Identification and Characterization of Four Novel Viruses in Balclutha incisa
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Sequencing
2.2. Genome Assembly and Virus Discovery
2.3. Determination of Viral Genomes
2.4. Genome Annotation and Phylogenetic Analysis
3. Results
3.1. Virome Analysis of B. incisa
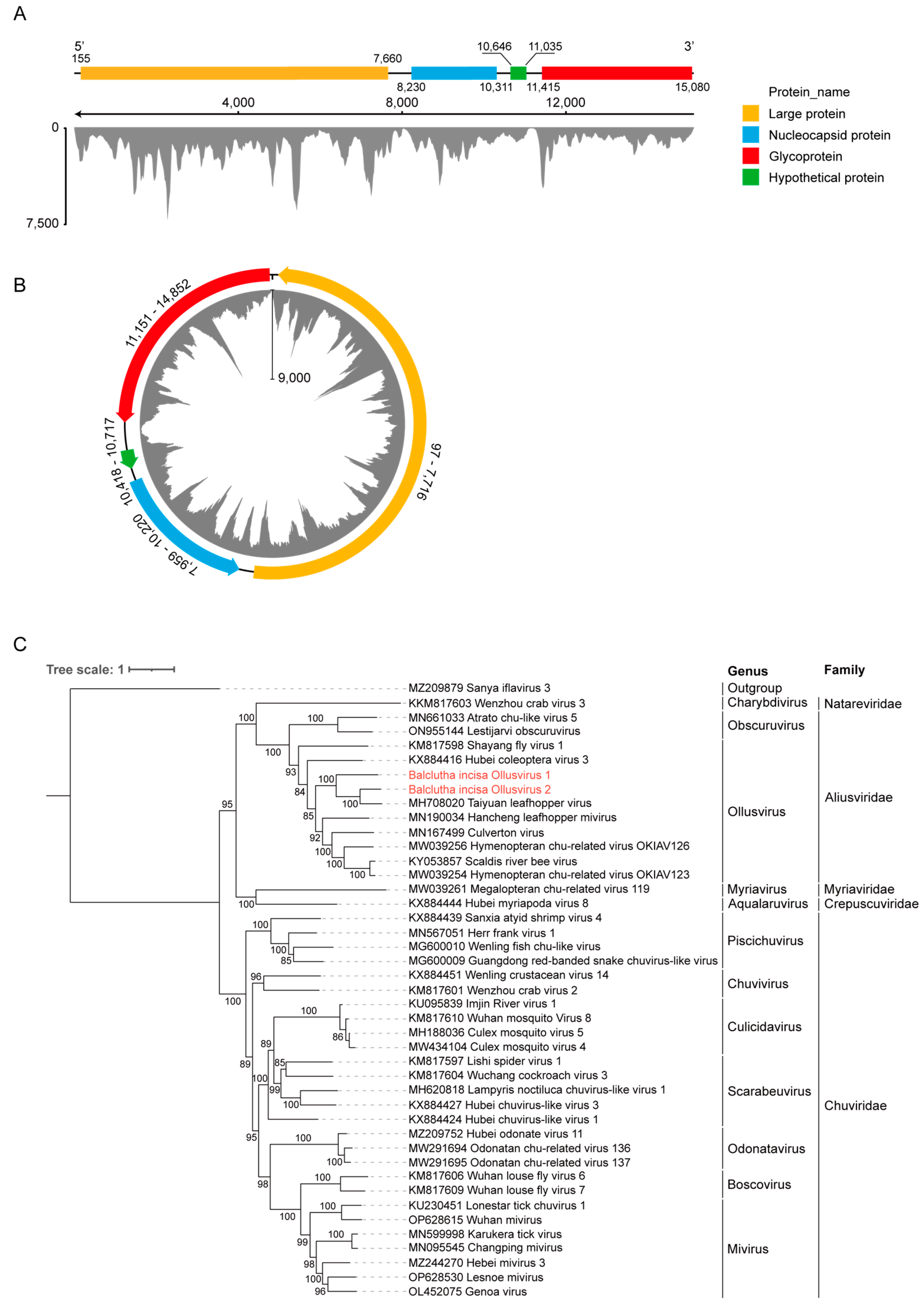
3.2. Identification of Two New Members Belonging to the Family Aliusviridae
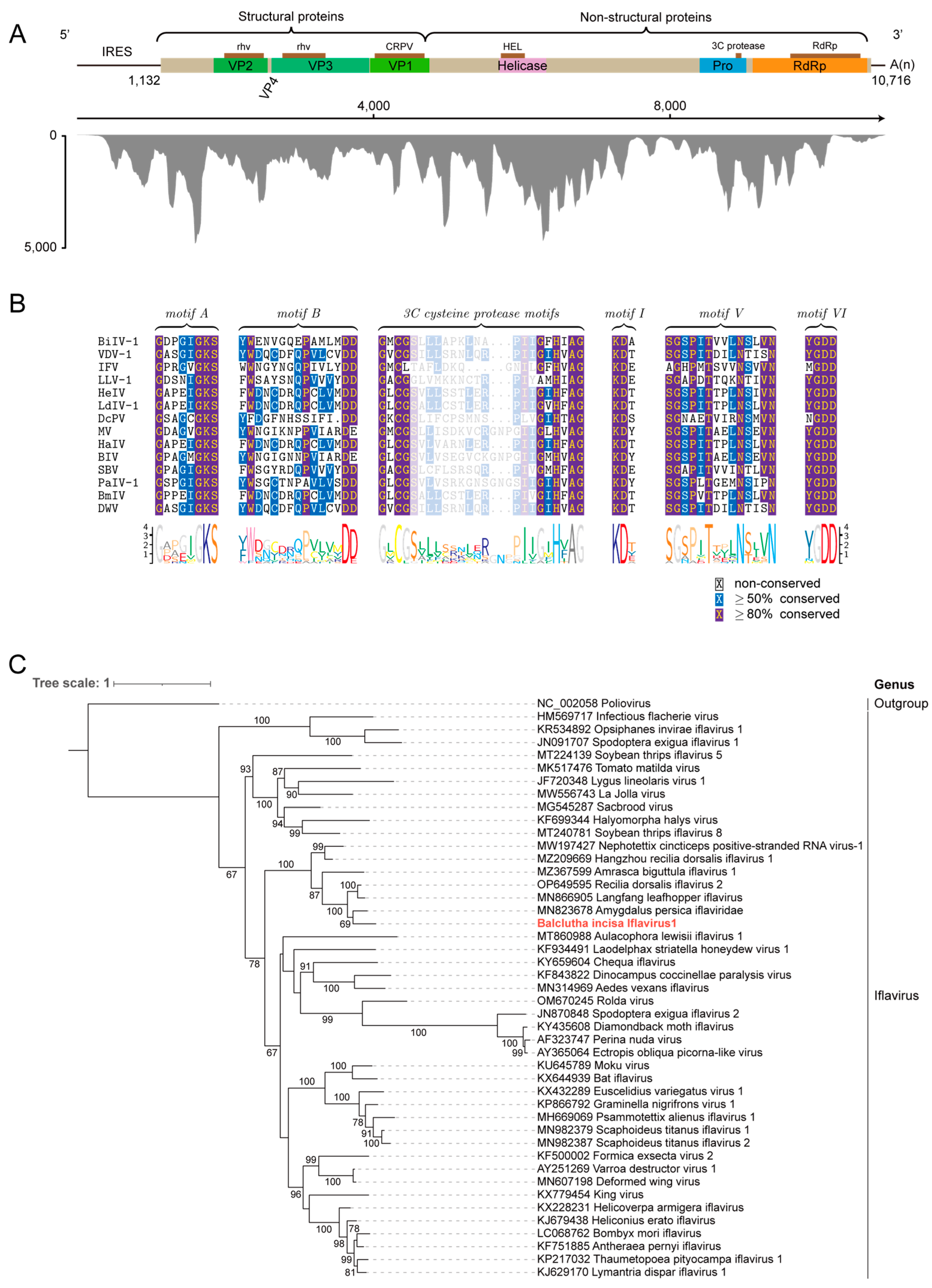
3.3. Identification of a Novel Member Belonging to the Family Iflaviridae
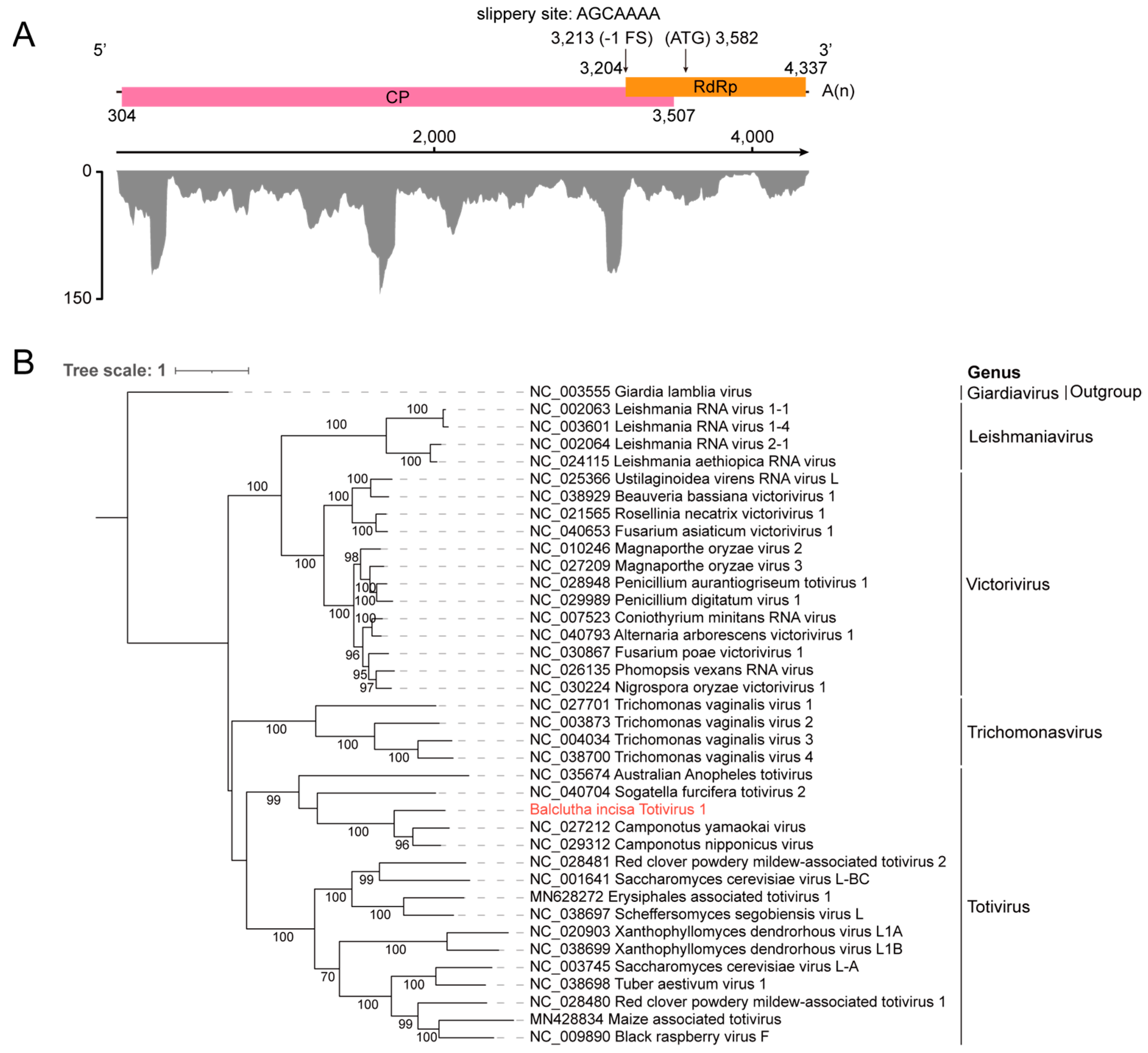
3.4. Identification of a New Member Belonging to the Family Totiviridae
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Prasad, R.; Shivay, Y.S.; Kumar, D. Current Status, Challenges, and Opportunities in Rice Production. In Rice Production Worldwide; Chauhan, B.S., Jabran, K., Mahajan, G., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 1–32. ISBN 978-3-319-47516-5. [Google Scholar]
- Elert, E. Rice by the Numbers: A Good Grain. Nature 2014, 514, S50–S51. [Google Scholar] [CrossRef] [PubMed]
- Hibino, H. Biology and Epidemiology of Rice Viruses. Annu. Rev. Phytopathol. 1996, 34, 249–274. [Google Scholar] [CrossRef]
- Wang, P.; Liu, J.; Lyu, Y.; Huang, Z.; Zhang, X.; Sun, B.; Li, P.; Jing, X.; Li, H.; Zhang, C. A Review of Vector-Borne Rice Viruses. Viruses 2022, 14, 2258. [Google Scholar] [CrossRef]
- Fujita, D.; Kohli, A.; Horgan, F. Rice Resistance to Planthoppers and Leafhoppers. Crit. Rev. Plant Sci. 2013, 32, 162–191. [Google Scholar] [CrossRef]
- Wu, J.-G.; Yang, G.-Y.; Zhao, S.-S.; Zhang, S.; Qin, B.-X.; Zhu, Y.-S.; Xie, H.-T.; Chang, Q.; Wang, L.; Hu, J.; et al. Current Rice Production Is Highly Vulnerable to Insect-Borne Viral Diseases. Natl. Sci. Rev. 2022, 9, nwac131. [Google Scholar] [CrossRef]
- Zhang, S.; Li, L.; Wang, X.; Zhou, G. Transmission of Rice Stripe Virus Acquired From Frozen Infected Leaves by the Small Brown Planthopper (Laodelphax striatellus Fallen). J. Virol. Methods 2007, 146, 359–362. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Andika, I.B.; Shen, J.; Lv, Y.; Ji, Y.; Sun, L.; Chen, J. Characterization of rice black-streaked dwarf virus-and rice stripe virus-derived siRNAs in singly and doubly infected insect vector Laodelphax striatellus. PLoS ONE 2013, 8, e66007. [Google Scholar] [CrossRef]
- Li, L.; Li, H.; Dong, H.; Wang, X.; Zhou, G. Transmission by Laodelphax striatellus Fallen of Rice Black-Streaked Dwarf Virus from Frozen Infected Rice Leaves to Healthy Plants of Rice and Maize. J. Phytopathol. 2011, 159, 1–5. [Google Scholar] [CrossRef]
- Nakasuji, F. Epidemiological Study on Rice Dwarf Virus Transmitted by the Green Rice Leafhopper Nephotettix cincticeps. JARQ 1974, 8, 84–91. [Google Scholar]
- Jia, W.; Wang, F.; Li, J.; Chang, X.; Yang, Y.; Yao, H.; Bao, Y.; Song, Q.; Ye, G. A Novel Iflavirus was Discovered in Green Rice Leafhopper Nephotettix Cincticeps and its Proliferation was Inhibited by Infection of Rice Dwarf Virus. Front. Microbiol. 2021, 11, 621141. [Google Scholar] [CrossRef]
- Pu, L.; Xie, G.; Ji, C.; Ling, B.; Zhang, M.; Xu, D.; Zhou, G. Transmission Characteristics of Southern Rice Black-streaked Dwarf Virus by Rice Planthoppers. Crop Prot. 2012, 41, 71–76. [Google Scholar] [CrossRef]
- Zhang, P.; Liu, W.; Cao, M.; Massart, S.; Wang, X. Two Novel Totiviruses in the White-backed Planthopper, Sogatella Furcifera. J. Gen. Virol. 2018, 99, 710–716. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.-N.; Ye, Z.-X.; Chen, M.-N.; Ren, P.-P.; Ning, C.; Sun, Z.-T.; Chen, J.-P.; Zhang, C.-X.; Li, J.-M.; Mao, Q. Identification and Characterization of Three Novel Solemo-like Viruses in the White-Backed Planthopper, Sogatella furcifera. Insects 2024, 15, 394. [Google Scholar] [CrossRef]
- Koudamiloro, A.; Nwilene, F.E.; Togola, A.; Akogbeto, M. Insect vectors of rice yellow mottle virus. J. Insects 2015, 2015, 721751. [Google Scholar] [CrossRef]
- Lozano, I.; Morales, F. Molecular characterisation of Rice stripe necrosis virus as a new species of the genus Benyvirus. Eur. J. Plant Pathol. 2009, 124, 673–680. [Google Scholar] [CrossRef]
- Gallet, R.; Michalakis, Y.; Blanc, S. Vector-Transmission of Plant Viruses and Constraints Imposed by Virus-Vector Interactions. Curr. Opin. Virol. 2018, 33, 144–150. [Google Scholar] [CrossRef]
- Narhardiyati, M.; Bailey, W. Biology and Natural Enemies of the Leafhopper Balclutha incisa (Matsumura) (Hemiptera: Cicadellidae: Deltocephalinae) in South-Western Australia. Aust. J. Entomol. 2005, 44, 104–109. [Google Scholar] [CrossRef]
- Khatri, I.; Rustamani, M.A.; Wagan, M.S.; Nizamani, S.M. Two Economically Important Leafhoppers Cicadulina bipunctata (Melichar) and Balclutha incisa (Matsumura) (Hemiptera: Cicadellidae: Deltocephalinae: Macrostelini) From Tando Jam, Pakistan. Pak. J. Zool. 2011, 43, 747–750. [Google Scholar]
- Abu Alloush, A.; Bianco, P.; Amashah, S.; Busato, E.; Mahasneh, A.; AlShoubaki, M.; Alma, A.; Tedeschi, R.; Quaglino, F. Identification of Four Distinct ‘Candidatus Phytoplasma’ Species in Pomegranate Trees Showing Witches’ Broom, Little Leaf and Yellowing in Jordan, and Preliminary Insights on Their Putative Insect Vectors and Reservoir Plants. Ann. Appl. Biol. 2022, 182, 159–170. [Google Scholar] [CrossRef]
- Salem, N.M.; Quaglino, F.; Abdeen, A.; Casati, P.; Bulgari, D.; Alma, A.; Bianco, P.A. First Report of “Candidatus Phytoplasma Solani” Strains Associated with Grapevine Bois Noir in Jordan. Plant Dis. 2013, 97, 1505. [Google Scholar] [CrossRef]
- Cassedy, A.; Parle-McDermott, A.; O’Kennedy, R. Virus Detection: A Review of the Current and Emerging Molecular and Immunological Methods. Front. Mol. Biosci. 2021, 8, 637559. [Google Scholar] [CrossRef] [PubMed]
- Hematian, A.; Sadeghifard, N.; Mohebi, R.; Taherikalani, M.; Nasrolahi, A.; Amraei, M.; Ghafourian, S. Traditional and Modern Cell Culture in Virus Diagnosis. Osong Public Health Res. Perspect. 2016, 7, 77–82. [Google Scholar] [CrossRef]
- Miller, R.R.; Montoya, V.; Gardy, J.L.; Patrick, D.M.; Tang, P. Metagenomics for Pathogen Detection in Public Health. Genome Med. 2013, 5, 81. [Google Scholar] [CrossRef]
- Simmonds, P.; Adams, M.J.; Benkő, M.; Breitbart, M.; Brister, J.R.; Carstens, E.B.; Davison, A.J.; Delwart, E.; Gorbalenya, A.E.; Harrach, B.; et al. Virus Taxonomy in the Age of Metagenomics. Nat. Rev. Microbiol. 2017, 15, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-M.; Sadiq, S.; Tian, J.-H.; Chen, X.; Lin, X.-D.; Shen, J.-J.; Chen, H.; Hao, Z.-Y.; Wille, M.; Zhou, Z.-C.; et al. RNA Viromes from Terrestrial Sites across China Expand Environmental Viral Diversity. Nat. Microbiol. 2022, 7, 1312–1323. [Google Scholar] [CrossRef] [PubMed]
- Li, C.-X.; Shi, M.; Tian, J.-H.; Lin, X.-D.; Kang, Y.-J.; Chen, L.-J.; Qin, X.-C.; Xu, J.; Holmes, E.C.; Zhang, Y.-Z. Unprecedented Genomic Diversity of RNA Viruses in Arthropods Reveals the Ancestry of Negative-Sense RNA Viruses. eLife 2015, 4, e05378. [Google Scholar] [CrossRef]
- Shi, M.; Lin, X.-D.; Tian, J.-H.; Chen, L.-J.; Chen, X.; Li, C.-X.; Qin, X.-C.; Li, J.; Cao, J.-P.; Eden, J.-S.; et al. Redefining the Invertebrate RNA Virosphere. Nature 2016, 540, 539–543. [Google Scholar] [CrossRef] [PubMed]
- Liang, G.; Bushman, F.D. The Human Virome: Assembly, Composition and Host Interactions. Nat. Rev. Microbiol. 2021, 19, 514–527. [Google Scholar] [CrossRef] [PubMed]
- Martino, C.; Dilmore, A.H.; Burcham, Z.M.; Metcalf, J.L.; Jeste, D.; Knight, R. Microbiota Succession throughout Life from the Cradle to the Grave. Nat. Rev. Microbiol. 2022, 20, 707–720. [Google Scholar] [CrossRef]
- Edwards, R.A.; Rohwer, F. Viral Metagenomics. Nat. Rev. Microbiol. 2005, 3, 504–510. [Google Scholar] [CrossRef]
- Di Paola, N.; Dheilly, N.M.; Junglen, S.; Paraskevopoulou, S.; Postler, T.S.; Shi, M.; Kuhn, J.H. Jingchuvirales: A New Taxonomical Framework for a Rapidly Expanding Order of Unusual Monjiviricete Viruses Broadly Distributed Among Arthropod Subphyla. Appl. Environ. Microbiol. 2022, 88, e01954-21. [Google Scholar] [CrossRef]
- Kuhn, J.H.; Dheilly, N.M.; Junglen, S.; Paraskevopoulou, S.; Shi, M.; Di Paola, N. ICTV Virus Taxonomy Profile: Jingchuvirales 2023. J. Gen. Virol. 2023, 104, 001924. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, P.T.T.; Culverwell, C.L.; Suvanto, M.T.; Korhonen, E.M.; Uusitalo, R.; Vapalahti, O.; Smura, T.; Huhtamo, E. Characterisation of the RNA Virome of Nine Ochlerotatus Species in Finland. Viruses 2022, 14, 1489. [Google Scholar] [CrossRef]
- Hong, H.; Ye, Z.; Lu, G.; Feng, K.; Zhang, M.; Sun, X.; Han, Z.; Jiang, S.; Wu, B.; Yin, X.; et al. Characterisation of a Novel Insect-Specific Virus Discovered in Rice Thrips, Haplothrips aculeatus. Insects 2024, 15, 303. [Google Scholar] [CrossRef]
- Valles, S.M.; Chen, Y.; Firth, A.E.; Guérin, D.A.; Hashimoto, Y.; Herrero, S.; de Miranda, J.R.; Ryabov, E.; ICTV Report Consortium. ICTV Virus Taxonomy Profile: Iflaviridae. J. Gen. Virol. 2017, 98, 527–528. [Google Scholar] [CrossRef]
- Van Oers, M.M. Insect Virology. In Genomics and Biology of Ifaviruses; Asgari, S., Johnson, K., Eds.; Caister Academic Press: Norfolk, UK, 2010; pp. 231–250. [Google Scholar]
- Ghabrial, S.A.; Nibert, M.L. Victorivirus, a New Genus of Fungal Viruses in the Family Totiviridae. Arch. Virol. 2009, 154, 373–379. [Google Scholar] [CrossRef]
- Chen, S.; Cao, L.; Huang, Q.; Qian, Y.; Zhou, X. The Complete Genome Sequence of a Novel Maize-Associated Totivirus. Arch. Virol. 2016, 161, 487–490. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An Ultra-Fast All-in-One FASTQ Preprocessor. Bioinformatic 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-Length Transcriptome Assembly from RNA-Seq Data without a Reference Genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and Applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Soding, J.; Biegert, A.; Lupas, A.N. The HHpred Interactive Server for Protein Homology Detection and Structure Prediction. Nucleic Acids Res. 2005, 33, W244–W248. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, T.; Yamada, K.D.; Tomii, K.; Katoh, K. Parallelization of MAFFT for Large-Scale Multiple Sequence Alignments. Bioinformatics 2018, 34, 2490–2492. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL): An Online Tool for Phylogenetic Tree Display and Annotation. Bioinformatics 2007, 23, 127–128. [Google Scholar] [CrossRef]
- Zheng, L.; Lu, X.; Liang, X.; Jiang, S.; Zhao, J.; Zhan, G.; Liu, P.; Wu, J.; Kang, Z. Molecular Characterization of Novel Totivirus-like Double-Stranded RNAs from Puccinia striiformis f. sp. Tritici, the Causal Agent of Wheat Stripe Rust. Front. Microbiol. 2017, 8, 1960. [Google Scholar]
- Villordo, S.M.; Alvarez, D.E.; Gamarnik, A.V. A Balance between Circular and Linear Forms of the Dengue Virus Genome Is Crucial for Viral Replication. RNA 2010, 16, 2325–2335. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Liu, Y.; Liu, W.; Cao, M.; Wang, X. Full genome sequence of a novel iflavirus from the leafhopper Psammotettix alienus. Arch. Virol. 2019, 164, 309–311. [Google Scholar] [CrossRef] [PubMed]
- Ma, R.; Nie, B.; Chen, J.; Lv, K.; Xiao, J.; Liu, R. Full genome sequence of a novel iflavirus from the leafhopper Recilia dorsalis. Arch. Virol. 2022, 167, 1593–1596. [Google Scholar] [CrossRef]
- Xiao, J.; Zhu, K.; Ma, R.; Zhang, C.; Lv, K.; Ge, D.; Liu, R. Full genome sequence of a novel iflavirus from the aster leafhopper Macrosteles fascifrons. Virus Genes 2023, 59, 338–342. [Google Scholar] [CrossRef] [PubMed]
- De Miranda, J.R.; Genersch, E. Deformed Wing Virus. J. Invertebr. Pathol. 2010, 103, S48–S61. [Google Scholar] [CrossRef] [PubMed]
- Wei, R.; Cao, L.; Feng, Y.; Chen, Y.; Chen, G.; Zheng, H. Sacbrood Virus: A Growing Threat to Honeybees and Wild Pollinators. Viruses 2022, 14, 1871. [Google Scholar] [CrossRef]
- Burnham, P.A.; Alger, S.A.; Case, B.; Boncristiani, H.; Hébert-Dufresne, L.; Brody, A.K. Flowers as Dirty Doorknobs: Deformed Wing Virus Sransmitted between Apis mellifera and Bombus Impatiens through Shared Flowers. J. Appl. Ecol. 2021, 58, 2065–2074. [Google Scholar] [CrossRef]
- Saqib, M.; Wylie, S.J.; Jones, M.G.K. Serendipitous Identification of a New Iflavirus-like Virus Infecting Tomato and its Subsequent Characterization. Plant. Pathol. 2015, 64, 519–527. [Google Scholar] [CrossRef]
- Yang, S.; Mao, Q.; Wang, Y.; He, J.; Yang, J.; Chen, X.; Xiao, Y.; He, Y.; Zhao, M.; Lu, J.; et al. Expanding Known Viral Diversity in Plants: Virome of 161 Species alongside an Ancient Canal. Environ. Microbiome 2022, 17, 58. [Google Scholar] [CrossRef] [PubMed]
- Goodman, R.P.; Ghabrial, S.A.; Fichorova, R.N.; Nibert, M.L. Trichomonasvirus: A New Genus of Protozoan Viruses in the Family Totiviridae. Arch. Virol. 2011, 156, 171–179. [Google Scholar] [CrossRef]
- Huang, Y.; Guo, X.; Zhang, S.; Zhao, Q.; Sun, Q.; Zhou, H.; Zhang, J.; Tong, Y. Discovery of Two Novel Totiviruses from Culex Tritaeniorhynchus Classifiable in a Aistinct Clade with Arthropod-infecting Viruses within the Family Totiviridae. Arch. Virol. 2018, 163, 2899–2902. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiao, J.; Yang, G.; Liu, R.; Ge, D. Identification and Characterization of Four Novel Viruses in Balclutha incisa. Insects 2024, 15, 772. https://doi.org/10.3390/insects15100772
Xiao J, Yang G, Liu R, Ge D. Identification and Characterization of Four Novel Viruses in Balclutha incisa. Insects. 2024; 15(10):772. https://doi.org/10.3390/insects15100772
Chicago/Turabian StyleXiao, Jiajing, Guang Yang, Renyi Liu, and Danfeng Ge. 2024. "Identification and Characterization of Four Novel Viruses in Balclutha incisa" Insects 15, no. 10: 772. https://doi.org/10.3390/insects15100772






