Bridging the Gap between Field Experiments and Machine Learning: The EC H2020 B-GOOD Project as a Case Study towards Automated Predictive Health Monitoring of Honey Bee Colonies
Abstract
:Simple Summary
Abstract
1. Introduction
2. The B-GOOD Project as a Case Study for a Standard Health Monitoring Method of Honey Bee Colonies and the Development of a Health Status Index (HSI)
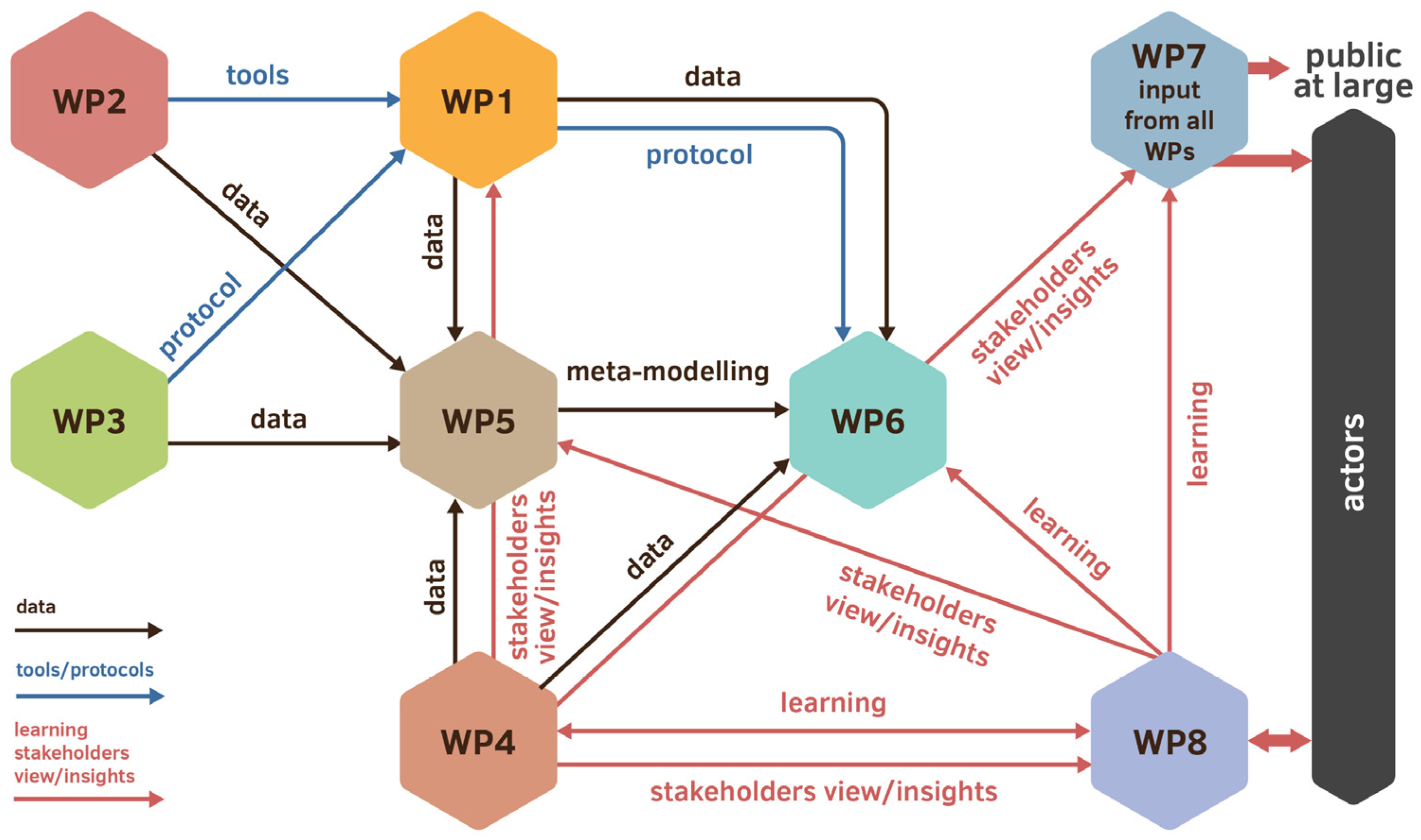
3. Infrastructure for Data Collection
- More bee health (semi)automated monitoring in a bee and user-friendly fashion.
- A ready to use end product of B-GOOD, validated by end users (primarily beekeepers) with a decreased amount of support.
- A larger coverage of the EU territory with increased diversity of bees, hives, and business models, and a variety of environments (ecotypes) where the monitoring of the bee health took place.
- Current health: No clinical symptoms of disease were detected by visual inspection and supported by laboratory analysis. In addition, when food resources are available, there should be brood in all stadia (BIAS) and foraging activity when weather permits it. When there are no food resources available, or foraging activity is hampered, there should be sufficient storage of resources for survival until this down period ends.
- Future health: Able to survive the winter or other long period of low resource availability, and to reproduce or to be willing to reproduce during the growing season.
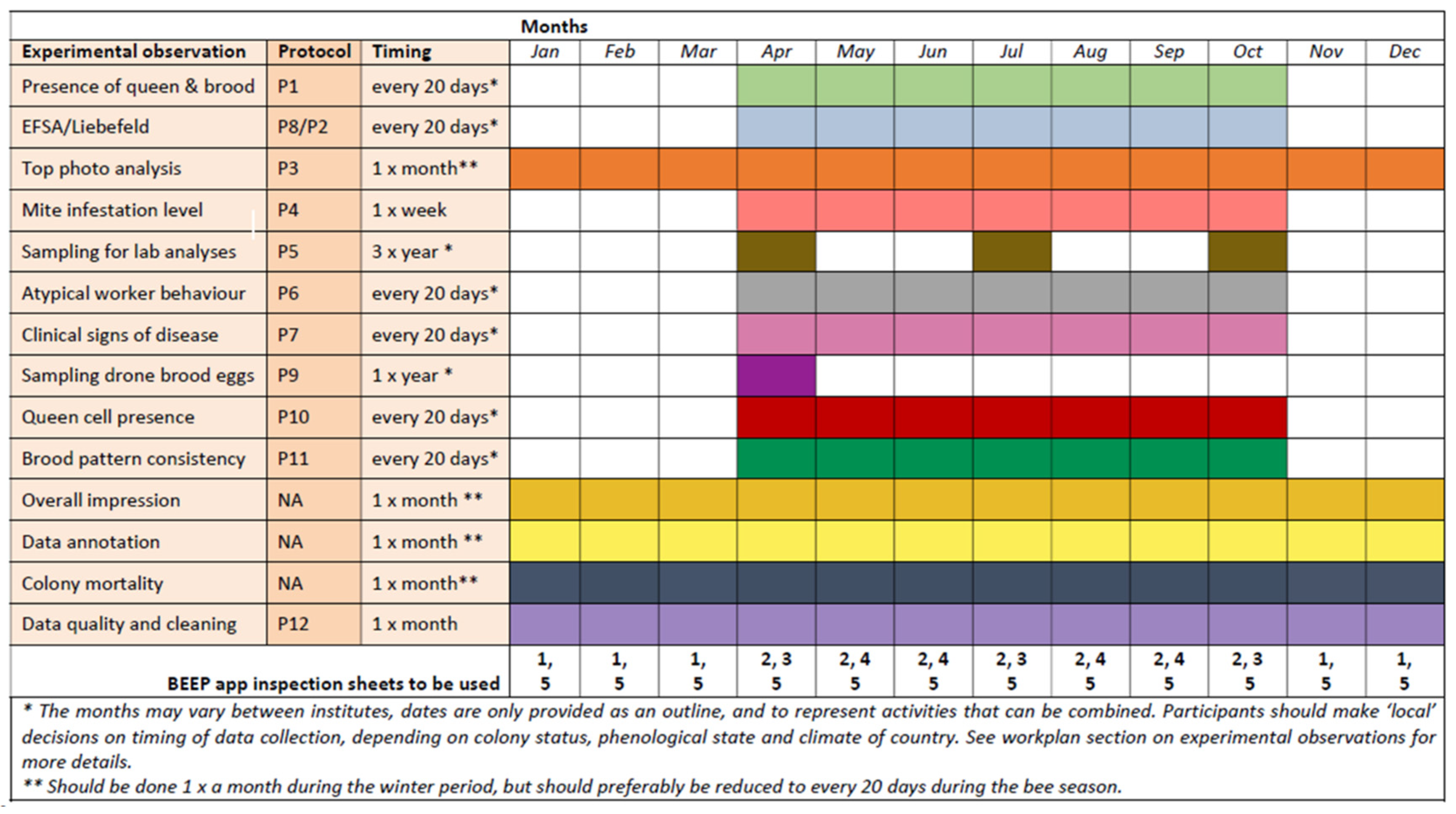
4. Data Collected
5. Harmonization and Standardization of the Workflow
- The guidance and standardization of beekeeping decreased.
- The number of protocols and invasiveness for the honey bee colonies decreased.
- The readability and user-friendliness of the protocols increased.
- The reliance on automated sensors and digital logging of management actions increased.
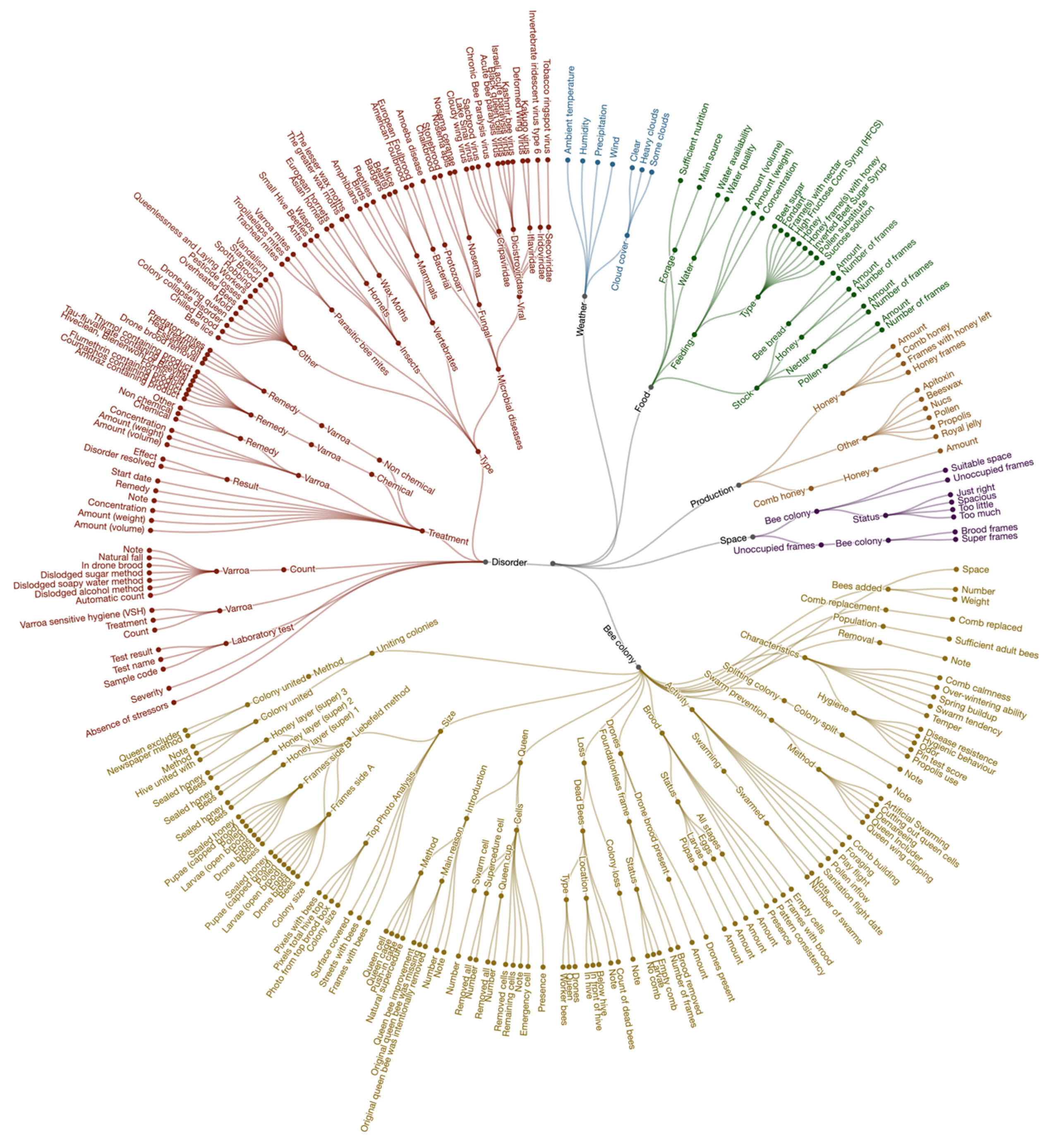
6. Standardization and Implementation of Data Language
- It enabled researchers to select and collect the data they needed for analysis.
- It helped data collectors enter high-quality data by using standard options.
- It facilitated structured data storage, enabling data comparison between research participants, colonies, locations, and potential meta-analysis research studies.
- Apiary
- ▪
- Hive
- o
- Queen
- -
- Inspection
- -
- Bee colony
- -
- Disorder
- -
- Food
- -
- Overall
- -
- Production
- -
- Weather
- o
- Device
- -
- Measurement
- Category name (in English and translated)
- Parent (the identifier of the parent category)
- Definition (the meaning of the category name in English plus the source)
- Input type (the kind of input field, such as an option list, smiley, or number)
- Physical quantity (the unit)
- Long or short description (long meaning a data collection instruction with optional visuals, such as a top photo analysis [see protocol Tier 1, P3 in Supplementary Information S1]; short meaning a sentence).
7. Managing High-Quality Data in an Open Science Platform
8. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aizen, M.A.; Harder, L.D. The global stock of domesticated honey bees is growing slower than agricultural demand for pollination. Curr. Biol. 2009, 19, 915–918. [Google Scholar] [CrossRef]
- Goulson, D.; Nicholls, E.; Botías, C.; Rotheray, E. Bee declines driven by combined stress from parasites, pesticides, and lack of flowers. Science 2015, 347, 1255957. [Google Scholar] [CrossRef] [PubMed]
- Jacques, A.; Laurent, M.; EPILOBEE Consortium; Ribière-Chabert, M.; Saussac, M.; Bougeard, S.; Budge, G.E.; Hendrikx, P.; Chauzat, M.-P. A pan-European epidemiological study reveals honey bee colony survival depends on beekeeper education and disease control. PLoS ONE 2017, 12, e0172591. [Google Scholar] [CrossRef] [PubMed]
- Potts, S.G.; Biesmeijer, J.C.; Kremen, C.; Neumann, P.; Schweiger, O.; Kunin, W.E. Global pollinator declines: Trends, impacts and drivers. Trends Ecol. Evol. 2010, 25, 345–353. [Google Scholar] [CrossRef] [PubMed]
- Potts, S.G.; Roberts, S.P.; Dean, R.; Marris, G.; Brown, M.A.; Jones, R.; Neumann, P.; Settele, J. Declines of managed honey bees and beekeepers in Europe. J. Apic. Res. 2010, 49, 15–22. [Google Scholar] [CrossRef]
- Martinello, M.; Manzinello, C.; Dainese, N.; Giuliato, I.; Gallina, A.; Mutinelli, F. The honey bee: An active bio sampler of environmental pollution and a possible warning biomarker for human health. Appl. Sci. 2021, 11, 6481. [Google Scholar] [CrossRef]
- Francis, R.M.; Nielsen, S.L.; Kryger, P. Varroa-virus interaction in collapsing honey bee colonies. PLoS ONE 2013, 8, e57540. [Google Scholar] [CrossRef]
- Genersch, E.; von der Ohe, W.; Kaatz, H.; Schroeder, A.; Otten Ch Büchler, R.; Berg, S.; Ritter, W.; Mühlen, W.; Gisder, S.; Meixner, M.; et al. The German bee monitoring project: A long-term study to understand periodically high winter losses of honey bee colonies. Apidologie 2010, 41, 332–352. [Google Scholar] [CrossRef]
- Meixner, M.D.; Francis, R.M.; Gajda, A.; Kryger, P.; Andonov, S.; Uzunov, A.; Topolska, G.; Costa, C.; Amiri, E.; Berg, S.; et al. Occurrence of parasites and pathogens in honey bee colonies used in a European genotype-environment interactions experiment. J. Apic. Res. 2014, 53, 215–229. [Google Scholar] [CrossRef]
- Olate-Olave, V.R.; Verde, M.; Vallejos, L.; Perez Raymonda, L.; Cortese, M.C.; Doorn, M. Bee health and productivity in Apis mellifera, a consequence of multiple factors. Vet. Sci. 2021, 8, 76. [Google Scholar] [CrossRef]
- Tsvetkov, N.; Samson-Robert, O.; Sood, K.; Patel, H.S.; Malena, D.A.; Gajiwala, P.H.; Maciukiewicz, P.; Fournier, V.; Zayed, A. Chronic exposure to neonicotinoids reduces honey bee health near corn crops. Science 2017, 356, 1395–1397. [Google Scholar] [CrossRef] [PubMed]
- van der Zee, R.; Gray, A.; Pisa, L.; de Rijk, T. An observational study of honey bee colony winter losses and their association with Varroa destructor, neonicotinoids and other risk factors. PLoS ONE 2015, 10, e0131611. [Google Scholar] [CrossRef] [PubMed]
- Blanken, L.J.; van Langevelde, F.; van Dooremalen, C. Interaction between Varroa destructor and imidacloprid reduces flight capacity of honey bees. Proc. R. Soc. B 2015, 282, 20151738. [Google Scholar] [CrossRef] [PubMed]
- Mariani, F.; Maggi, M.D.; Porrini, M.P.; Fuselli, S.; Caraballo, G.; Brasesco, C.; Barrios, C.; Principal, J.; Eguaras, M. Parasitic interactions between Nosema spp. and Varroa destructor in Apis mellifera colonies. Zootec. Trop. 2012, 30, 81–90. [Google Scholar]
- Pettis, J.S.; vanEngelsdorp, D.; Johnson, J.; Dively, G. Pesticide exposure in honey bees results in increased levels of the gut pathogen Nosema. Naturwissenschaften 2012, 99, 153–158. [Google Scholar] [CrossRef]
- van Dooremalen, C.; Cornelissen, B.; Poleij-Hok-Ahin, C.; Blacquière, T. Single and interactive effects of Varroa destructor, Nosema spp., and imidacloprid on honey bee colonies (Apis mellifera). Ecosphere 2018, 9, e02378. [Google Scholar] [CrossRef]
- Evans, J.D.; Spivak, M. Socialized medicine: Individual and communal disease barriers in honey bees. J. Invertebr. Pathol. 2010, 103, S62–S72. [Google Scholar] [CrossRef]
- Seeley, T.D.; Visscher, P.K. Survival of honey bees in cold climates: The critical timing of colony growth and reproduction. Ecol. Entomol. 1985, 10, 81–88. [Google Scholar] [CrossRef]
- Meikle, W.G.; Holst, N. Application of continuous monitoring of honey bee colonies. Apidologie 2015, 46, 10–22. [Google Scholar] [CrossRef]
- Ulgezen, Z.N.; van Dooremalen, C.; van Langevelde, F. Understanding social resilience in honey bee colonies. Curr. Res. Insect Sci. 2021, 1, 100021. [Google Scholar] [CrossRef]
- Braga, A.R.; Gomes, D.G.; Freitas, B.M.; Cazier, J.A. A cluster-classification method for accurate mining of seasonal honey bee patterns. Ecol. Inform. 2020, 59, 101107. [Google Scholar] [CrossRef]
- Braga, A.R.; Gomes, D.G.; Rogers, R.; Hassler, E.E.; Freitas, B.M.; Cazier, J.A. A method for mining combined data from in-hive sensors, weather and apiary inspections to forecast the health status of honey bee colonies. Comput. Electron. Agric. 2020, 169, 105161. [Google Scholar] [CrossRef]
- Cecchi, S.; Spinsante, S.; Terenzi, A.; Orcioni, S. A smart sensor-based measurement system for advanced bee hive monitoring. Sensors 2020, 20, 2726. [Google Scholar] [CrossRef] [PubMed]
- Meikle, W.G.; Weiss, M.; Maes, P.W.; Fitz, W.; Snyder, L.A.; Sheehan, T.; Mott, B.M.; Anderson, K.E. Internal hive temperature as a means of monitoring honey bee colony health in a migratory beekeeping operation before and during winter. Apidologie 2017, 48, 666–680. [Google Scholar] [CrossRef]
- Metlek, S.; Kayaalp, K. Detection of bee diseases with a hybrid deep learning method. J. Fac. Eng. Archit. Gazi Univ. 2021, 36, 1715–1731. [Google Scholar]
- EFSA European Food Safety Authority. EFSA Panel on Animal Health and Welfare (AHAW) [Scientific opinion]. Assess. Health Status Manag. Honey Bee Colon. (HEALTHY-B): A Toolbox Facil. Harmon. Data Collect. EFSA J. 2016, 14, e04578. [Google Scholar] [CrossRef]
- Duan, X.; Wallis, D.; Hatjina, F.; Simon-Delso, N.; Bruun Jensen, A.; Topping, C.J. ApisRAM formal model description. EFSA Support. Publ. 2022, 19, 7184E. [Google Scholar] [CrossRef]
- de Graaf, D.C.; Bencsik, M.; De Smet, L.; Neumann, P.; Schoonman, M.; Sousa, J.P.; Topping, C.J.; Verbeke, W.; Williams, J.; van Dooremalen, C. B-GOOD: Giving Beekeeping Guidance by cOmputatiOnal-assisted Decision making. Res. Ideas Outcomes 2022, 8, e84129. [Google Scholar] [CrossRef]
- Claeys Boúúaert, D.; De Smet, L.; Brunain, M.; Dahle, B.; Blacquière, T.; Dalmon, A.; Dezmirean, D.; Elen, D.; Filipi, J.; Giurgiu, A.; et al. Dynamics in vertical transmission of viruses in naturally selected and traditionally managed honey bee colonies across Europe. bioRxiv 2010, 103, S62–S72. [Google Scholar] [CrossRef]
- Huber, M.; van Vliet, M.; Giezenberg, M.; Winkens, B.; Heerkens, Y.; Dagnelie, P.C.; Knottnerus, J.A. Towards A ‘Patient-Centered’ Oper. New Dyn. Concept Health: A Mix. Methods Study. BMJ Open 2016, 6, e010091. [Google Scholar] [CrossRef]
- Nowak, M.A.; Tarnita, C.E.; Wilson, E.O. The evolution of eusociality. Nature 2010, 466, 1057–1062. [Google Scholar] [CrossRef] [PubMed]
- Büchler, R.; Costa, C.; Hatjina, F.; Andonov, S.; Meixner, M.D.; Le Conte, Y.; Uzunov, A.; Berg, S.; Bienkowska, M.; Bouga, M. The influence of genetic origin and its interaction with environmental effects on the survival of Apis mellifera L. colonies in Europe. J. Apic. Res. 2014, 53, 205–214. [Google Scholar] [CrossRef]
- Hatjina, F.; Costa, C.; Büchler, R.; Uzunov, A.; Drazic, M.; Filipi, J.; Charistos, L.; Ruottinen, L.; Andonov, S.; Meixner, M.D. Population dynamics of European honey bee genotypes under different environmental conditions. J. Apic. Res. 2014, 53, 233–247. [Google Scholar] [CrossRef]
- Meixner, M.D.; Kryger, P.; Costa, C. Effects of genotype, environment, and their interactions on honey bee health in Europe. Curr. Opin. Insect Sci. 2015, 10, 177–184. [Google Scholar] [CrossRef] [PubMed]
- Reese, H.W. The learning-by-doing principle. Behav. Dev. Bull. 2011, 17, 1–19. [Google Scholar] [CrossRef]
- Delaplane, K.S.; van der Steen, J.; Guzman-Novoa, E. Standard methods for estimating strength parameters of Apis mellifera colonies. J. Apic. Res. 2013, 52, 1–12. [Google Scholar] [CrossRef]
- Human, H.; Brodschneider, R.; Dietemann, V.; Dively, G.; Ellis, J.D.; Forsgren, E.; Fries, I.; Hatjina, F.; Hu, F.-L.; Jaffé, R. Miscellaneous standard methods for Apis mellifera research. J. Apic. Res. 2013, 52, 1–53. [Google Scholar] [CrossRef]
- van Dooremalen, C.; Gerritsen, L.; Cornelissen, B.; van der Steen JJ, M.; van Langevelde, F.; Blacquière, T. Winter survival of individual honey bees and honey bee colonies depends on level of Varroa destructor infestation. PLoS ONE 2012, 7, e36285. [Google Scholar] [CrossRef]
- van Dooremalen, C.; van Langevelde, F. Can colony size of honey bees (Apis mellifera) be used as predictor for colony losses due to Varroa destructor during winter? Agriculture 2021, 11, 529. [Google Scholar] [CrossRef]
- Büchler, R.; Andonov, S.; Bienefeld, K.; Costa, C.; Hatjina, F.; Kezic, N. Standard methods for rearing and selection of Apis mellifera queens. J. Apic. Res. 2013, 52, 1–30. [Google Scholar] [CrossRef]
- Dietemann, V.; Nazzi, F.; Martin, S.J.; Anderson, D.L.; Locke, B.; Delaplane, K.S.; Wauquiez, Q.; Tannahill, C.; Frey, E.; Ziegelmann, B.; et al. Standard methods for varroa research. J. Apic. Res. 2013, 52, 1–54. [Google Scholar] [CrossRef]
- OIE World Organization for Animal Health. Varroosis of honey bees. Terr. Anim. Health Code 2021, 1, 1–13. [Google Scholar]
- Matthijs, S.; De Waele, V.; Vandenberge, V.; Verhoeven, B.; Evers, J.; Brunain, M.; Saegerman, C.; De Winter PJ, J.; Roles, S.; de Graaf, D.C.; et al. Nationwide screening for bee viruses and parasites in Belgian honey bees. Viruses 2020, 12, 890. [Google Scholar] [CrossRef]
- Schäfer, M.O.; Horenk, J.; Wylezich, C. Molecular Detection of Malpighamoeba mellificae in Honey Bees. Vet. Sci. 2022, 9, 148. [Google Scholar] [CrossRef] [PubMed]
- Blanchard, P.; Ribière, M.; Celle, O.; Lallemand, P.; Schurr, F.; Olivier, V.; Iscache, A.L.; Faucon, J.P. Evaluation of a real-time two-step RT-PCR assay for quantitation of Chronic bee paralysis virus (CBPV) genome in experimentally-infected bee tissues and in life stages of a symptomatic colony. J. Virol. Methods 2007, 141, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Budge, G.E.; Barrett, B.; Jones, B.; Pietravalle, S.; Marris, G.; Chantawannakul, P.; Thwaites, R.; Hall, J.; Cuthbertson AG, S.; Brown, M.A. The occurrence of Melissococcus plutonius in healthy colonies of Apis mellifera and the efficacy of European foulbrood control measures. J. Invertebr. Pathol. 2010, 105, 164–170. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Evans, J.D.; Zhou, L.; Boncristiani, H.; Kimura, K.; Xiao, T.; Litkowski, A.M.; Pettis, J.S. Asymmetrical coexistence of Nosema ceranae and Nosema apis in honey bees. J. Invertebr. Pathol. 2009, 101, 204–209. [Google Scholar] [CrossRef] [PubMed]
- Ciglenečki, U.J.; Toplak, I. Development of a real-time RT-PCR assay with TaqMan probe for specific detection of acute bee paralysis virus. J. Virol. Methods 2012, 184, 63–68. [Google Scholar] [CrossRef]
- Dainat, B.; Grossar, D.; Ecoffey, B.; Haldemann, C. Triplex real-time PCR method for the qualitative detection of European and American foulbrood in honeybee. J. Microbiol. Methods 2018, 146, 61–63. [Google Scholar] [CrossRef]
- Schurr, F.; Tison, A.; Militano, L.; Cheviron, N.; Sircoulomb, F.; Rivière, M.-P.; Ribière-Chabert, M.; Thiéry, R.; Dubois, E. Validation of quantitative real-time RT-PCR assays for the detection of six honey bee viruses. J. Virol. Methods 2019, 270, 70–78. [Google Scholar] [CrossRef]
- Chantawannakul, P.; Ward, L.; Boonham, N.; Brown, M. A scientific note on the detection of honey bee viruses using real-time PCR (TaqMan) in Varroa mites collected from a Thai honey bee (Apis mellifera) apiary. J. Invertebr. Pathol. 2006, 91, 69–73. [Google Scholar] [CrossRef] [PubMed]
- Blanchard, P.; Guillot, S.; Antùnez, K.; Köglberger, H.; Kryger, P.; de Miranda, J.R.; Franco, S.; Chauzat, M.-P.; Thiéry, R.; Ribière, M. Development and validation of a real-time two-step RT-qPCR TaqMan® assay for quantitation of Sacbrood virus (SBV) and its application to a field survey of symptomatic honey bee colonies. J. Virol. Methods 2014, 197, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Ritter, W.; Akratanakul, P. Honey Bee Diseases and Pests: A Practical Guide. Food and Agriculture Organization of the United Nations. Available online: https://iucat.iu.edu/iue/7517626 (accessed on 1 June 2021).


| Variable | Category | Data/Units | Tier 1 | Tier 2 | Tier 3 |
|---|---|---|---|---|---|
| Weight | Automated data | kg | 15 min | 15 min | 15 min |
| Ambient temperature | Automated data | °C (Celsius degrees) | 15 min | 15 min | 15 min |
| In-hive temperature | Automated data | °C (Celsius degrees) | 15 min | 15 min | 15 min |
| Sound | Automated data | Frequency count (122–583 Hz) | 15 min | 15 min | 15 min |
| Battery | Automated data | Volt | 15 min | 15 min | 15 min |
| Signal strength (data transmission) | Automated data | dBm | 15 min | 15 min | 15 min |
| Signal noise (data transmission) | Automated data | dB | 15 min | 15 min | 15 min |
| Sufficient adult bees | Data annotation | Yes/no | 7–30 days | 7–30 days | 7–30 days |
| Brood in all stages | Data annotation | Yes/no | 7–30 days | 7–30 days | 7–30 days |
| Presence of queen | Data annotation | Yes/no | 7–30 days | 7–30 days | 7–30 days |
| Suitable space | Data annotation | Yes/no | 7–30 days | 7–30 days | 7–30 days |
| Absence of stressors | Data annotation | Yes/no | 7–30 days | 7–30 days | 7–30 days |
| Sufficient nutrition | Data annotation | Yes/no | 7–30 days | 7–30 days | 7–30 days |
| General impression | Experimental observation | Good, average, bad (smileys) | 7–30 days | 7–30 days | 7–30 days |
| Eggs | Experimental observation | Estimated number of cells | Every 21 days (beekeeping season) | NA | NA |
| Larvae | Experimental observation | Estimated number of cells | Every 21 days (beekeeping season) | NA | NA |
| Bees | Experimental observation | Estimated number of cells | Every 21 days (beekeeping season) | NA | NA |
| Pollen | Experimental observation | Estimated number of cells | Every 21 days (beekeeping season) | NA | NA |
| Sealed honey | Experimental observation | Estimated number of cells | Every 21 days (beekeeping season) | NA | NA |
| Pupae (capped brood) | Experimental observation | Estimated number of cells | Every 21 days (beekeeping season) | NA | NA |
| Drone brood | Experimental observation | Estimated number of cells | Every 21 days (beekeeping season) | NA | NA |
| Atypical behavior | Experimental observation | Yes/no | Every 21 days (beekeeping season) | NA | NA |
| Colony loss | Experimental observation | Yes/no | When necessary | When necessary | When necessary |
| Dead bees | Experimental observation | Yes/no | Every 21 days (beekeeping season) | Every 30 days (beekeeping season) | NA |
| Varroa natural fall | Experimental observation | mites/day | Once a week | optional | NA |
| Clinical signs of disease | Experimental observation | Categorized by type | Every 21 days (beekeeping season) | Every 30 days (beekeeping season) | NA |
| Presence of eggs | Experimental observation | Yes/no | Every 21 days (beekeeping season) | Every 30 days (beekeeping season) | Every 30 days (beekeeping season) |
| Presence of larvae | Experimental observation | Yes/no | Every 21 days (beekeeping season) | Every 30 days (beekeeping season) | Every 30 days (beekeeping season) |
| Presence of pupae | Experimental observation | Yes/no | Every 21 days (beekeeping season) | Every 30 days (beekeeping season) | Every 30 days (beekeeping season) |
| Queen presence | Experimental observation | Yes/no | Every 21 days (beekeeping season) | Every 30 days (beekeeping season) | Every 30 days (beekeeping season) |
| Top photo analysis | Experimental observation | Estimated number of bees | Every 30 days during winter, and every 21 days during beekeeping season | Every 30 days | NA |
| Queen cell presence | Experimental observation | Yes/no | Every 21 days (beekeeping season) * | Every 30 days (beekeeping season) ** | NA |
| Queen cell type | Experimental observation | Supersedure, emergency, cup, swarm | Every 21 days (beekeeping season) * | Every 30 days (beekeeping season) ** | NA |
| Brood pattern | Experimental observation | Brood spottiness rating. Scale 1–5 | Every 21 days (beekeeping season) * | NA | NA |
| Drone presence | Experimental observation | Yes/no | NA | Every 30 days (swarming season) ** | NA |
| Suppressed in ovo virus infection | Lab analyses | PCR data | Once every queen * | NA | NA |
| Viral diversity Deformed wing virus | Lab analyses | Sequencing data | Select number of samples within each country ** | NA | NA |
| Varroa destructor | Lab analyses | Mites/100 bees | 3 times a year (spring, summer, fall) | 3 times a year (spring, summer, fall) | 3 times a year (spring, summer, fall) |
| Deformed wing virus A | Lab analyses | PCR data | 3 times a year (spring, summer, fall) | 3 times a year (spring, summer, fall) | 3 times a year (spring, summer, fall) |
| Deformed wing virus B | Lab analyses | PCR data | 3 times a year (spring, summer, fall) | 3 times a year (spring, summer, fall) | 3 times a year (spring, summer, fall) |
| Acute bee paralysis virus | Lab analyses | PCR data | 2 times a year (spring, fall) | 2 times a year (spring, fall) | 2 times a year (spring, fall) |
| Chronic bee paralysis virus | Lab analyses | PCR data | 2 times a year (spring, fall) | 2 times a year (spring, fall) | 2 times a year (spring, fall) |
| American Foulbrood | Lab analyses | PCR data | Once a year (fall) | Once a year (fall) | Once a year (fall) |
| European Foulbrood | Lab analyses | PCR data | Once a year (fall) | Once a year (fall) | Once a year (fall) |
| Nosema ceranae | Lab analyses | PCR data | 2 times a year (spring, summer) | 2 times a year (spring, summer) | 2 times a year (spring, summer) |
| Nosema apis | Lab analyses | PCR data | 2 times a year (spring, summer) | 2 times a year (spring, summer) | 2 times a year (spring, summer) |
| Sacbrood virus | Lab analyses | PCR data | 3 times a year (spring, summer, fall) | 3 times a year (spring, summer, fall) | 3 times a year (spring, summer, fall) |
| Black queen cell virus | Lab analyses | PCR data | 3 times a year (spring, summer, fall) | 3 times a year (spring, summer, fall) | 3 times a year (spring, summer, fall) |
| Malpighamoeba mellificae | Lab analyses | PCR data | 3 times a year (spring, summer, fall) | 3 times a year (spring, summer, fall) | 3 times a year (spring, summer, fall) |
| Foundationless frame | Management actions | Yes/no | When necessary | When necessary | When necessary |
| Drone brood removal | Management actions | Yes/no | When necessary | When necessary | When necessary |
| Brood layers | Management actions | Number | When necessary | When necessary | When necessary |
| Frames per layer | Management actions | Number | When necessary | When necessary | When necessary |
| Honey super | Management actions | Number | When necessary | When necessary | When necessary |
| Comb replaced | Management actions | Number | When necessary | When necessary | When necessary |
| Nutrition/sugar feeding | Management actions | Weight/volume | When necessary | When necessary | When necessary |
| Swarming prevention | Management actions | Method | When necessary | When necessary | When necessary |
| Queen introduction | Management actions | Reason and method | When necessary | When necessary | When necessary |
| Queen marking | Management actions | Colour | When necessary | When necessary | When necessary |
| Queen cell removal | Management actions | Number | When necessary | When necessary | When necessary |
| Colony split | Management actions | Yes/no | When necessary | When necessary | When necessary |
| Colony united | Management actions | Yes/no | When necessary | When necessary | When necessary |
| Colony feeding | Management actions | Volume/weight | When necessary | When necessary | When necessary |
| Honey harvest | Management actions | Weight/volume | When necessary | When necessary | When necessary |
| Varroa treatment | Management actions | Method | When necessary | When necessary | When necessary |
| Temperature | Weather (from weather service) | °C (Celsius degrees) | 15 min | NA | NA |
| Wind speed | Weather (from weather service) | m/s | 15 min | NA | NA |
| Humidity | Weather (from weather service) | % RH | 15 min | NA | NA |
| Rainfall | Weather (from weather service) | mm/h | 15 min | NA | NA |
| Label | Protocol | Description | Tier 1/ Researchers | Tier 2/ Beekeepers | Tier 3 ****/ Beekeepers |
|---|---|---|---|---|---|
| P1 | Queen & BIAS | Finding the Queen and checking Brood In All Stages | 20210129 | 20220513 | 20220225 |
| P2 * | Liebefeld | How to apply the Liebefeld method for counting bees, amount of brood and food resources | 20220513 | ||
| P3 | Top Photo Analysis | Analyses of colony sizes by taking pictures of brood from the top | 20220513 | 20220513 | |
| P4 | Varroa | Counting natural Varroa mitefall to measure mite infestation level | 20220513 | ||
| P5 | Lab analyses | How to sample bees sent for Lab analysis for diagnostic purposes | 20220513 | 20220513 ** | 20220225 |
| P6 | Atypical behaviour | Visually assess colony behaviour | 20220204 | ||
| P7 | Clinical signs | How to visually check colonies for clinical signs of disease | 20220204 | 20220202 | |
| P8 * | EFSA protocol | Performing the EFSA to estimate colony size, amount of brood and food resources | 20220513 | ||
| P9 | Drone eggs | How to collect drone eggs | 20220513 | ||
| P10 | Queen cell presence | Checking for colony cells and explaining the four different queen cell types | 20220204 | 20220224 | |
| P11 | Brood pattern | How to measure brood pattern consistency | 20220204 | ||
| P12 | Data quality | Checking and cleaning up data on the BEEP app | 20220204 | ||
| P13 | Drone Presence | Checking colonies for presence of drone brood | 20220513 *** |
| Target | Primers (5′-3′) | Probe (5′-3′) | Reference |
|---|---|---|---|
| DWV A | Fwd: GCGGCTAAGATTGTAAATTG Rev: GTGACTAGCATAACCATGATTA | CCTTGACCAGTAGACACAGCATC | [50] |
| DWV B | Fwd: GGTCTGAAGCGAAAATAG Rev: CTAGCATATCCATGATTATAAAC | CCTTGTCCAGTAGATACAGCATCACA | [50] |
| ABPV | Fwd: CATATTGGCGAGCCACTATG Rev: CTACCAGGTTCAAAGAAAATTTC | ATAGTTAAAACAGCTTTTCACACTGG | [48] |
| CBPV | Fwd: CGCAAGTACGCCTTGATAAAGAAC Rev: ACTACTAGAAACTCGTCGCTTCG | TCAAGAACGAGACCACCGCCAAGTTC | [45] |
| Nosema apis | Fwd: CCATTGCCGGATAAGAGAGT Rev: CCACCAAAAACTCCCAAGAG | ATAGTGAGGCTCTATCACTCCGCTG | [47] |
| Nosema ceranae | Fwd: CGGATAAAAGAGTCCGTTACC Rev: TGAGCAGGGTTCTAGGGAT | CGTTACCCTTCGGGGAATCTTC | [47] |
| Melissococcus plutonius (EFB) | Fwd: TGTTGTTAGAGAAGAATAGGGGAA Rev: CGTGGCTTTCTGGTTAGA | AGAGTAACTGTTTTCCTCGTGACGGT | [46] |
| Paenibacillus larvae (AFB) | Fwd: TACGCTTTTCGATTCTCTG Rev: GTCTGTACTGAACCAAGTC | ATCTGCTTCCACTTGTTCACTCACCA | [49] |
| BQCV | Fwd: GGTGCGGGAGATGATATGGA Rev: GCCGTCTGAGATGCATGAATAC | TTTCCATCTTTATCGGTACGCCGCC | [51] |
| SBV | Fwd: AACGTCCACTACACCGAAATGTC Rev: ACACTGCGCGTCTAACATTCC | TGATGAGAGTGGACGAAGA | [52] |
| Malpighamoeba mellificae | Fwd: TATACAGATTGTGTAAAAGCG Rev: TTAGCCTCTATCTAACCTACC | TACAAGAGGATCTGCCCTATCAACTAT | [44] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
van Dooremalen, C.; Ulgezen, Z.N.; Dall’Olio, R.; Godeau, U.; Duan, X.; Sousa, J.P.; Schäfer, M.O.; Beaurepaire, A.; van Gennip, P.; Schoonman, M.; et al. Bridging the Gap between Field Experiments and Machine Learning: The EC H2020 B-GOOD Project as a Case Study towards Automated Predictive Health Monitoring of Honey Bee Colonies. Insects 2024, 15, 76. https://doi.org/10.3390/insects15010076
van Dooremalen C, Ulgezen ZN, Dall’Olio R, Godeau U, Duan X, Sousa JP, Schäfer MO, Beaurepaire A, van Gennip P, Schoonman M, et al. Bridging the Gap between Field Experiments and Machine Learning: The EC H2020 B-GOOD Project as a Case Study towards Automated Predictive Health Monitoring of Honey Bee Colonies. Insects. 2024; 15(1):76. https://doi.org/10.3390/insects15010076
Chicago/Turabian Stylevan Dooremalen, Coby, Zeynep N. Ulgezen, Raffaele Dall’Olio, Ugoline Godeau, Xiaodong Duan, José Paulo Sousa, Marc O. Schäfer, Alexis Beaurepaire, Pim van Gennip, Marten Schoonman, and et al. 2024. "Bridging the Gap between Field Experiments and Machine Learning: The EC H2020 B-GOOD Project as a Case Study towards Automated Predictive Health Monitoring of Honey Bee Colonies" Insects 15, no. 1: 76. https://doi.org/10.3390/insects15010076
APA Stylevan Dooremalen, C., Ulgezen, Z. N., Dall’Olio, R., Godeau, U., Duan, X., Sousa, J. P., Schäfer, M. O., Beaurepaire, A., van Gennip, P., Schoonman, M., Flener, C., Matthijs, S., Claeys Boúúaert, D., Verbeke, W., Freshley, D., Valkenburg, D.-J., van den Bosch, T., Schaafsma, F., Peters, J., ... de Graaf, D. C. (2024). Bridging the Gap between Field Experiments and Machine Learning: The EC H2020 B-GOOD Project as a Case Study towards Automated Predictive Health Monitoring of Honey Bee Colonies. Insects, 15(1), 76. https://doi.org/10.3390/insects15010076











