Molecular Markers for Analyses of Genetic Diversity within the Anastrepha fraterculus Complex with Emphasis on Argentine Populations
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples and DNA Extraction
2.2. ITS2 Marker Characterisation
2.3. Data Analyses: Variability and Differentiation
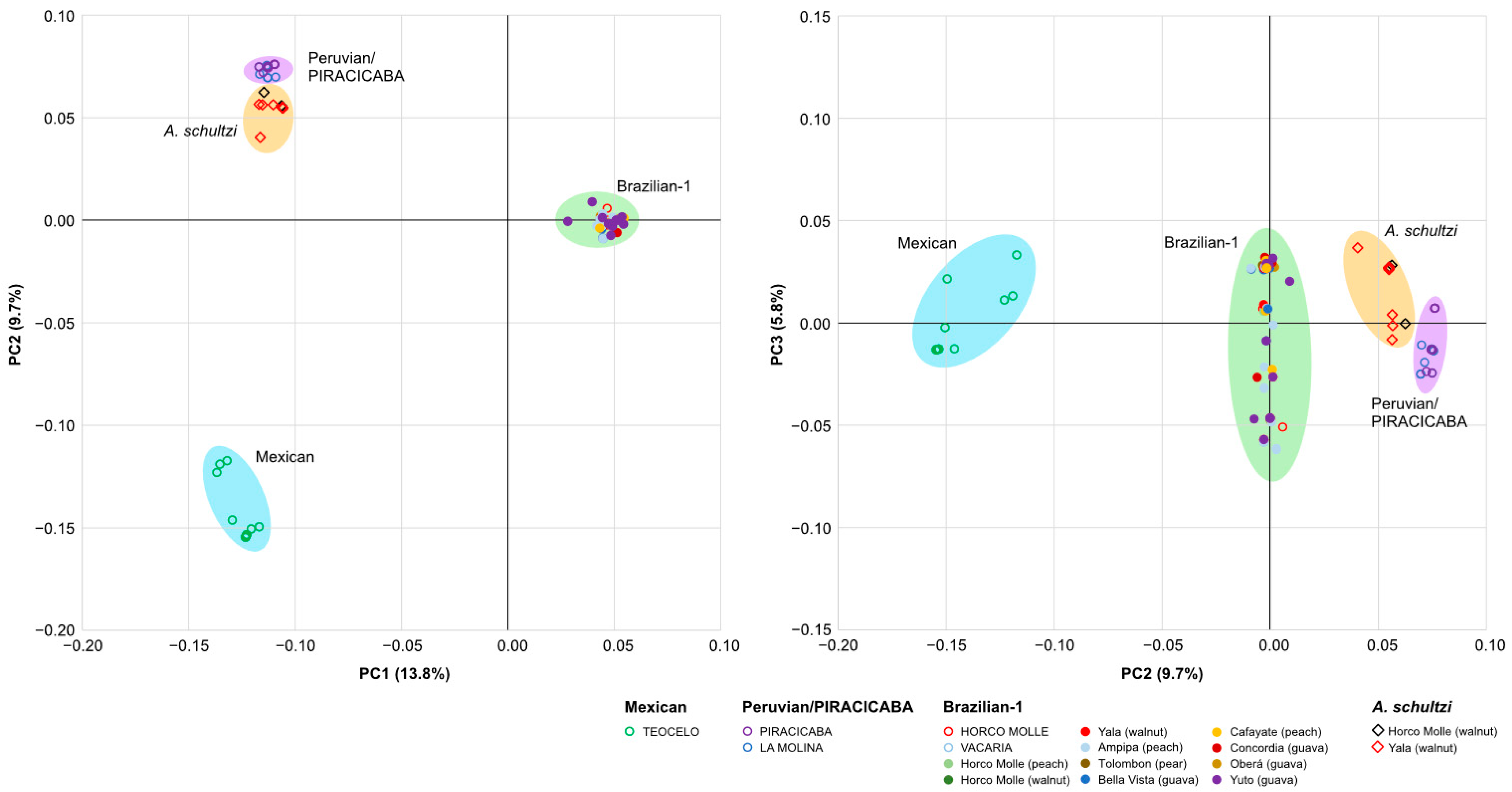
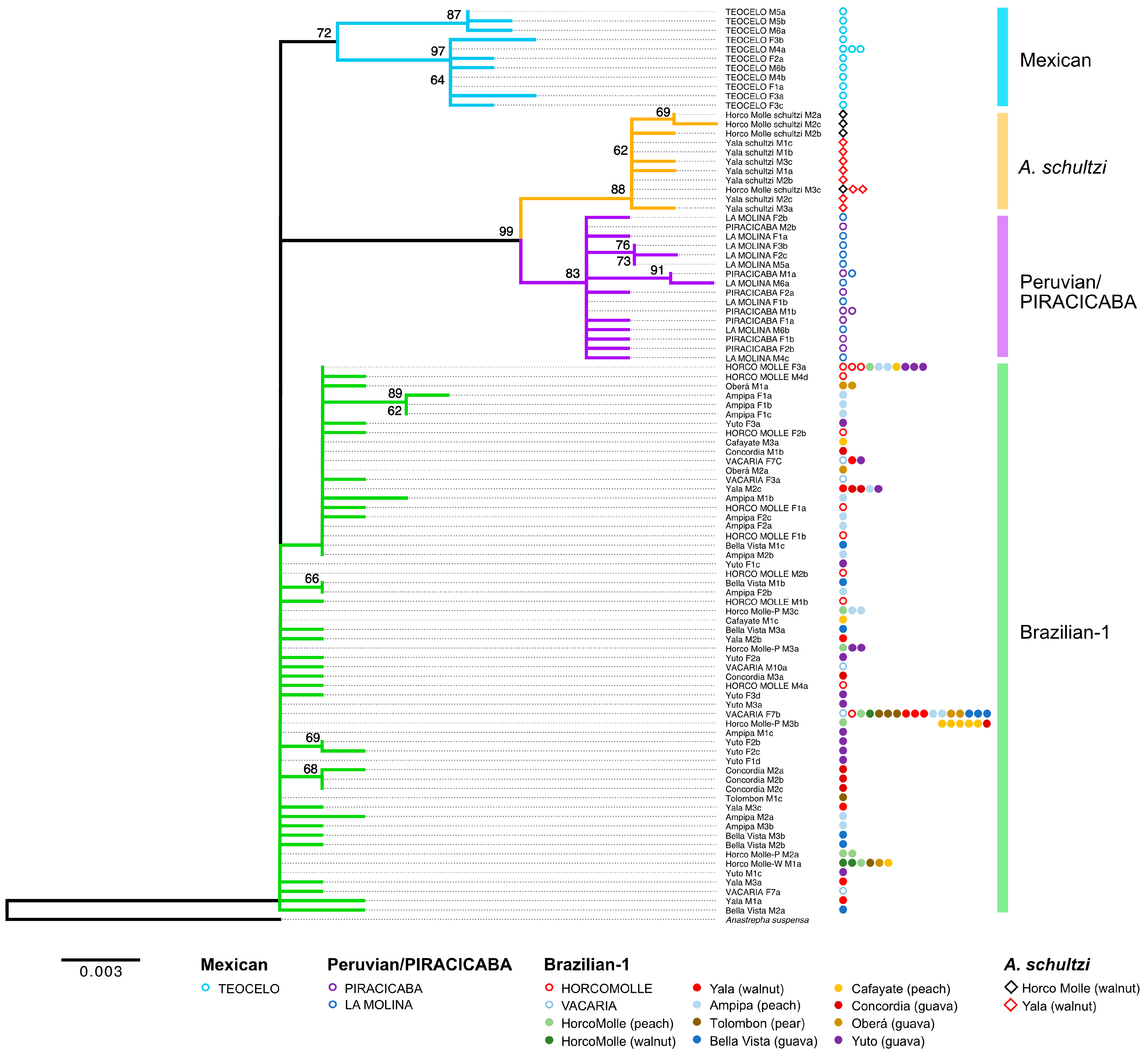
3. Results
3.1. Characterisation of the ITS2 Locus
3.2. ITS2 Sequence Variability
3.3. Sample Differentiation
4. Discussion
4.1. Much Hidden Variation Is Present among and within the Morphotypes
4.2. Genetic Differentiation and Morphotype Classification
4.3. Implications for Pest Control
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Norrbom, A.L.; Zucchi, R.A.; Hernández-Ortiz, V. Phylogeny of the genera Anastrepha and Toxotrypana (Trypetinae: Toxotrypanini) based on morphology. In Fruit Flies (Tephritidae): Phylogeny and Evolution of Behavior; Aluja, M., Norrbom, A., Eds.; CRC Press: Boca Raton, FL, USA, 1999; pp. 299–342. [Google Scholar]
- Norrbom, A.L.; Muller, A.; Gangadin, A.; Sutton, B.D.; Rodriguez, E.J.; Savaris, M.; Lampert, S.; Clavijo, P.A.R.; Steck, G.J.; Moore, M.R.; et al. New species and host plants of Anastrepha (Diptera: Tephritidae) primarily from Suriname and Par, Brazil. Zootaxa 2021, 5044, 1–74. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Ortiz, V.; Bartolucci, A.F.; Morales-Valles, P.; Frías, D.; Selivon, D. Cryptic species of the Anastrepha fraterculus complex (Diptera: Tephritidae): A multivariate approach for the recognition of South American morphotypes. Ann. Entomol. Soc. Am. 2012, 105, 305–318. [Google Scholar] [CrossRef]
- Steck, G. Biochemical systematics and population genetic-structure of Anastrepha fraterculus and related species (Diptera, Tephritidae). Ann. Entomol. Soc. Am. 1991, 84, 10–28. [Google Scholar] [CrossRef]
- Steck, G.J. Taxonomic status of Anastrepha fraterculus In The South American Fruit Fly, Anastrepha fraterculus (Wied.): Advances in Artificial Rearing, Taxonomic Status and Biological Studies; IAEA Tech-Doc 1064; International Atomic Energy Agency: Vienna, Austria, 1999; pp. 13–20. [Google Scholar]
- Stone, A. The fruit flies of the genus Anastrepha. U.S. Dep. Agric. Misc. Publ. 1942, 439, 1–112. [Google Scholar]
- Norrbom, A.L. Host plant database for Anastrepha and Toxotrypana (Diptera: Tephritidae: Toxotrypanini). In Diptera Data Dissemination Disk (CD-ROM) 2; North American Dipterist’s Society: Washington, DC, USA, 2004. [Google Scholar]
- Prezotto, L.F.; Perondini, A.L.P.; Hernández-Ortiz, V.; Frías, D.; Selivon, D. What can integrated analysis of morphological and genetic data still reveal about the Anastrepha fraterculus (Diptera: Tephritidae) cryptic species complex? Insects 2019, 10, 408. [Google Scholar] [CrossRef]
- Selivon, D.; Perondini, A.L.P. Eggshell morphology in two cryptic species of the Anastrepha fraterculus complex (Diptera: Tephritidae). Ann. Entomol. Soc. Am. 1998, 91, 473–478. [Google Scholar] [CrossRef]
- Selivon, D.; Vretos, C.; Fontes, L.; Perondini, A.L.P. New variant forms in the Anastrepha fraterculus complex. In Proceedings of the 6th International Symposium on Fruit Flies of Economic Importance, Stellenbosh, South Africa, 6–10 May 2002; Barnes, B.N., Ed.; Isteg Scientific Publications: Irene, South Africa, 2004; pp. 253–258. [Google Scholar]
- Selivon, D.; Perondini, A.L.P.; Morgante, J.S. A genetic-morphological characterization of two cryptic species of the Anastrepha fraterculus complex (Diptera: Tephritidae). Ann. Entomol. Soc. Am. 2005, 98, 367–381. [Google Scholar] [CrossRef]
- Selivon, D.; Perondini, A.L.P.; Hernandez-Ortiz, V.; doVal, F.C.; Camacho, A.; Gomes, F.R.; Prezotto, L.F. Genetical, morphological, behavioral, and ecological traits support the existence of three Brazilian species of the Anastrepha fraterculus complex of cryptic species. Front. Ecol. Evol. 2022, 10, 836608. [Google Scholar] [CrossRef]
- Rull, J.; Abraham, S.; Kovaleski, A.; Segura, D.F.; Mendoza, M.; Liendo, M.C.; Vera, M.T. Evolution of pre-zygotic and post-zygotic barriers to gene flow among three cryptic species within the Anastrepha fraterculus complex. Entomol. Exp. Et Appl. 2013, 148, 213–222. [Google Scholar] [CrossRef]
- Hendrichs, J.; Vera, M.T.; De Meyer, M.; Clarke, A.R. Resolving cryptic species complexes of major tephritid pests. Zookeys 2015, 540, 5–39. [Google Scholar] [CrossRef]
- Dias, V.S.; Silva, J.G.; Lima, K.M.; Petitinga, C.S.C.D.; Hernández-Ortiz, V.; Laumann, R.A.; Paranhos, B.J.; Uramoto, K.; Zucchi, R.A.; Joachim-Bravo, I.S. An integrative multidisciplinary approach to understanding cryptic divergence in Brazilian species of the Anastrepha fraterculus complex (Diptera: Tephritidae). Biol. J. Linn. Soc. 2016, 117, 725–746. [Google Scholar] [CrossRef]
- Vera, M.T.; Cáceres, C.; Wornoayporn, V.; Islam, A.; Robinson, A.S.; De La Vega, M.H.; Hendrichs, J.; Cayol, J.P. Mating incompatibility among populations of the South American fruit fly Anastrepha fraterculus (Diptera: Tephritidae). Ann. Entomol. Soc. Am. 2006, 99, 387–397. [Google Scholar] [CrossRef]
- Devescovi, F.; Abraham, S.; Roriz, A.K.P.; Nolazco, N.; Castañeda, R.; Tadeo, E.; Cáceres, C.; Segura, D.F.; Vera, M.T.; Joachim-Bravo, I.; et al. Ongoing speciation within the Anastrepha fraterculus cryptic species complex: The case of the Andean morphotype. Entomol. Exp. Appl. 2014, 152, 238–247. [Google Scholar] [CrossRef]
- Sutton, B.D.; Steck, G.J.; Norrbom, A.L.; Rodriguez, E.J.; Srivastava, P.; Alvarado, N.N.; Colque, F.; Landa, E.Y.; Sánchez, J.J.; Quisberth, E.; et al. Nuclear ribosomal internal transcribed spacer 1 (ITS1) variation in the Anastrepha fraterculus cryptic species complex (Diptera, Tephritidae) of the Andean region. Zookeys 2015, 540, 175–191. [Google Scholar] [CrossRef]
- Hebert, P.D.; Cywinska, A.; Ball, S.L.; deWaard, J.R. Biological identifications through DNA barcodes. Proc. Biol. Sci. 2003, 270, 313–321. [Google Scholar] [CrossRef]
- Barr, N.B.; Ruiz-Arce, R.; Farris, R.E.; Silva, J.G.; Lima, K.M.; Dutra, V.S.; Ronchi-Teles, B.; Kerr, P.H.; Norrbom, A.L.; Nolazco, N.; et al. Identifying Anastrepha (Diptera; Tephritidae) species using DNA barcodes. J. Econ. Entomol. 2018, 111, 405–421. [Google Scholar] [CrossRef]
- Bartolini, I.; Rivera, J.; Nolazco, N.; Olórtegui, A. Towards the implementation of a DNA barcode library for the identification of Peruvian species of Anastrepha (Diptera: Tephritidae). PLoS ONE 2020, 15, e0228136. [Google Scholar] [CrossRef]
- Scally, M.; Into, F.; Thomas, D.B.; Ruiz-Arce, R.; Barr, N.B.; Schuenzel, E.L. Resolution of inter and intra-species relationships of the West Indian fruit fly Anastrepha obliqua. Mol. Phylogenet. Evol. 2016, 101, 286–293. [Google Scholar] [CrossRef]
- Barr, N.; Ruiz-Arce, R.; Obregón, O.; Shatters, R.; Norrbom, A.L.; Nolazco, N.; Thomas, D. Diagnostic characters within ITS2 DNA support molecular identification of Anastrepha suspensa (Diptera: Tephritidae). Fla. Entomol. 2017, 100, 182–185. [Google Scholar] [CrossRef]
- Freilij, D.; Vilardi, J.C.; Gómez-Cendra, P. Rapid cryptic divergence of the Anastrepha fraterculus complex in the Late Pleistocene: A phylogeographical-ecological approach. Biol. J. Linn. Soc. 2024, 142, 21–37. [Google Scholar] [CrossRef]
- Blanchard, E.E. Especies Argentinas del género Anastrepha Schiner (sens. lat.) (Diptera: Tephritidae). Rev. Invest. Agric. Buenos Aires 1961, 15, 281–342. [Google Scholar]
- Baruffi, L.; Damiani, G.; Guglielmino, C.R.; Bandi, C.; Malacrida, A.R.; Gasperi, G. Polymorphism within and between populations of Ceratitis capitata: Comparison between RAPD and multilocus enzyme electrophoresis data. Heredity 1995, 74 Pt 4, 425–437. [Google Scholar] [CrossRef] [PubMed]
- Fritz, A.H. Sequence analysis of nuclear rDNA of Anastrepha suspensa. Ann. Entomol. Soc. Am. 2006, 99, 369–373. [Google Scholar] [CrossRef]
- Douglas, L.J.; Haymer, D.S. Ribosomal ITS1 polymorphisms in Ceratitis capitata and Ceratitis rosa (Diptera: Tephritidae). Ann. Entomol. Soc. Am. 2001, 94, 726–731. [Google Scholar] [CrossRef]
- Wiegmann, B.M.; Trautwein, M.D.; Winkler, I.S.; Barr, N.B.; Kim, J.W.; Lambkin, C.; Bertone, M.A.; Cassel, B.K.; Bayless, K.M.; Heimberg, A.M.; et al. Episodic radiations in the fly tree of life. Proc. Natl. Acad. Sci. USA 2011, 108, 5690–5695. [Google Scholar] [CrossRef]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—New capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef]
- Nei, M. Molecular Evolutionary Genetics; Columbia University Press: New York, NY, USA, 1987. [Google Scholar]
- Excoffier, L.; Lischer, H.E. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Konishi, T.; Matsukuma, S.; Fuji, H.; Nakamura, D.; Satou, N.; Okano, K. Principal Component Analysis applied directly to Sequence Matrix. Sci. Rep. 2019, 9, 19297. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Darriba, D.; Posada, D.; Kozlov, A.M.; Stamatakis, A.; Morel, B.; Flouri, T. ModelTest-NG: A new and scalable tool for the selection of DNA and protein evolutionary models. Mol. Biol. Evol. 2020, 37, 291–294. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, M.; Kishino, H.; Yano, T. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 1985, 22, 160–174. [Google Scholar] [CrossRef]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef]
- Huelsenbeck, J.P.; Larget, B.; Alfaro, M.E. Bayesian phylogenetic model selection using reversible jump Markov chain Monte Carlo. Mol. Biol. Evol. 2004, 21, 1123–1133. [Google Scholar] [CrossRef]
- Manni, M.; Lima, K.M.; Guglielmino, C.R.; Lanzavecchia, S.B.; Juri, M.; Vera, T.; Cladera, J.; Scolari, F.; Gomulski, L.; Bonizzoni, M.; et al. Relevant genetic differentiation among Brazilian populations of Anastrepha fraterculus (Diptera, Tephritidae). Zookeys 2015, 540, 157–173. [Google Scholar] [CrossRef]
- Schliserman, P.; Ovruski, S.; Colin, C.; Norrbom, A.L.; Aluja, M. First report of Juglans australis (Juglandaceae) as a natural host plant for Anastrepha schultzi (Diptera: Tephritidae) with notes on probable parasitism by Doryctobracon areolatus, D-Brasiliensis, Opius bellus (Braconidae) and Aganaspis pelleranoi (Figitidae). Fla. Entomol. 2004, 87, 597–599. [Google Scholar] [CrossRef]
- Rosetti, N.; Remis, M.I. Spatial genetic structure and mitochondrial DNA phylogeography of Argentinean populations of the grasshopper Dichroplus elongatus. PLoS ONE 2012, 7, e40807. [Google Scholar] [CrossRef]
- Roriz, A.K.P.; Japyassú, H.F.; Cáceres, C.; Vera, M.T.; Joachim-Bravo, I.S. Pheromone emission patterns and courtship sequences across distinct populations within Anastrepha fraterculus (Diptera-Tephritidae) cryptic species complex. Bull. Entomol. Res. 2019, 109, 408–417. [Google Scholar] [CrossRef]
- Congrains, C.; Dupuis, J.R.; Rodriguez, E.J.; Norrbom, A.L.; Steck, G.; Sutton, B.; Nolazco, N.; de Brito, R.A.; Geib, S.M. Phylogenomic analysis provides diagnostic tools for the identification of Anastrepha fraterculus (Diptera: Tephritidae) species complex. Evol. Appl. 2023, 16, 1598–1618. [Google Scholar] [CrossRef] [PubMed]
- Diaz, F.; Luís, A.; Lima, A.; Nakamura, A.M.; Fernandes, F.; Sobrinho, J.I.; De Brito, R.A. Evidence for introgression among three species of the Anastrepha fraterculus group, a radiating species complex of fruit flies. Front. Genet. 2018, 10, 359. [Google Scholar] [CrossRef] [PubMed]
- Paredes-Esquivel, C.C.; Townson, H. Functional constraints and evolutionary dynamics of the repeats in the rDNA internal transcribed spacer 2 of members of the Anopheles barbirostris group. Parasit Vectors 2014, 7, 106. [Google Scholar] [CrossRef] [PubMed]
- Wilkerson, R.C.; Reinert, J.F.; Li, C. Ribosomal DNA ITS2 sequences differentiate six species in the Anopheles crucians complex (Diptera: Culicidae). J. Med. Entomol. 2004, 41, 392–401. [Google Scholar] [CrossRef]
- Gomulski, L.; Meiswinkel, R.; Delécolle, J.; Goffredo, M.; Gasperi, G. Phylogenetic relationships of the subgenus Avaritia Fox, 1955 including Culicoides obsoletus (Diptera, Ceratopogonidae) in Italy based on internal transcribed spacer 2 ribosomal DNA sequences. Syst. Entomol. 2005, 30, 619–631. [Google Scholar] [CrossRef]
- Boykin, L.; Schutze, M.; Krosch, M.; Chomic, A.; Chapman, T.; Englezou, A.; Armstrong, K.; Clarke, A.; Hailstones, D.; Cameron, S. Multi-gene phylogenetic analysis of south-east Asian pest members of the Bactrocera dorsalis species complex (Diptera: Tephritidae) does not support current taxonomy. J. Appl. Entomol. 2014, 138, 235–253. [Google Scholar] [CrossRef]



| Sample Name | Locality | Province | Coordinates | Host Fruit | Collection Year | Morphotype | |
|---|---|---|---|---|---|---|---|
| A. fraterculus | Horco Molle-W | Horco Molle | Tucumán | 26°48′10″ S 65°19′50″ W | Walnut | 2019 | Brazilian 1 |
| Horco Molle-P | Horco Molle | Tucumán | 26°48′10″ S 65°19′50″ W | Peach | 2019 | Brazilian 1 | |
| Ampimpa-P | Ampimpa | Tucumán | 26°36′44″ S 65°50′31″ W | Peach | 2021 | Brazilian 1 | |
| Tolombón-Pr | Tolombón | Salta | 26°11′04″ S 65°56′25″ W | Pear | 2019 | Brazilian 1 | |
| Cafayate-P | Cafayate | Salta | 24°44′13″ S 65°23′21″ W | Peach | 2019 | Brazilian 1 | |
| Yala-W | Yala | Jujuy | 24°07′11″ S 65°24′17″ W | Walnut | 2021 | Brazilian 1 | |
| Yuto-G | Yuto | Jujuy | 23°35′01″ S 64°30′25″ W | Guava | 2021 | Brazilian 1 | |
| Concordia-G | Concordia | Entre Ríos | 31°22′37″ S 58°01′01″ W | Guava | 2019 | Brazilian 1 | |
| Bella Vista-G | Bella Vista | Corrientes | 28°30′46″ S 59°02′31″ W | Guava | 2019 | Brazilian 1 | |
| Oberá-G | Oberá | Misiones | 27°39′01″ S 55°24′10″ W | Guava | 2019 | Brazilian 1 | |
| A. schultzi | Horco Molle-W | Horco Molle | Tucumán | 26°47′34″ S 65°18′58″ W | Walnut | 2019 | |
| Yala-W | Yala | Jujuy | 24°07′11″ S 65°24′17″ W | Walnut | 2021 |
| Type | Sample | Morphotype | Individuals Analysed | Sequences Analysed | Haplotypes | Length (bp) 1 | % GC | Haplotype Diversity ± SD | Nucleotide Diversity ± SD |
|---|---|---|---|---|---|---|---|---|---|
| A. fraterculus | TEOCELO | Mexican | 6 | 13 | 11 | 580–598 | 20.54 | 0.962 ± 0.050 | 0.020 ± 0.011 |
| (lab strains) | LA MOLINA | Peruvian | 6 | 10 | 10 | 599–603 | 20.10 | 1.000 ± 0.045 | 0.008 ± 0.005 |
| VACARIA | Brazilian 1 | 3 | 5 | 5 | 601–604 | 20.12 | 1.000 ± 0.127 | 0.008 ± 0.006 | |
| HORCO MOLLE | Brazilian 1 | 6 | 11 | 9 | 602–605 | 20.15 | 0.946 ± 0.066 | 0.006 ± 0.004 | |
| PIRACICABA | Brazilian 1 | 4 | 8 | 7 | 599–603 | 20.08 | 0.964 ± 0.077 | 0.005 ± 0.003 | |
| A. fraterculus | Horco Molle-W | Brazilian 1 | 1 | 3 | 2 | 602–603 | 20.09 | 0.667 ± 0.314 | 0.001 ± 0.001 |
| (wild samples) | Horco Molle-P | Brazilian 1 | 3 | 8 | 7 | 601–607 | 20.04 | 0.964 ± 0.077 | 0.007 ± 0.004 |
| Ampimpa-P | Brazilian 1 | 6 | 18 | 15 | 602–605 | 20.11 | 0.980 ± 0.024 | 0.007 ± 0.004 | |
| Tolombón-Pr | Brazilian 1 | 2 | 5 | 3 | 599–603 | 20.10 | 0.700 ± 0.218 | 0.003 ± 0.003 | |
| Cafayate-P | Brazilian 1 | 3 | 9 | 5 | 600–603 | 20.09 | 0.722 ± 0.159 | 0.004 ± 0.002 | |
| Yala-W | Brazilian 1 | 3 | 9 | 7 | 601–606 | 20.13 | 0.917 ± 0.092 | 0.006 ± 0.004 | |
| Yuto-G | Brazilian 1 | 6 | 16 | 13 | 600–605 | 20.12 | 0.967 ± 0.036 | 0.007 ± 0.004 | |
| Concordia-G | Brazilian 1 | 3 | 8 | 7 | 600–603 | 20.07 | 0.964 ± 0.077 | 0.007 ± 0.004 | |
| Bella Vista-G | Brazilian 1 | 3 | 9 | 7 | 601–604 | 20.14 | 0.917 ± 0.092 | 0.006 ± 0.004 | |
| Oberá-G | Brazilian 1 | 2 | 6 | 4 | 602–603 | 20.14 | 0.867 ± 0.129 | 0.006 ± 0.004 | |
| A. schultzi | Horco Molle-W | 2 | 4 | 4 | 596–600 | 20.52 | 1.000 ± 0.177 | 0.006 ± 0.005 | |
| (wild samples) | Yala-W | 3 | 9 | 8 | 587–601 | 20.50 | 0.972 ± 0.064 | 0.008 ± 0.005 | |
| Total | 62 | 151 | 97 2 |
| Source of Variation | d.f. | Sum of Squares | Variance Components | Percent of Variation | p |
|---|---|---|---|---|---|
| Among samples | 14 | 702.459 | 5.274 | 69.83 | <0.00001 |
| Within samples | 123 | 280.353 | 2.280 | 30.17 | |
| Total | 137 | 982.812 | 7.554 |
| Source of Variation | d.f. | Sum of Squares | Variance Components | Percent of Variation | p |
|---|---|---|---|---|---|
| Among samples | 14 | 702.459 | 5.107 | 67.63 | <0.00001 |
| Within samples | 42 | 150.353 | 0.839 | 11.11 | <0.00001 |
| Within individuals | 81 | 130.000 | 1.605 | 21.25 | <0.00001 |
| Total | 137 | 982.812 | 7.552 |
| Source of Variation | d.f. | Sum of Squares | Variance Components | Percent of Variation | p |
|---|---|---|---|---|---|
| Among morphotypes | 2 | 514.798 | 11.814 | 75.58 | <0.01271 |
| Within morphotypes | 12 | 187.660 | 1.537 | 9.83 | <0.00001 |
| Within samples | 123 | 280.353 | 2.279 | 14.58 | <0.00001 |
| Total | 137 | 982.812 | 15.229 |
| Source of Variation | d.f. | Sum of Squares | Variance Components | Percent of Variation | p |
|---|---|---|---|---|---|
| Among morphotypes | 2 | 669.375 | 12.895 | 84.67 | <0.00001 |
| Within morphotypes | 12 | 33.084 | 0.055 | 0.36 | 0.01760 |
| Within samples | 123 | 280.353 | 2.279 | 14.97 | <0.00001 |
| Total | 137 | 982.812 | 15.229 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gomulski, L.M.; Vera, M.T.; Lanzavecchia, S.B.; Piccinno, R.; Fiorenza, G.; De Luca, D.; Carrizo, B.N.; Bouvet, J.P.R.; Viana, V.A.; Cárceres, C.; et al. Molecular Markers for Analyses of Genetic Diversity within the Anastrepha fraterculus Complex with Emphasis on Argentine Populations. Insects 2024, 15, 748. https://doi.org/10.3390/insects15100748
Gomulski LM, Vera MT, Lanzavecchia SB, Piccinno R, Fiorenza G, De Luca D, Carrizo BN, Bouvet JPR, Viana VA, Cárceres C, et al. Molecular Markers for Analyses of Genetic Diversity within the Anastrepha fraterculus Complex with Emphasis on Argentine Populations. Insects. 2024; 15(10):748. https://doi.org/10.3390/insects15100748
Chicago/Turabian StyleGomulski, Ludvik M., María Teresa Vera, Silvia B. Lanzavecchia, Riccardo Piccinno, Giulia Fiorenza, Daniel De Luca, Beatriz N. Carrizo, Juan Pedro R. Bouvet, Valeria A. Viana, Carlos Cárceres, and et al. 2024. "Molecular Markers for Analyses of Genetic Diversity within the Anastrepha fraterculus Complex with Emphasis on Argentine Populations" Insects 15, no. 10: 748. https://doi.org/10.3390/insects15100748
APA StyleGomulski, L. M., Vera, M. T., Lanzavecchia, S. B., Piccinno, R., Fiorenza, G., De Luca, D., Carrizo, B. N., Bouvet, J. P. R., Viana, V. A., Cárceres, C., Enkerlin, W., Malacrida, A. R., & Gasperi, G. (2024). Molecular Markers for Analyses of Genetic Diversity within the Anastrepha fraterculus Complex with Emphasis on Argentine Populations. Insects, 15(10), 748. https://doi.org/10.3390/insects15100748







