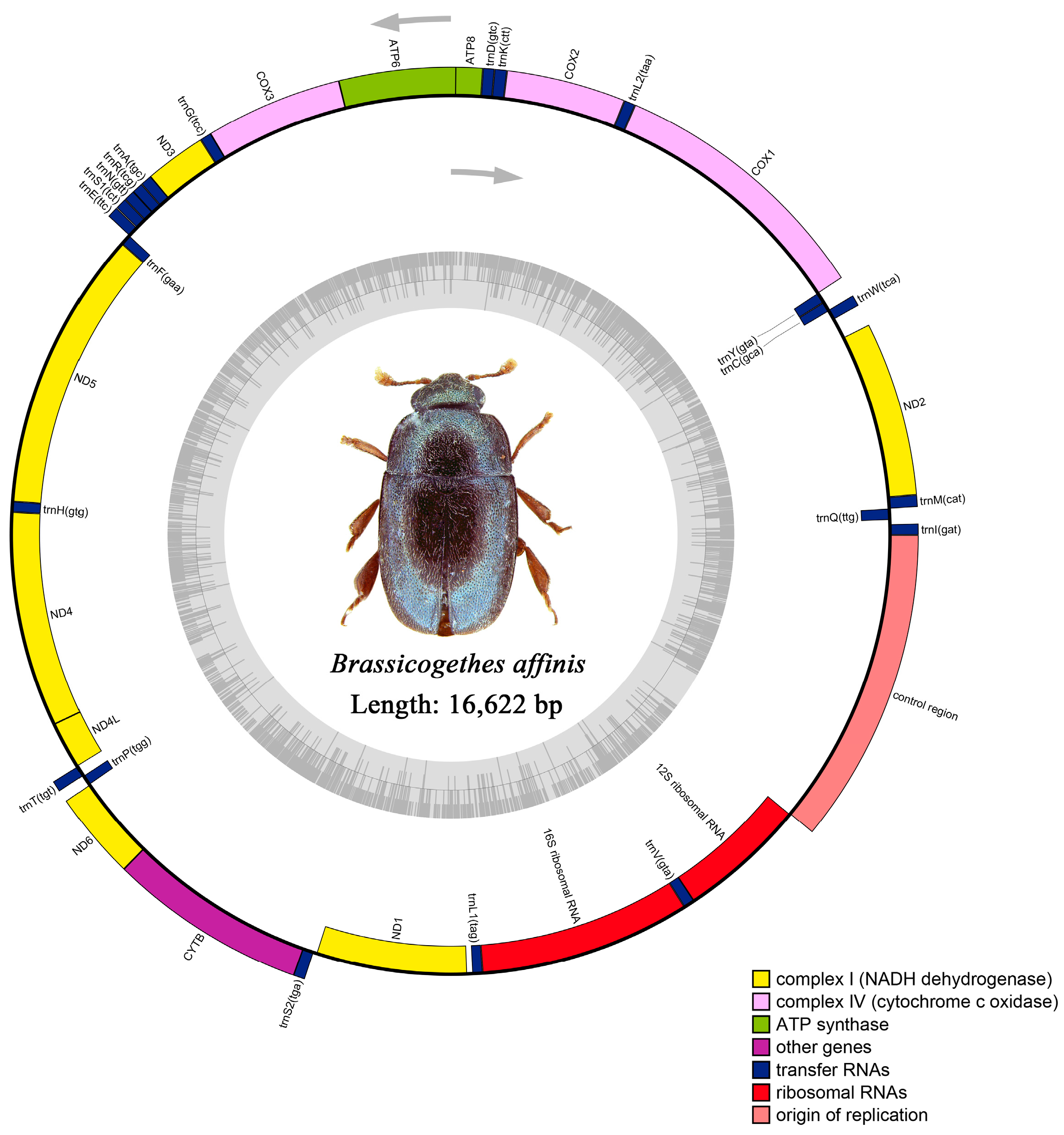
The raw data of M. tschungseni and B. affinis were 6.21 gb and 4.65 gb, respectively. The complete mitochondrial genomes of M. tschungseni (GenBank accession number: ON782471) and B. affinis (GenBank accession number: ON782472) were 15,783 bp (Figure 1) and 16,622 bp (Figure 2), respectively. These two mitochondrial genomes showed the same gene arrangement as the other mitochondrial genomes of Nitidulidae. They contained the complete set of 37 genes (13 PCGs, 22 tRNAs, and 2 rRNAs) and a control region (A + T-rich region). The differences in the sequence length among Nitidulidae are mainly determined by the length of the control region and the length of the intergenic spacers between some tRNAs. The majority strand (J-strand) encoded most of the genes, including 9 PCGs (nad2, cox1, cox2, atp8, atp6, cox3, nad3, nad6, and cytb) and 14 tRNAs (trnI, trnM, trnW, trnL2, trnK, trnD, trnG, trnA, trnR, trnN, trnS1, trnE, trnT, and trnS2), while the minority strand (N-strand) encoded other genes, including 4 PCGs (nad5, nad4, nad4L, and nad1), 8 tRNAs (trnQ, trnC, trnY, trnF, trnH, trnP, trnL1, and trnV), and 2 rRNAs (rrnL and rrnS) (Table 2). Additionally, seven intergenic spacers were found in the mitochondrial genomes of M. tschungseni (113 bp in total) and B. affinis (200 bp in total) (Table 2). The longest intergenic spacer in M. tschungseni was between trnY and cox1 (41 bp), and the longest in B. affinis was between nad2 and trnW (122 bp) (Table 2). A total of 13 and 12 overlapping regions were found in the mitochondrial genomes of M. tschungseni (36 bp in total) and B. affinis (26 bp in total), respectively, and the longest overlapping regions were between trnW and trnC (8 bp), between nad4L and trnT (8 bp) in M. tschungseni, and between trnW and trnC (8 bp) in B. affinis (Table 2).
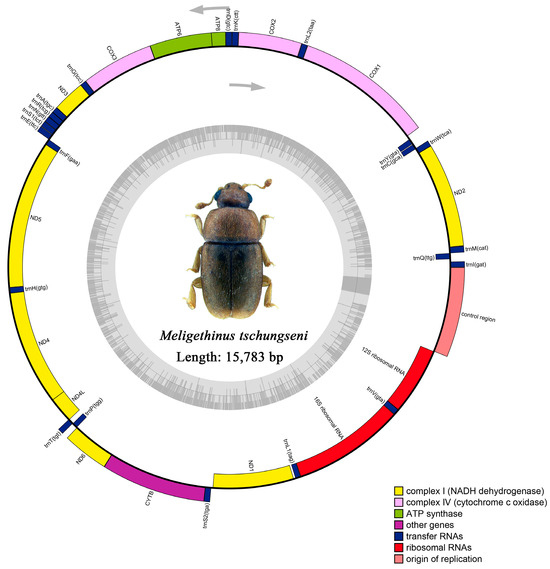
Figure 1.
Circle map of the complete mitochondrial genome of Meligethinus tschungseni.
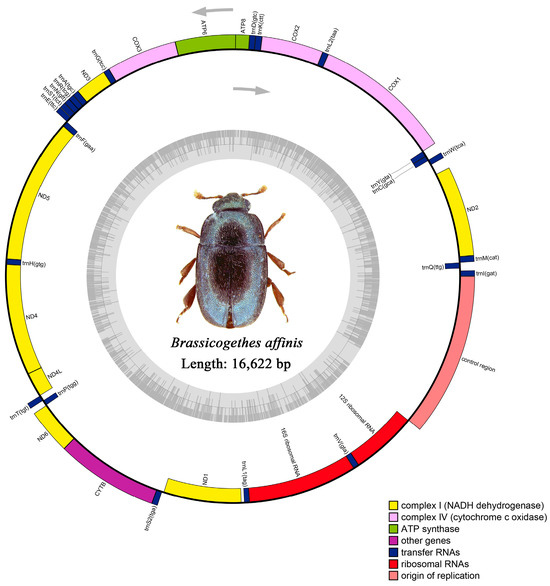
Figure 2.
Circle map of the complete mitochondrial genome of Brassicogethes affinis.

Table 2.
Mitogenomic organization of Meligethinus tschungseni and Brassicogethes affinis.
The AT contents of the mitochondrial genomes of M. tschungseni and B. affinis were 77% and 76.7%, respectively (Table 3 and Table 4), which were significantly higher than the GC content (Table 4). In addition, most of the known species of Nitidulidae showed a positive AT skew and negative GC skew in the mitochondrial genomes (Table 3), and M. tschungseni and B. affinis in this study also showed a positive AT skew and negative GC skew (Table 3 and Table 4), which indicated a higher content of A than T and a higher content of C than G in the mitochondrial genomes (Table 4).

Table 3.
Nucleotide composition of mitochondrial genomes of 10 species of Nitidulidae: Carpophilus pilosellus (C1.); Carpophilus dimidiatus (C2.); Epuraea guttata (E1.); Epuraea sp. (E2.); Xenostrongylus variegatus (X.); Omosita colon (O.); Aethina tumida (A.); Meligethinus tschungseni (M.); Brassicogethes affinis (B.); Teucriogethes sp. (T.).

Table 4.
Nucleotide composition of mitochondrial genomes of Meligethinus tschungseni and Brassicogethes affinis.
3.2. Protein-Coding Genes (PCGs) and Codon Usage
The total lengths of 13 PCGs in the mitochondrial genomes of M. tschungseni and B. affinis were 11,196 bp and 11,097 bp, respectively (Table 4), both of which contained seven NADH dehydrogenase subunits (nad1, nad2, nad3, nad4, nad5, nad6, nad4L), three cytochrome c oxidase subunits (cox1, cox2, cox3), two ATPase subunits (atp6, atp8), and one cytochrome b gene (cytb) (Figure 1 and Figure 2, Table 5). The AT contents of 13 PCGs of M. tschungseni and B. affinis were 77.2% and 75.8%, respectively (Table 4). And both species showed a negative AT skew and a negative GC skew (Table 4), which indicated a higher content of T than A and a higher content of C than G. The AT contents of the third codon (91.1%, 87.4%) of M. tschungseni and B. affinis were much higher than that of the first codon (71.9%, 70.9%) and the second codon (68.7%, 68.8%) (Table 4). Other than nad1 in the mitochondrial genomes of Epuraea guttata, Omosita colon, and Aethina tumida starting with TTG, the other PCGs of Nitidulidae in this study were typical start codons ATN (ATA, ATT, ATG, and ATC) (Table 5). Except for cox1, cox2, and nad5, all 10 species of Nitidulidae terminated with a single T, cox3 and nad4 always terminated with a single T, and atp8 in Xenostrongylus variegatus terminated with a single T. The other PCGs in the mitochondrial genomes of the 10 species of Nitidulidae in this study all terminated with a stop codon TAA or TAG. Among them, cox1, cox2, nad4, and nad5 in the mitochondrial genomes of M. tschungseni and B. affinis also terminated with a single T (Table 5). This incomplete stop codon is common in insects and can be converted to a complete stop codon through post-transcriptional polyadenylation [63].

Table 5.
Start and stop codons of the mitochondrial genomes of 10 species of Nitidulidae: Carpophilus pilosellus (C1.); Carpophilus dimidiatus (C2.); Epuraea guttata (E1.); Epuraea sp. (E2.); Xenostrongylus variegatus (X.); Omosita colon (O.); Aethina tumida (A.); Meligethinus tschungseni (M.); Brassicogethes affinis (B.); Teucriogethes sp. (T.).
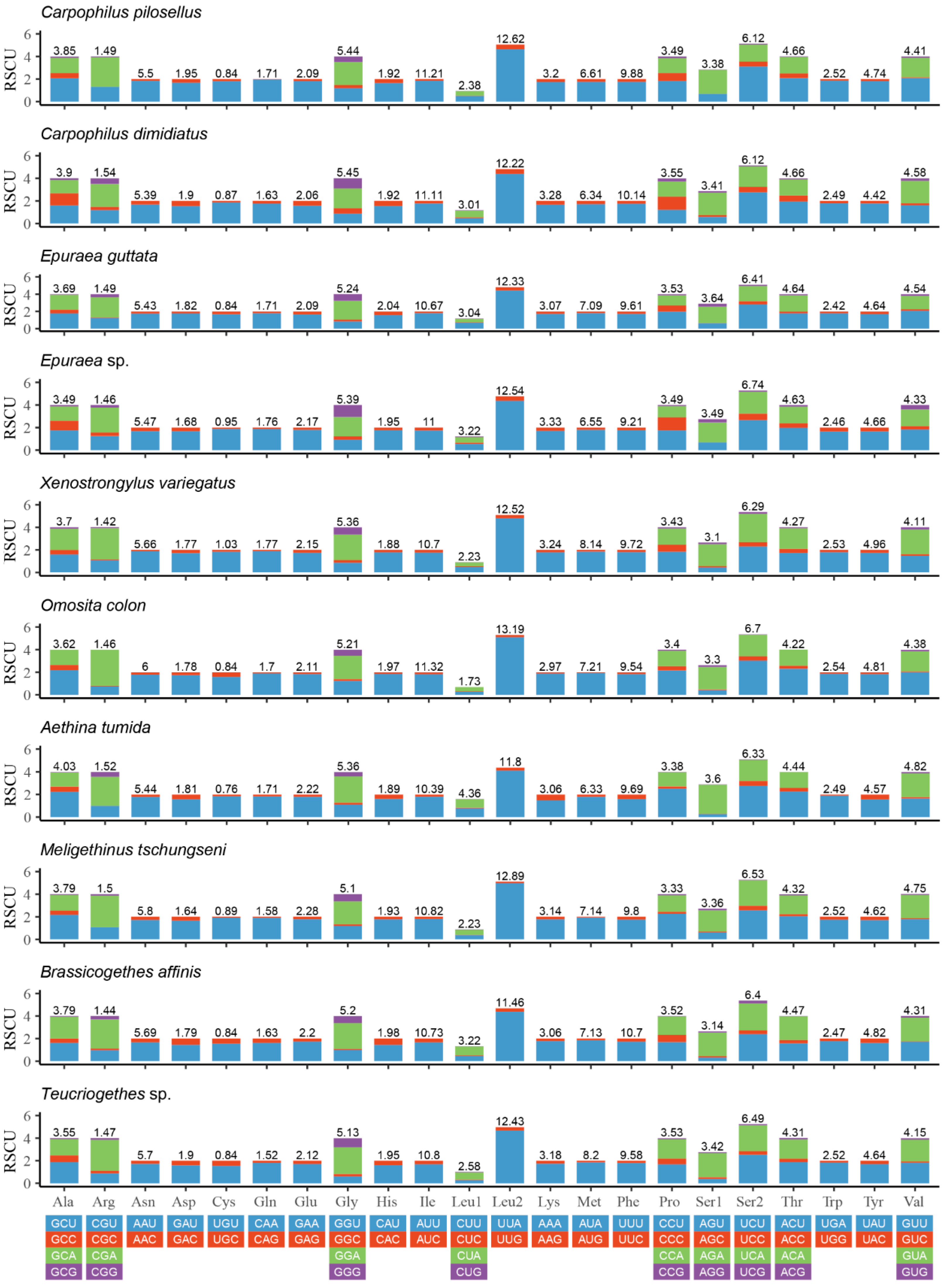
The relative synonymous codon usage (RSCU) of PCGs of the known 10 species of Nitidulidae is shown in Figure 3. UUA (Leu2), AUU (Ile), UUU (Phe), and AUA (Met) were commonly used codons in Nitidulidae. These codons all consisted of A or U (Figure 3), which may be one of the reasons for the higher AT contents of the PCGs in Nitidulidae (Table 3).
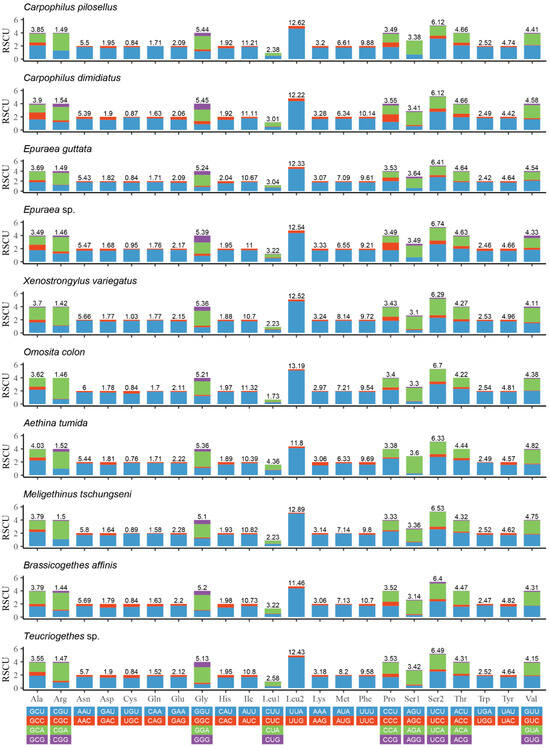
Figure 3.
Relative synonymous codon usage (RSCU) of the PCGs of 10 species of Nitidulidae. The numbers above the bar graph indicate the frequency of amino acids.
3.3. Transfer and Ribosomal RNA Genes
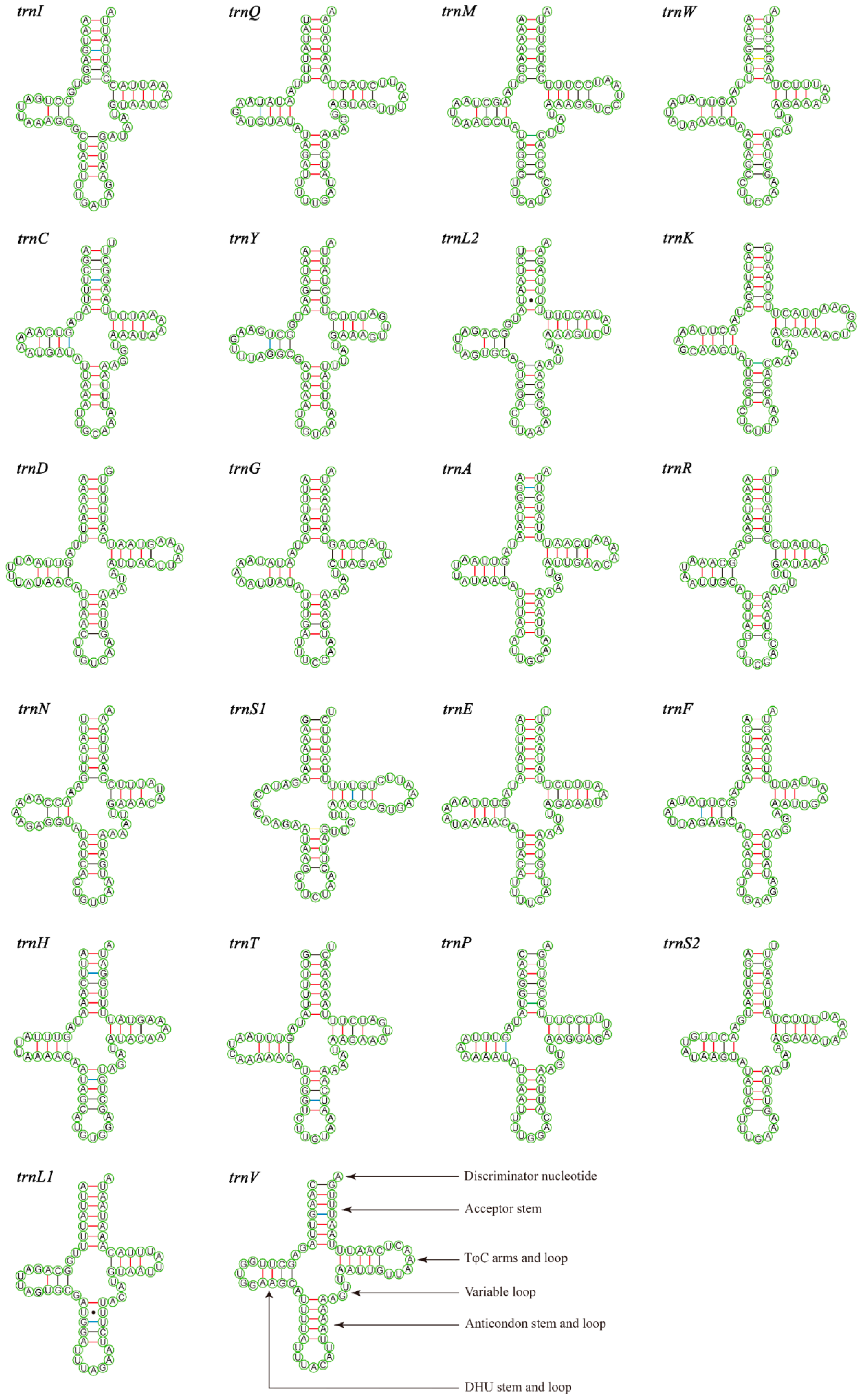
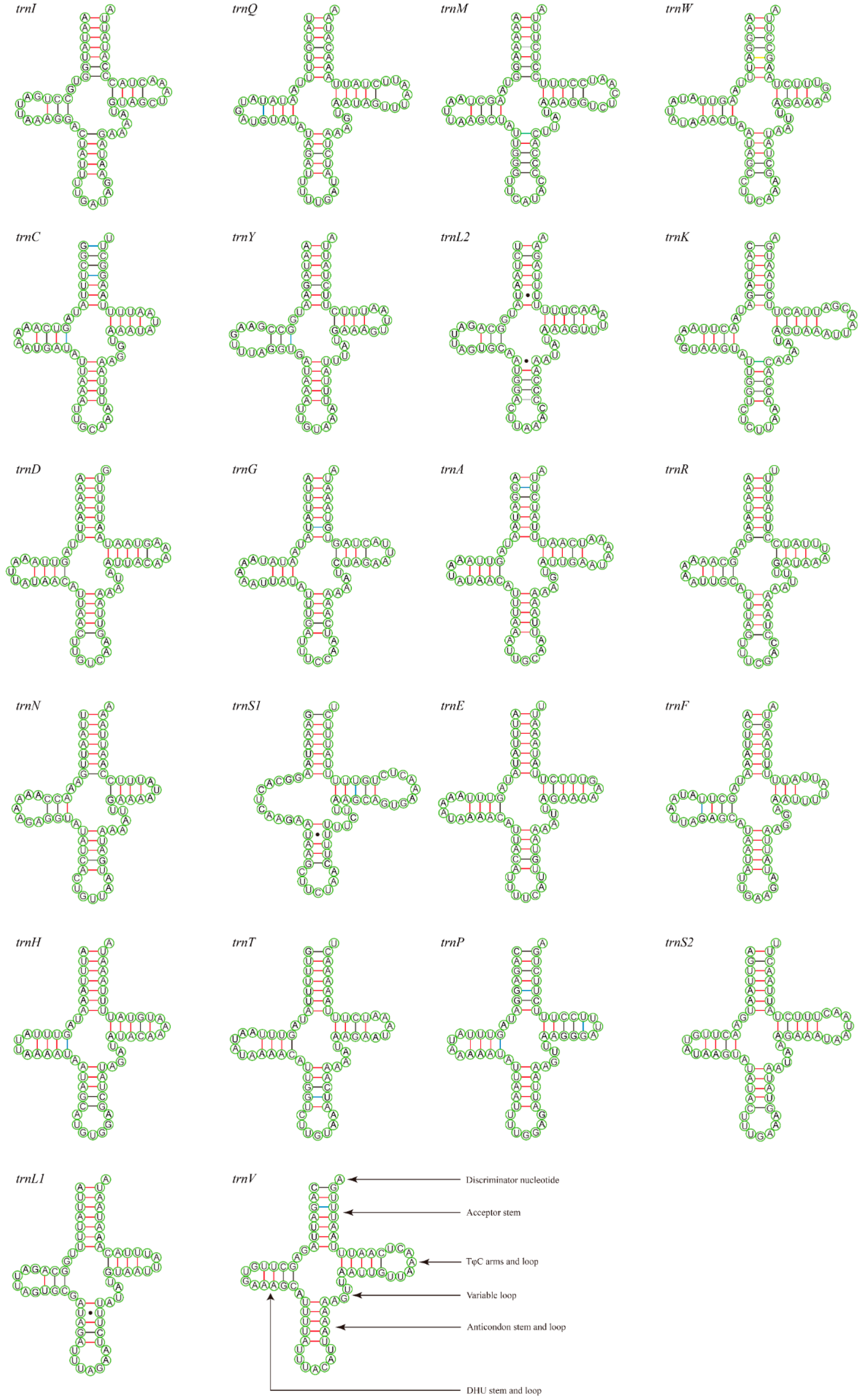
The total lengths of the 22 tRNAs of M. tschungseni and B. affinis were 1455 bp and 1451 bp, respectively; within the known tRNA length range of the mitochondrial genomes of Nitidulidae (Table 3 and Table 4). The AT contents of the tRNAs of M. tschungseni and B. affinis were 78% and 79%, respectively (Table 3 and Table 4). In addition, the tRNAs of both species showed a positive AT skew and positive GC skew (Table 4), indicating more A than T and more G than C. Except for trnS1, which showed a reduced dihydrouridine (DHU) arm, the other 21 tRNAs of M. tschungseni and B. affinis consisted of “four arms” and “four loops”, they could fold into the typical clover-leaf structure, and the amino-acid arm (14 bp) and the anticodon loop (7 bp) were highly conserved (Figure 4 and Figure 5). The DHU arm had three or four base pairs and the TφC arm had 3–5 base pairs in both M. tschungseni and B. affinis (Figure 4 and Figure 5). The lengths of the DHU loop in M. tschungseni and B. affinis were 3–8 bases and 4–8 bases, respectively. The lengths of the TφC loop of both M. tschungseni and B. affinis were 3–9 bases (Figure 4 and Figure 5). There were five types (G-U, C-U, A-C, A-G, U-U) of a total of 24 mismatched base pairs in M. tschungseni and six types (G-U, C-U, A-C, A-G, U-U, A-A) of 24 mismatched base pairs in B. affinis (Figure 4 and Figure 5).
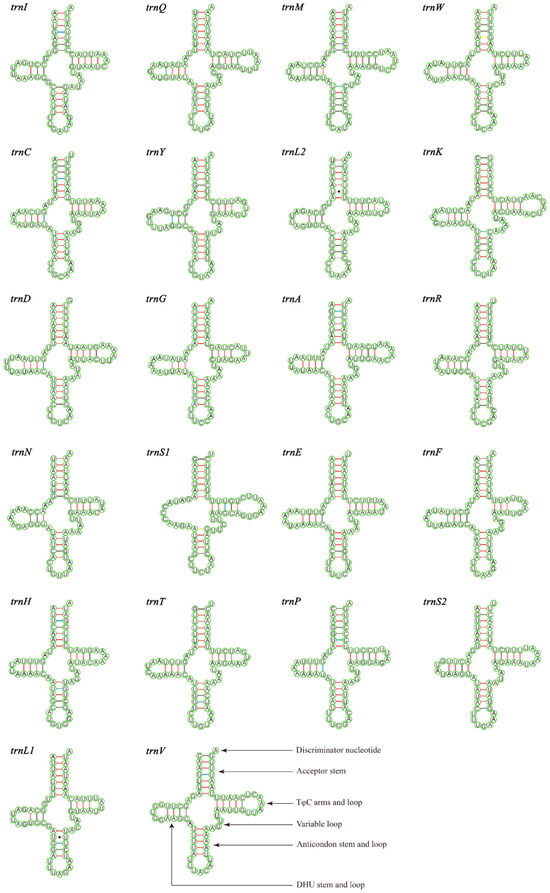
Figure 4.
Predicted secondary structure for the tRNAs of Meligethinus tschungseni (A-U, G-C regular paired keys marked with red and black lines, respectively; G-U, C-U, A-C, A-G mismatched keys marked with blue, green, gray, and yellow lines, respectively; U-U mismatched keys marked with solid black dots).
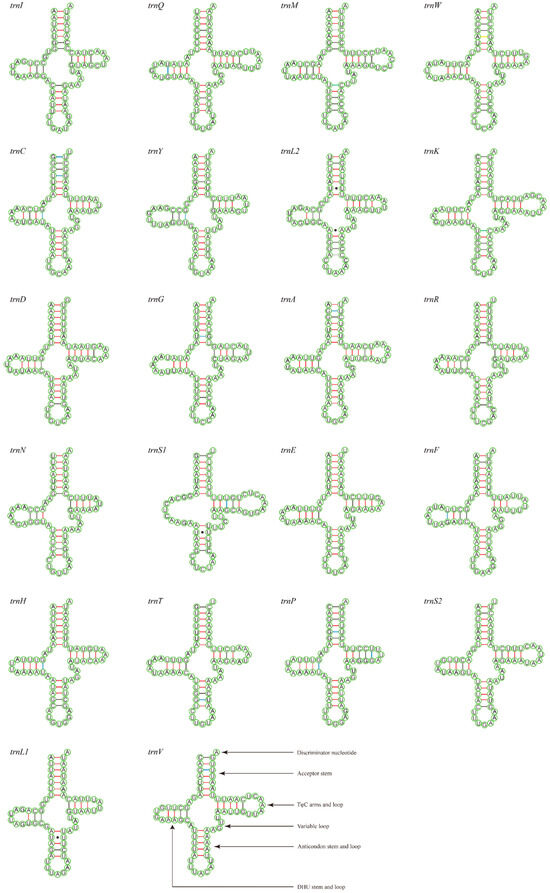
Figure 5.
Predicted secondary structure for the tRNAs of Brassicogethes affinis (A-U, G-C regular paired keys marked with red and black lines, respectively; G-U, C-U, A-C, A-G mismatched keys marked with blue, green, gray, and yellow lines, respectively; U-U, A-A mismatched keys marked with solid black dots).
In M. tschungseni and B. affinis, the total lengths of the two rRNAs were 2073 bp and 2075 bp, respectively (Table 3 and Table 4). The rrnL were all 1292 bp in these two species and were located between trnL1 and trnV. The rrnS were 781 bp and 783 bp, respectively, and were located between trnV and the control region (Figure 1 and Figure 2, Table 2). The AT contents of the rRNAs of M. tschungseni and B. affinis were both high, 81.5% and 79.3%, respectively, and both showed a negative AT skew and positive GC skew, indicating a higher content of T than A and a higher content of G than C (Table 3 and Table 4).
3.4. Control Region
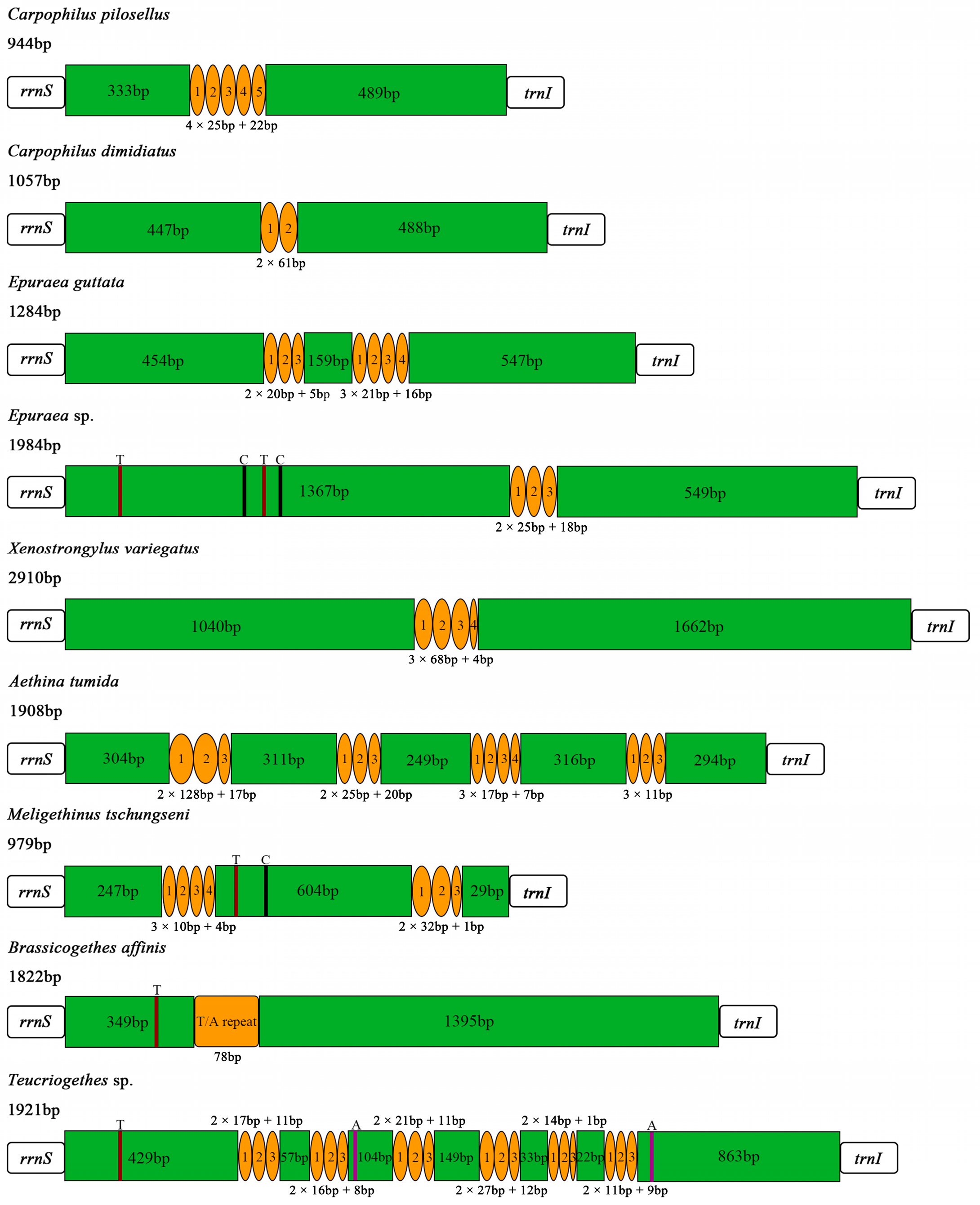
The control region (A + T-rich region) was located between rrnS and trnI, with lengths of 979 bp and 1822 bp in M. tschungseni and B. affinis, respectively (Figure 1 and Figure 2, Table 2, Table 3 and Table 4), within the known control region lengths of the mitochondrial genomes of Nitidulidae (Table 3) [64]. The AT contents were 62.5% and 76.1%, respectively (Table 3 and Table 4). Both species showed a negative AT skew and negative GC skew (Table 4), indicating a higher content of T than A and a higher content of C than G. The number of tandem repeat regions in the control region greatly varied among the 10 species of Nitidulidae mitochondrial genomes. Among them, Omosita colon had no tandem repeat region, Carpophilus pilosellus, C. dimidiatus, Epuraea sp., Xenostrongylus variegatus, and B. affinis had one tandem repeat region, E. guttata and M. tschungseni had two tandem repeat regions, and Aethina tumida and Teucriogethes sp. had four and six tandem repeat regions, respectively (Figure 6).
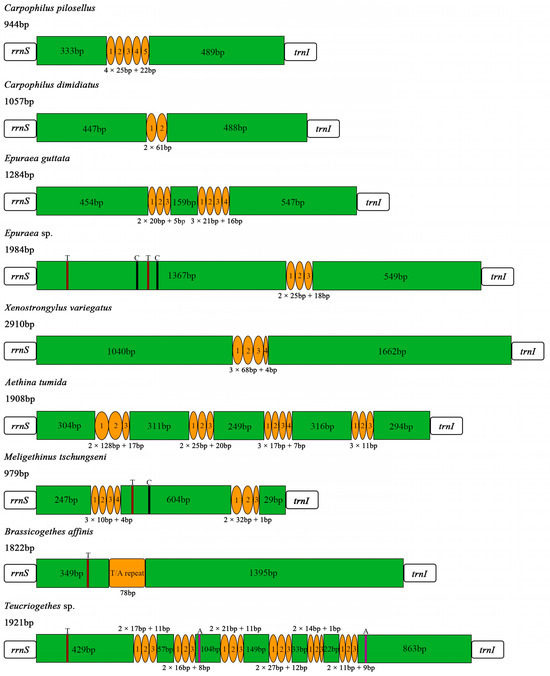
Figure 6.
Structure of the control regions in the Nitidulidae mitochondrial genomes. Orange circles and box represent tandem repeat regions, and green boxes represent non-repeat regions. The brown, black, and purple regions represent poly (T), poly (C), and poly (A), respectively.
3.5. Nucleotide Diversity and Genetic Distance
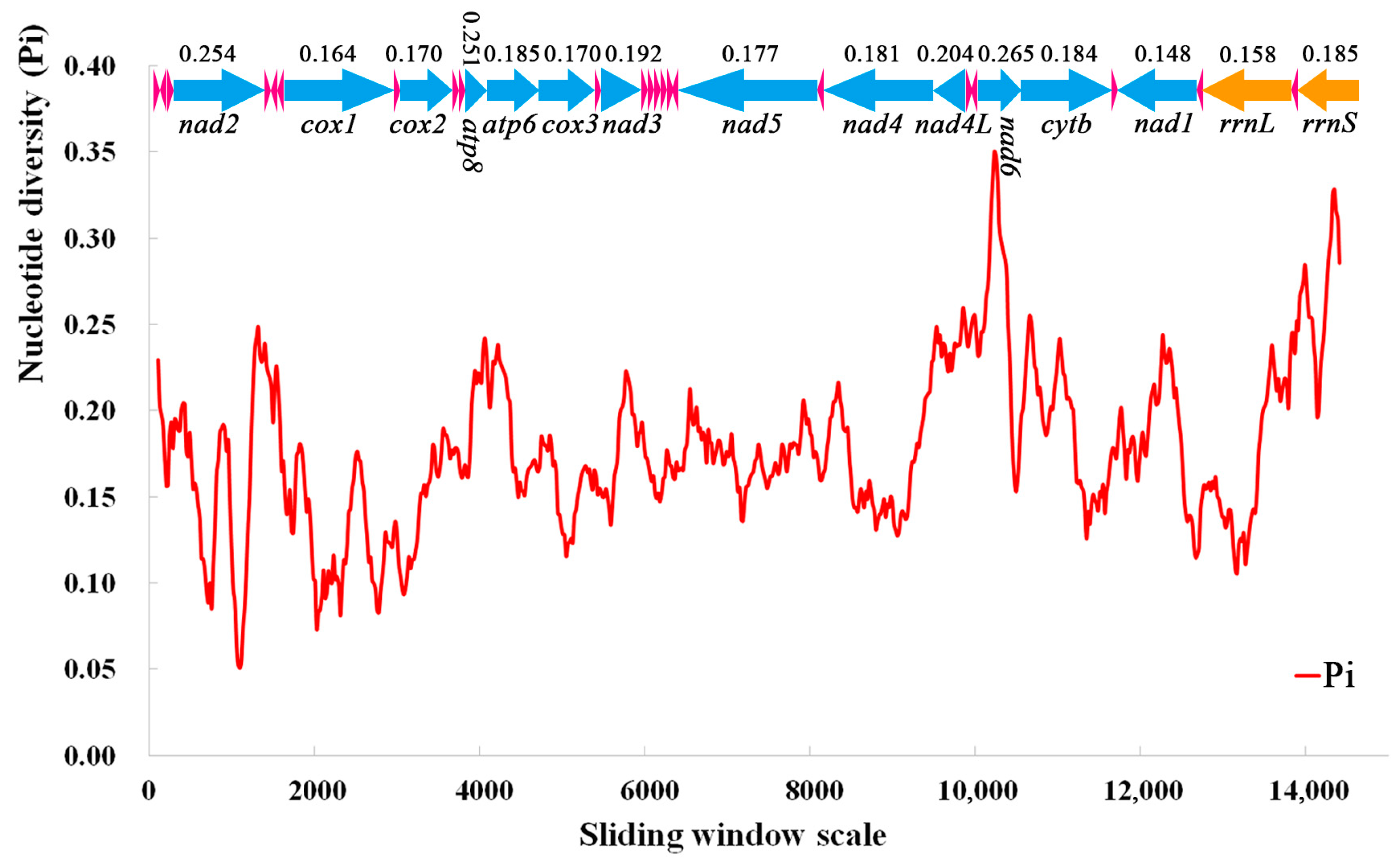
A sliding window analysis was used to study the nucleotide diversity of 13 PCGs, 22 tRNAs, and 2 rRNAs in the mitochondrial genomes of 10 species of Nitidulidae (Figure 7). The nucleotide diversity values ranged from 0.148 (nad1) to 0.265 (nad6). The nad6 (0.265), nad2 (0.254), and atp8 (0.251) genes had a higher nucleotide diversity, indicating that these genes had a high variability in Nitidulidae. On the contrary, nad1 (0.148) and cox1 (0.164) had a lower nucleotide diversity; therefore, nad1 and cox1 were conserved genes in Nitidulidae.
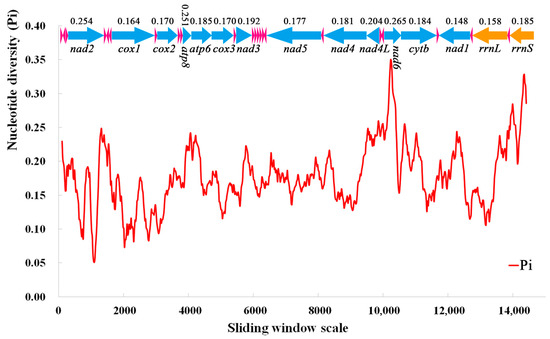
Figure 7.
Sliding window analysis of 13 PCGs, 22 tRNAs, and 2 rRNAs in the mitochondrial genomes of 10 species of Nitidulidae. The red line represents the nucleotide diversity (Pi) value (window size = 200 bp, step size = 20 bp); the arrows represent the direction of gene coding—above the arrow is the Pi value of each gene, and below the arrow is the name of each gene; the blue arrows represent 13 PCGs, the pink arrows represent 22 tRNAs, and the orange arrows represent two rRNAs.
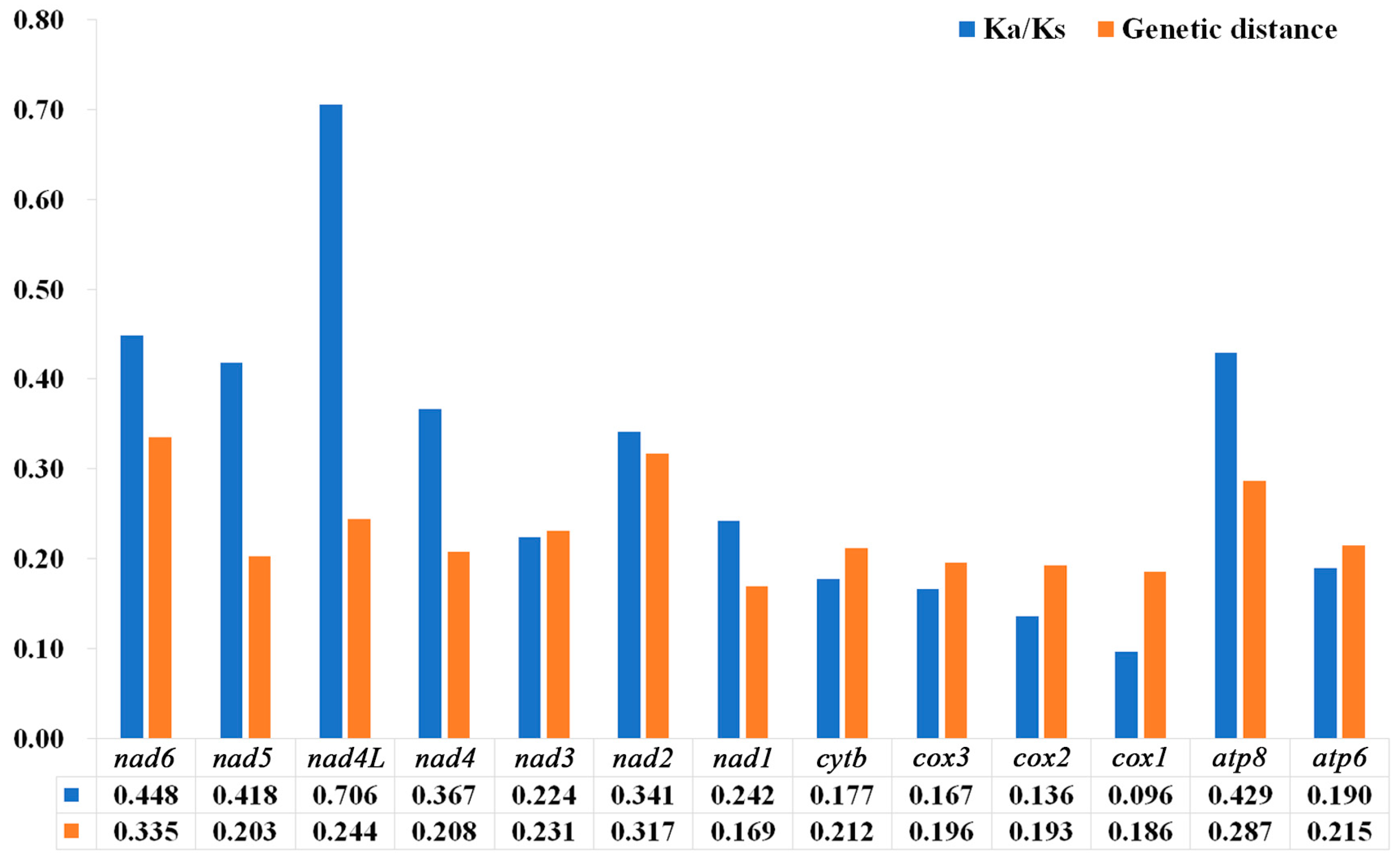
The results showed that the Ka/Ks values of the 13 PCGs were between 0.096–0.706, and the Ka/Ks values were all less than 1 (Figure 8), representing all the genes that evolved under purifying selection. Furthermore, cox1 (0.096) had the lowest Ka/Ks value, the lowest evolution rate, and exhibited the strongest purifying selection. In contrast, nad4L (0.706) and nad6 (0.448) showed higher Ka/Ks values than the other PCGs, and they exhibited relaxed purifying selection. The results of the pairwise genetic distances of the 13 PCGs of the 10 species of Nitidulidae are shown in Figure 8. cox1 (0.186) and nad1 (0.169) evolved relatively slowly, while nad6 (0.335) and nad2 (0.317) evolved relatively quickly.
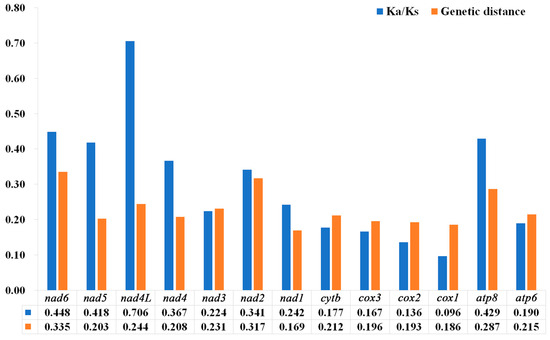
Figure 8.
Genetic distances and ratios of non-synonymous (Ka) to synonymous (Ks) substitution rates of 13 PCGs among 10 species of Nitidulidae. The average value for each PCG is shown below the gene name.
A nucleotide diversity analysis is the key to designing species-specific markers, and it aids in the molecular identification of species that are difficult to identify based on morphology [65,66,67]. Generally, cox1 can be used as a potential marker for species identification and has been widely used in insect classification [68]. While in this study, cox1 was the most conserved gene in the mitochondrial genomes of Nitidulidae, nad6 had the fastest evolutionary rate compared with the other PCGs. Therefore, nad6 may be more suitable as a barcode gene for species identification of Nitidulidae.
3.6. Phylogenetic Analysis
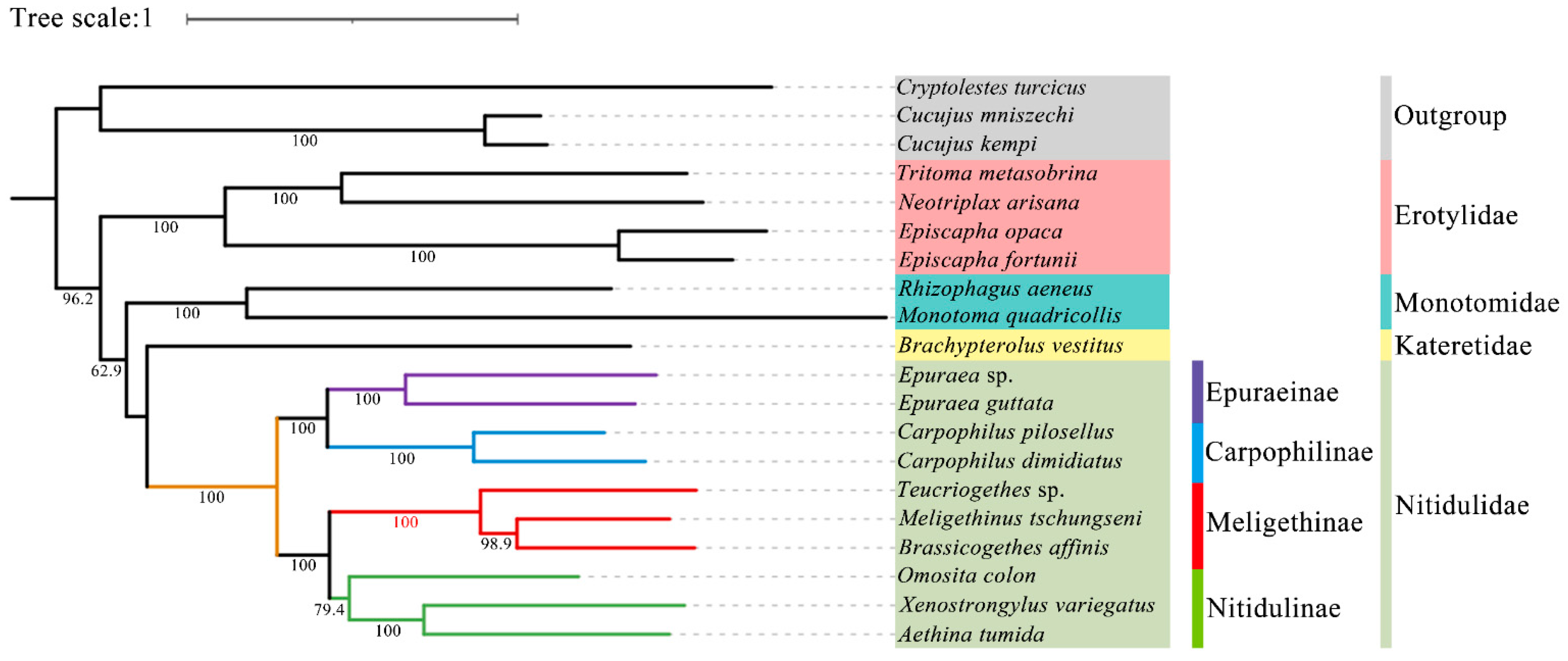
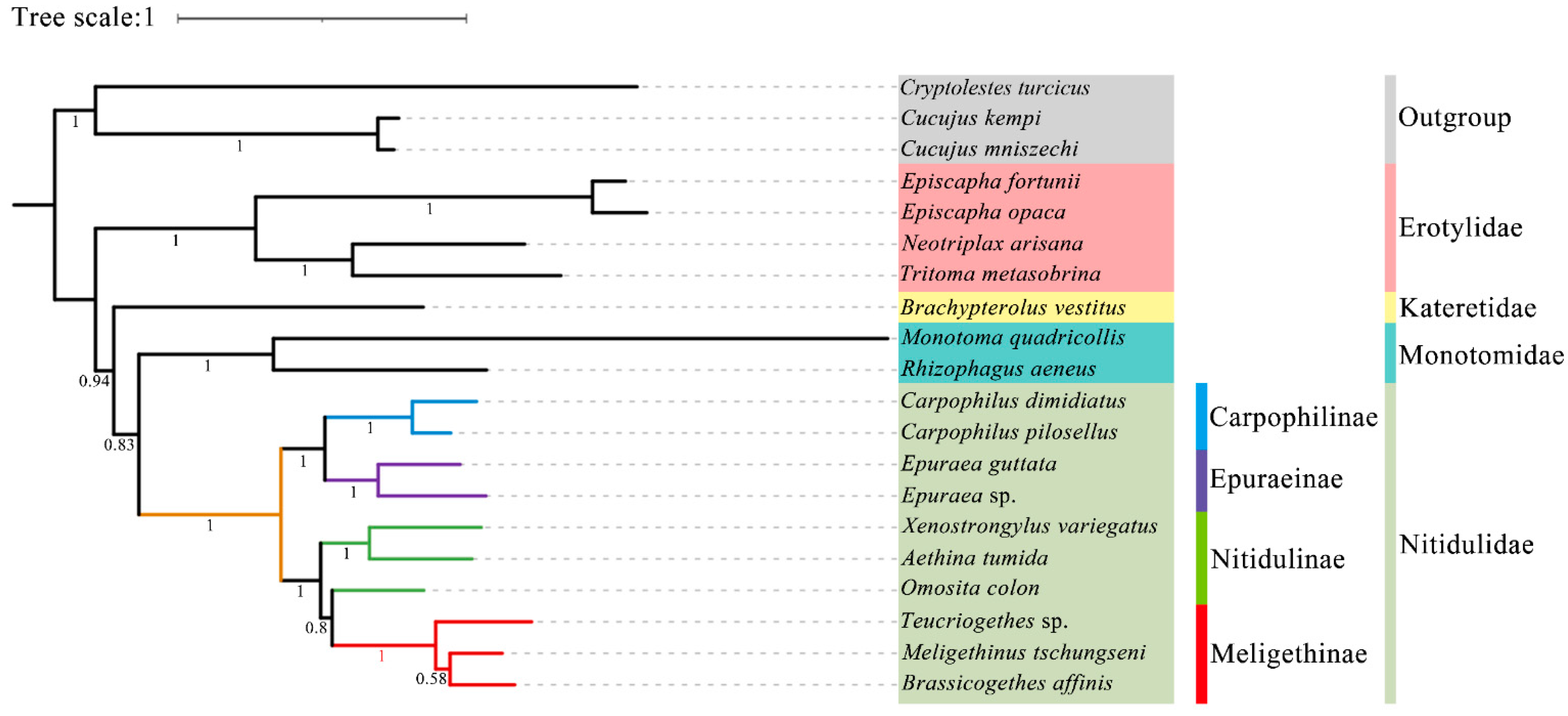
In this study, all the sites of the nucleotide substitution saturation test (Table S1) in the Gblocks showed that the index of substitution saturation (Iss) was less than the critical Iss based on a symmetrical tree (Iss.cSym) and p < 0.05. Table S2 lists the best model and optimal partitioning strategy selected for the four datasets of the ML analysis by ModelFinder. Four datasets (PCG12, PCG123, PCG12R, PCG123R) were used to construct ML trees and PhyloBayes trees for 20 species of Coleoptera, and a total of eight phylogenetic trees were obtained (Figure 9 and Figure 10 and Figures S1–S6).
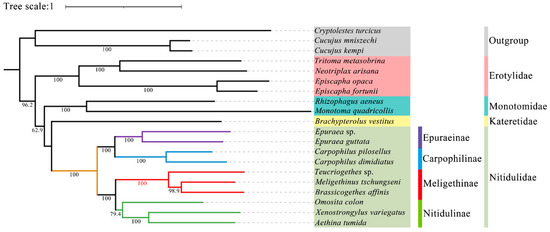
Figure 9.
Phylogenetic tree generated based on the ML analysis of the PCG123 dataset under the best model. The bootstrap support values of the corresponding nodes are represented by Arabic numerals.
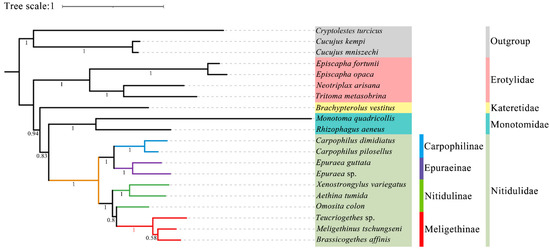
Figure 10.
Phylogenetic tree generated based on the PhyloBayes analysis of the PCG123 dataset under the site-heterogeneous mixture CAT + GTR substitution model. The Bayesian posterior probabilities of the corresponding nodes are represented by Arabic numerals.
Although the topological structure of these eight trees are not exactly same, they all strongly support (ML bootstrap support values (BS) = 100, Bayesian posterior probabilities (BPP) = 1) that the four subfamilies (Nitidulinae, Meligethinae, Carpophilinae, Epuraeinae) of Nitidulidae formed a monophyletic group based on the available complete mitochondrial genomes (Figure 9 and Figure 10 and Figures S1–S6). We further assumed that Nitidulidae is monophyletic, which is consistent with the results of studies based on morphological and molecular data analysis [1,3,27,28,29,30]. Regarding the sister group relationship of Nitidulidae, based on the PCG123 dataset analyzed using ML, the topological structure of the tree supported that the sister group relationship of Nitidulidae is Nitidulidae + Kateretidae (Figure 9), which is consistent with previous studies based on morphological characteristics [7,31,32], short molecular fragments [2,25,28], and fossil data [3]. Another seven topological trees showed that the sister group relationship of Nitidulidae is Nitidulidae + Monotomidae, but this clade has a BS of < 69 and BPP of < 0.85 (Figure 10 and Figures S1–S6). Previous studies have shown that clades with a of BS 50–69 or BPP of 0.85–0.89 are considered weakly supported, and clades with a BS of < 50 or BPP of < 0.85 are considered unsupported [69]; therefore, the sister group relationship of Nitidulidae + Monotomidae is untenable. Previously, Chen et al. [29] first proposed a sister group relationship of Nitidulidae + Monotomidae based on the complete mitochondrial genomes of 17 species. However, this clade had a low BS in ML trees. This study further supported that the sister group relationship of Nitidulidae is Nitidulidae + Kateretidae; however, considering that there is only one complete mitochondrial genome sequence of Kateretidae in GenBank, in future studies, we will strive to sequence more mitochondrial genomes of Kateretidae and Nitidulidae to explore the sister group relationship of Nitidulidae more clearly.
More importantly, although the mitochondrial genomes of all the genera in Meligethinae have not yet been obtained, this study attempted to analyze the taxonomic status and sister group relationship of Meligethinae based on the complete mitochondrial genomes of three genera and three species. The topological structures of all eight trees (Figure 9 and Figure 10 and Figures S1–S6) clearly showed that three genera (Meligethinus, Brassicogethes, Teucriogethes) of Meligethinae clustered into a single clade. Therefore, we further assumed that Meligethinae is monophyletic (BS = 100, BPP = 1), which is consistent with the studies based on adult morphological characteristics, biological habits, and short molecular fragments by Kirejtshuk et al. [34,35,36,37], Trizzino et al. [38], Audisio et al. [39], and Lee et al. [1]. Regarding the sister group relationship of Meligethinae, four ML trees supported (BS = 100) the sister group relationship of Meligethinae + Nitidulinae (Figure 9 and Figures S1–S3). However, the four BI trees showed that the zoosaprophagous Omosita colon [70] and the anthophagous Meligethinae [71] were abnormally clustered into one clade (Figure 10 and Figures S4–S5), but the BPP of this clade was <0.85, so this clade was considered unsupported [69]. In future studies, we plan to add new sequences of representative species of each subfamily of Nitidulidae, as well as of different and more distantly related genera and complexes of genera within Nitidulinae, to further explore the phylogenetic relationships of Meligethinae.
4. Conclusions
In this study, two complete mitochondrial genomes of Meligethinae (Meligethinus tschungseni and Brassicogethes affinis) were provided for the first time, and the mitochondrial genomes of 10 species among Nitidulidae were compared. The phylogenetic trees of 20 species of related families of Coleoptera were constructed to try to analyze the higher phylogeny of Nitidulidae. Based on the available complete mitochondrial genomes, this study confirmed that the four subfamilies (Nitidulinae, Meligethinae, Carpophilinae, Epuraeinae) of Nitidulidae formed a monophyletic group, further supporting that the sister group relationship of Nitidulidae is Nitidulidae + Kateretidae. This study also assumed that Meligethinae is monophyletic (BS = 100, BPP = 1) based on the complete mitochondrial genome, which was analyzed for the first time, and its sister group relationship is likely to be Meligethinae + Nitidulinae.
Considering that the representative genera used in this study do not cover all genera, in the future, it is necessary to sequence the complete mitochondrial genomes of more species for an in-depth molecular phylogenetic analysis of Nitidulidae. Furthermore, the phylogenetic analysis of Nitidulidae can be based on an integrative approach, such as combining the morphological characteristics of adults and larvae, mitochondrial genome data, nuclear genome data, and fossils, as well as biological information such as host plants, etc.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/insects15010057/s1, Table S1: Substitution saturation tests for PCGs, rRNAs, and tRNAs of mitochondrial genomes of Coleoptera; Table S2: The best model and optimal partitioning strategy selected for the four datasets of the ML analysis using ModelFinder; Figure S1: Phylogenetic tree produced from the ML analysis based on the PCG12 dataset; Figure S2: Phylogenetic tree produced from the ML analysis based on the PCG12R dataset; Figure S3: Phylogenetic tree produced from the ML analysis based on the PCG123R dataset; Figure S4: Phylogenetic tree produced from the PhyloBayes analysis based on the PCG12 dataset; Figure S5: Phylogenetic tree produced from the PhyloBayes analysis based on the PCG12R dataset; Figure S6: Phylogenetic tree produced from the PhyloBayes analysis based on the PCG123R dataset.
Author Contributions
Conceptualization, M.L. and P.A.; methodology, J.D., M.L., W.W. and A.D.G.; validation, J.D., S.S. and P.A.; investigation, J.D., M.L., S.S. and P.A.; data analysis, J.D. and M.L.; resources, W.W. and M.L.; writing—original draft preparation, J.D. and M.L.; writing—review and editing, A.D.G., S.S. and P.A.; funding acquisition, M.L. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the National Natural Science Foundation of China (No. 32000321), with funding awarded to the corresponding author, M.L.
Data Availability Statement
The newly sequenced mitochondrial genome in this study has been uploaded to GenBank (ON782471, ON782472, OR387485).
Acknowledgments
We appreciate the constructive comments of the anonymous reviewers and processing editors and their help in revising the manuscript.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Lee, M.H.; Lee, S.; Leschen, R.A.B.; Lee, S. Evolution of feeding habits of sap beetles (Coleoptera: Nitidulidae) and placement of Calonecrinae. Syst. Entomol. 2020, 45, 911–923. [Google Scholar] [CrossRef]
- Robertson, J.A.; Ślipiński, A.; Moulton, M.; Shockley, F.W.; Giorgi, A.; Lord, N.P.; Mckenna, D.D.; Tomaszewska, W.; Forrester, J.; Miller, K.B. Phylogeny and classification of Cucujoidea and the recognition of a new superfamily Coccinelloidea (Coleoptera: Cucujiformia). Syst. Entomol. 2015, 40, 745–778. [Google Scholar] [CrossRef]
- Cai, C.; Tihelka, E.; Giacomelli, M.; Lawrence, J.F.; Kundrata, R.; Yamamoto, S.; Thayer, M.K.; Newton, A.F.; Leschen, R.A.B.; Gimmel, M.L.; et al. Integrated phylogenomics and fossil data illuminate the evolution of beetles. R. Soc. Open Sci. 2022, 9, 211771. [Google Scholar] [CrossRef]
- Audisio, P. Fauna d’Italia 32. Coleoptera, Nitidulidae–Kateridae; Edizioni Calderini: Roma, Italy, 1993; Volume 32, p. 971. [Google Scholar]
- Audisio, P.; Cline, A.R.; De Biase, A.; Antonini, G.; Mancini, E.; Trizzino, M.; Costantini, L.; Strika, S.; Lamanna, F.; Cerretti, P. Preliminary re-examination of genus-level taxonomy of the pollen beetle subfamily Meligethinae (Coleoptera: Nitidulidae). Acta Entomol. Musei Natl. Pragae 2009, 49, 341–504. [Google Scholar]
- Audisio, P.; Sabatelli, S.; Jelínek, J. Revision of the pollen beetle genus Meligethes (Coleoptera: Nitidulidae). Fragm. Entomol. 2014, 46, 19–112. [Google Scholar] [CrossRef][Green Version]
- Jelínek, J.; Carlton, C.E.; Cline, A.R.; Leschen, R.A.B. Nitidulidae Latreille. In Handbook of Zoology. Arthropoda: Insecta. Coleoptera, 2 Morphology and Systematics (Elateroidea, Bostrichiformia, Cucujiformia Partim); Beutel, R.G., Leschen, R.A.B., Lawrence, J.F., Eds.; Walter De Gruyter: Berlin, Germany, 2010; pp. 390–407. [Google Scholar]
- Liu, M.; Yang, X.; Huang, M.; Jelínek, J.; Audisio, P. Four new species of Meligethes Stephens from China and additional data on other species of the genus (Coleoptera: Nitidulidae: Meligethinae). Zootaxa 2016, 4121, 101–116. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Huang, M.; Cline, A.R.; Audisio, P. Two new Lamiogethes Audisio & Cline from China (Coleoptera: Nitidulidae, Meligethinae). Fragm. Entomol. 2017, 49, 145–150. [Google Scholar]
- Liu, M.; Huang, M.; Cline, A.R.; Sabatelli, S.; Audisio, P. A new species of Meligethes Stephens from China and additional data on members of the M. chinensis species-complex (Coleoptera: Nitidulidae, Meligethinae). Fragm. Entomol. 2017, 49, 79–84. [Google Scholar] [CrossRef][Green Version]
- Liu, M.; Huang, M.; Cline, A.R.; Audisio, P. New and poorly known Meligethes Stephens from China, with bionomical data on some species (Coleoptera: Nitidulidae: Meligethinae). Zootaxa 2018, 4392, 546–566. [Google Scholar] [CrossRef]
- Liu, M.; Huang, M.; Cline, A.R.; Cardoli, P.; Audisio, P.; Sabatelli, S. Re-examination of the genus-level taxonomy of the pollen beetle subfamily Meligethinae—Part 1. Sagittogethes Audisio & Cline 2009 and allied genera; with description of a new genus (Coleoptera: Nitidulidae). Fragm. Entomol. 2020, 52, 119–135. [Google Scholar]
- Liu, M.; Yang, X.K.; Huang, M.; Cline, A.R.; Sabatelli, S.; Audisio, P. Five new species of Lamiogethes Audisio & Cline from China (Coleoptera: Nitidulidae: Meligethinae). Zootaxa 2020, 4728, 63–76. [Google Scholar]
- Liu, M.; Huang, M.; Cline, A.R.; Mancini, E.; Scaramuzzi, A.; Paradisi, S.; Audisio, P.; Badano, D.; Sabatelli, S. Rosaceae, Brassicaceae and pollen beetles: Exploring relationships and evolution in an anthophilous beetle lineage (Nitidulidae, Meligethes-complex of genera) using an integrative approach. Front. Zool. 2021, 18, 9. [Google Scholar] [CrossRef] [PubMed]
- Sabatelli, S.; Liu, M.; Badano, D.; Mancini, E.; Trizzino, M.; Cline, A.R.; Endrestol, A.; Huang, M.; Audisio, P. Molecular phylogeny and host-plant use (Lamiaceae) of the Thymogethes pollen beetles (Coleoptera). Zool. Scr. 2020, 49, 28–46. [Google Scholar] [CrossRef]
- Kirejtshuk, A.G. On recent knowledge on the sap-beetles (Coleoptera, Nitidulidae) of India. Zool. Inst. Russ. Acad. Sci. 1999, 1, 21–32. [Google Scholar]
- Krishnan, K.T.; Neumann, P.; Ahmad, A.H.; Pimid, M. A scientific note on the association of Haptoncus luteolus (Coleoptera: Nitidulidae) with colonies of multiple stingless bee species. Apidologie 2015, 46, 262–264. [Google Scholar] [CrossRef]
- Deng, G.R.; Zeng, Q.D.; Li, W.Q.; Huang, D.X.; Zhou, Z.H. The harm of Tricanus japonicus on Dictyophora indusiata and its control. Plant Prot. 2006, 32, 117–118. [Google Scholar]
- Lounsberry, Z.; Spiewok, S.; Pernal, S.F.; Sonstegard, T.S.; Hood, W.M.; Pettis, J.; Neumann, P.; Evans, J.D. Worldwide Diaspora of Aethina tumida (Coleoptera: Nitidulidae), a Nest Parasite of Honey Bees. Ann. Entomol. Soc. Am. 2010, 103, 671–677. [Google Scholar] [CrossRef]
- Bai, X.G.; Cao, Y.; Zhou, Y.X. Identification of seven species of larvae of Nitidulidae from China. J. Zhengzhou Grain Coll. 1992, 50–56. [Google Scholar] [CrossRef]
- Dobson, R.M. The Species of Carpophilus Stephens (Col. Nitidulidae) associated with Stored Products. Bull. Entomol. Res. 1954, 45, 389–402. [Google Scholar] [CrossRef]
- Nadel, H.; Pena, J.E. Identity, Behavior, and Efficacy of Nitidulid Beetles (Coleoptera: Nitidulidae) Pollinating Commercial Annona Species in Florida. Environ. Entomol. 1994, 23, 878–886. [Google Scholar] [CrossRef]
- Yang, Q.P.; Liu, W.C.; Huang, C. Statistics and analysis on oilseed rape losses caused by main diseases and insect pests in recent ten years. Plant Prot. 2018, 44, 24–30. [Google Scholar]
- He, C.G.; Wang, G.L.; Fan, Y.H.; Zou, Y.X.; Deng, Y.X.; Deng, H.Y. Study on a new pest of oilseed-Xenostrongylus variegatus. Acta Agric. Boreali Occident. Sin. 1998, 7, 18–23. [Google Scholar]
- Bocak, L.; Barton, C.; Crampton-Platt, A.; Chesters, D.; Ahrens, D.; Vogler, A.P. Building the Coleoptera tree-of-life for >8000 species: Composition of public DNA data and fit with Linnaean classification. Syst. Entomol. 2014, 39, 97–110. [Google Scholar] [CrossRef]
- Tang, P.; Li, M.; Feng, R.; Wang, J.; Liu, M.; Wang, Y.; Yuan, M. Phylogenetic relationships among superfamilies of Cucujiformia (Coleoptera: Polyphaga) inferred from mitogenomic data. Sci. Sinca 2019, 49, 163–171. [Google Scholar] [CrossRef]
- Lawrence, J.F.; Ślipiński, A.; Seago, A.E.; Thayer, M.K.; Newton, A.F.; Marvaldi, A.E. Phylogeny of the Coleoptera Based on Morphological Characters of Adults and Larvae. Ann. Zool. 2011, 61, 1–217. [Google Scholar] [CrossRef]
- Cline, A.R.; Smith, T.R.; Miller, K.; Moulton, M.; Whiting, M.; Audisio, P. Molecular phylogeny of Nitidulidae: Assessment of subfamilial and tribal classification and formalization of the family Cybocephalidae (Coleoptera: Cucujoidea). Syst. Entomol. 2014, 39, 758–772. [Google Scholar] [CrossRef]
- Chen, X.; Song, Q.; Huang, M. Characterization of the Complete Mitochondrial Genomes from Two Nitidulid Pests with Phylogenetic Implications. Insects 2020, 11, 779. [Google Scholar] [CrossRef]
- McKenna, D.D.; Shin, S.; Ahrens, D.; Balke, M.; Beza-Beza, C.; Clarke, D.J.; Donath, A.; Escalona, H.E.; Friedrich, F.; Letsch, H.; et al. The evolution and genomic basis of beetle diversity. Proc. Natl. Acad. Sci. USA 2019, 116, 24729–24737. [Google Scholar] [CrossRef]
- Leschen, R.A.; Lawrence, J.F.; Ślipiński, S. Classification of basal Cucujoidea (Coleoptera: Polyphaga): Cladistic analysis, keys and review of new families. Invertebr. Syst. 2005, 19, 17–73. [Google Scholar] [CrossRef]
- Cline, A.R.; Slipinski, S.A. Discolomatidae Horn, 1878. In Handbuch der Zoologie/Handbook of Zoology; Leschen, R.A.B., Beutel, R.G., Lawrence, J.F., Eds.; Band. Arthropoda: Insecta, Teilband; W. DeGruyter: Berlin, Germany, 2010; Volume IV, pp. 435–442. [Google Scholar]
- Li, Y.W.; Yu, L.; Zhang, Y.P. “Long-branch Attraction” artifact in phylogenetic reconstruction. Hereditas 2020, 29, 659–667. [Google Scholar] [CrossRef]
- Kirejtshuk, A.G. English translation in Entomological Review. Systematic position of the genus Calonecrus J. Thomas and notes on the phylogeny of the family Nitidulidae (Coleoptera). Entomologicheskoye Obozreniye. Russ. Engl. Transl. Entomol. Rev. 1982, 61, 117–129. [Google Scholar]
- Kirejtshuk, A.G. On polyphyly of the Carpophilinae with description of a new subfamily, Cillaeinae (Coleoptera: Nitidulidae). Coleopt. Bull. 1986, 40, 217–221. [Google Scholar]
- Kirejtshuk, A.G. New taxa of the Nitidulidae (Coleoptera) of the eastern hemisphere. Trudy Zool. Inst. Ross. Akad. Nauk 1995, 258, 3–50. [Google Scholar]
- Kirejtshuk, A.G. A current generic classification of sap beetles (Coleoptera, Nitidulidae). Zoosyst. Ross. 2008, 17, 107–122. [Google Scholar] [CrossRef]
- Trizzino, M.; Audisio, P.; Antonini, G.; De Biase, A.; Mancini, E. Comparative analysis of sequences and secondary structures of the rRNA internal transcribed spacer 2 (ITS2) in pollen beetles of the subfamily Meligethinae (Coleoptera, Nitidulidae): Potential use of slippage-derived sequences in molecular systematics. Mol. Phylogenet. Evol. 2009, 51, 215–226. [Google Scholar] [CrossRef]
- Audisio, P.; De Biase, A.; Trizzino, M.; Mancini, E.; Antini, G. A new species of Meligethes (Coleoptera: Nitidulidae: Meligethinae) of the M. lugubris complex from Sardinia. Zootaxa 2009, 2318, 386–393. [Google Scholar] [CrossRef]
- Song, F.; Li, H.; Jiang, P.; Zhou, X.; Liu, J.; Sun, C.; onVogler, A.P.; Cai, W. Capturing the Phylogeny of Holometabola with Mitochondrial Genome Data and Bayesian Site-Heterogeneous Mixture Models. Genome Biol. Evol. 2016, 8, 1411–1426. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, H.; Song, F.; Zhao, Y.; Wilson, J.J.; Cai, W. Higher-level phylogeny and evolutionary history of Pentatomomorpha (Hemiptera: Heteroptera) inferred from mitochondrial genome sequences. Syst. Entomol. 2019, 44, 810–819. [Google Scholar] [CrossRef]
- Li, Q.; Yang, L.; Xiang, D.; Wan, Y.; Wu, Q.; Huang, W.; Zhao, G. The complete mitochondrial genomes of two model ectomycorrhizal fungi (Laccaria): Features, intron dynamics and phylogenetic implications. Int. J. Biol. Macromol. 2020, 145, 974–984. [Google Scholar] [CrossRef]
- Du, Z.; Hasegawa, H.; Cooley, J.R.; Simon, C.; Yoshimura, J.; Cai, W.; Sota, T.; Li, H. Mitochondrial Genomics Reveals Shared Phylogeographic Patterns and Demographic History among Three Periodical Cicada Species Groups. Mol. Biol. Evol. 2019, 36, 1187–1200. [Google Scholar] [CrossRef]
- Dowton, M.; Cameron, S.L.; Dowavic, J.I.; Austin, A.D.; Whiting, M.F. Characterization of 67 Mitochondrial tRNA Gene Rearrangements in the Hymenoptera Suggests That Mitochondrial tRNA Gene Position Is Selectively Neutral. Mol. Biol. Evol. 2009, 26, 1607–1617. [Google Scholar] [CrossRef] [PubMed]
- Song, S.N.; Tang, P.; Wei, S.J.; Chen, X.X. Comparative and phylogenetic analysis of the mitochondrial genomes in basal hymenopterans. Sci. Rep. 2016, 6, 20972. [Google Scholar] [CrossRef] [PubMed]
- Mao, M.; Gibson, T.; Dowton, M. Higher-level phylogeny of the Hymenoptera inferred from mitochondrial genomes. Mol. Biol. Evol. 2015, 84, 34–43. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Lan, Y.; Xia, L.; Cui, M.; Sun, W.; Dong, Z.; Cao, Y. The First Complete Mitochondrial Genomes of Two Sibling Species from Nitidulid Beetles Pests. Insects 2020, 11, 24. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Wang, Y.; Wang, M.; Wang, Y.; Zhang, Y.; Wang, J. Characterization of the complete mitochondrial genome of Omosita colon (Coleoptera: Nitidulidae). Mitochondrial DNA B 2021, 6, 1547–1553. [Google Scholar] [CrossRef]
- Duquesne, V.; Delcont, A.; Huleux, A.; Beven, V.; Touzain, F.; Ribiere-Chabert, M. Complete Mitochondrial Genome Sequence of Aethina tumida (Coleoptera: Nitidulidae), a Beekeeping Pest. Genome Announc. 2017, 5, e01165-17. [Google Scholar] [CrossRef] [PubMed]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Bernt, M.; Donath, A.; Juhling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Putz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Biol. Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef]
- Lowe, T.M.; Chan, P.P. tRNAscan-SE On-line: Integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016, 44, W54–W57. [Google Scholar] [CrossRef]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef]
- Lohse, M.; Drechsel, O.; Kahlau, S.; Bock, R. OrganellarGenomeDRAW—A suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 2013, 41, W575–W581. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Gao, F.; Jakovlic, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Rozas, J.; Ferrer-Mata, A.; Sanchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sanchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.Y.; Liu, G.; Li, L.; Xin, T.; Lei, K.; Xia, B. The complete mitochondrial genome of Cryptolestes turcicus (Grouvelle) (Coleoptera: Laemophloeidae). Mitochondrial DNA A 2016, 27, 3701–3702. [Google Scholar] [CrossRef] [PubMed]
- Jin, M.; Zwick, A.; Ślipiński, A.; Marris, J.W.M.; Thomas, M.C.; Pang, H. A comprehensive phylogeny of flat bark beetles (Coleoptera: Cucujidae) with a revised classification and a new South American genus. Syst. Entomol. 2020, 45, 248–268. [Google Scholar] [CrossRef]
- Liu, J.; Wang, Y.; Zhang, R.; Shi, C.; Lu, W.; Li, J.; Bai, M. Three Complete Mitochondrial Genomes of Erotylidae (Coleoptera: Cucujoidea) with Higher Phylogenetic Analysis. Insects 2021, 12, 524. [Google Scholar] [CrossRef]
- Lam-Tung, N.; Schmidt, H.A.; von Haeseler, A.; Bui Quang, M. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef]
- Ojala, D.; Montoya, J.; Attardi, G. tRNA punctuation model of RNA processing in human mitochondria. Nature 1981, 290, 470–474. [Google Scholar] [CrossRef]
- Mancini, E.; De Biase, A.; Mariottini, P.; Bellini, A.; Audisio, P. Structure and evolution of the mitochondrial control region of the pollen beetle Meligethes thalassophilus (Coleoptera: Nitidulidae). Genome 2008, 51, 196–207. [Google Scholar] [CrossRef] [PubMed]
- Ye, F.; Easy, R.H.; King, S.D.; Cone, D.K.; You, P. Comparative analyses within Gyrodactylus (Platyhelminthes: Monogenea) mitochondrial genomes and conserved polymerase chain reaction primers for gyrodactylid mitochondrial DNA. J. Fish Dis. 2017, 40, 541–555. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Zhang, Z.; Niu, L.; Wang, Q.; Wang, C.; Lan, J.; Deng, J.; Fu, Y.; Nie, H.; Yan, N.; et al. The mitochondrial genome of Baylisascaris procyonis. PLoS ONE 2011, 6, e27066. [Google Scholar] [CrossRef] [PubMed]
- Jia, W.Z.; Yan, H.B.; Guo, A.J.; Zhu, X.Q.; Wang, Y.C.; Shi, W.G.; Chen, H.T.; Zhan, F.; Zhang, S.H.; Fu, B.Q.; et al. Complete mitochondrial genomes of Taenia multiceps, T. hydatigena and T. pisiformis: Additional molecular markers for a tapeworm genus of human and animal health significance. BMC Genom. 2010, 11, 447. [Google Scholar] [CrossRef] [PubMed]
- Hebert, P.D.N.; Ratnasingham, S.; deWaard, J.R. Barcoding animal life: Cytochrome c oxidase subunit 1 divergences among closely related species. R. Soc. 2003, 270 (Suppl. S1), S96–S99. [Google Scholar] [CrossRef]
- Cai, C.Y.; Wang, Y.L.; Liang, L.; Yin, Z.W.; Thayer, M.K.; Newton, A.F.; Zhou, Y.L. Congruence of morphological and molecular phylogenies of the rove beetle subfamily Staphylininae (Coleoptera: Staphylinidae). Sci. Rep. 2019, 9, 15137. [Google Scholar] [CrossRef]
- Cao, Y.K.; Huang, M. A SEM study of the antenna and mouthparts of Omosita colon (Linnaeus) (Coleoptera: Nitidulidae). Microsc. Res. Tech. 2016, 79, 1152–1164. [Google Scholar] [CrossRef]
- Crowson, R.A. A new genus of Boganiidae (Coleoptera) from Australia, with observations on glandular openings, cycad associations and geographical distribution in the family. Aust. J. Entomol. 1990, 29, 91–99. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).