Integrative Analyses of Transcriptomics and Metabolomics in Immune Response of Leguminivora glycinivorella Mats to Beauveria bassiana Infection
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Test Strains and Insects
2.2. Infection of L. glycinivorella with B. bassiana
2.3. Extraction and Transcriptome Sequencing of Total RNA from Soybean Heartworm
2.4. Widely Targeted Metabolomic Analysis of L. glycinivorella
2.5. Combined Analysis of the Transcriptome and Metabolome of Soybean Heartworm
2.6. Real-Time Fluorescence Quantitative PCR Detection
3. Results
3.1. Transcriptomic Analysis of the Immune Response of Soybean Heartworm Infected with B. bassiana
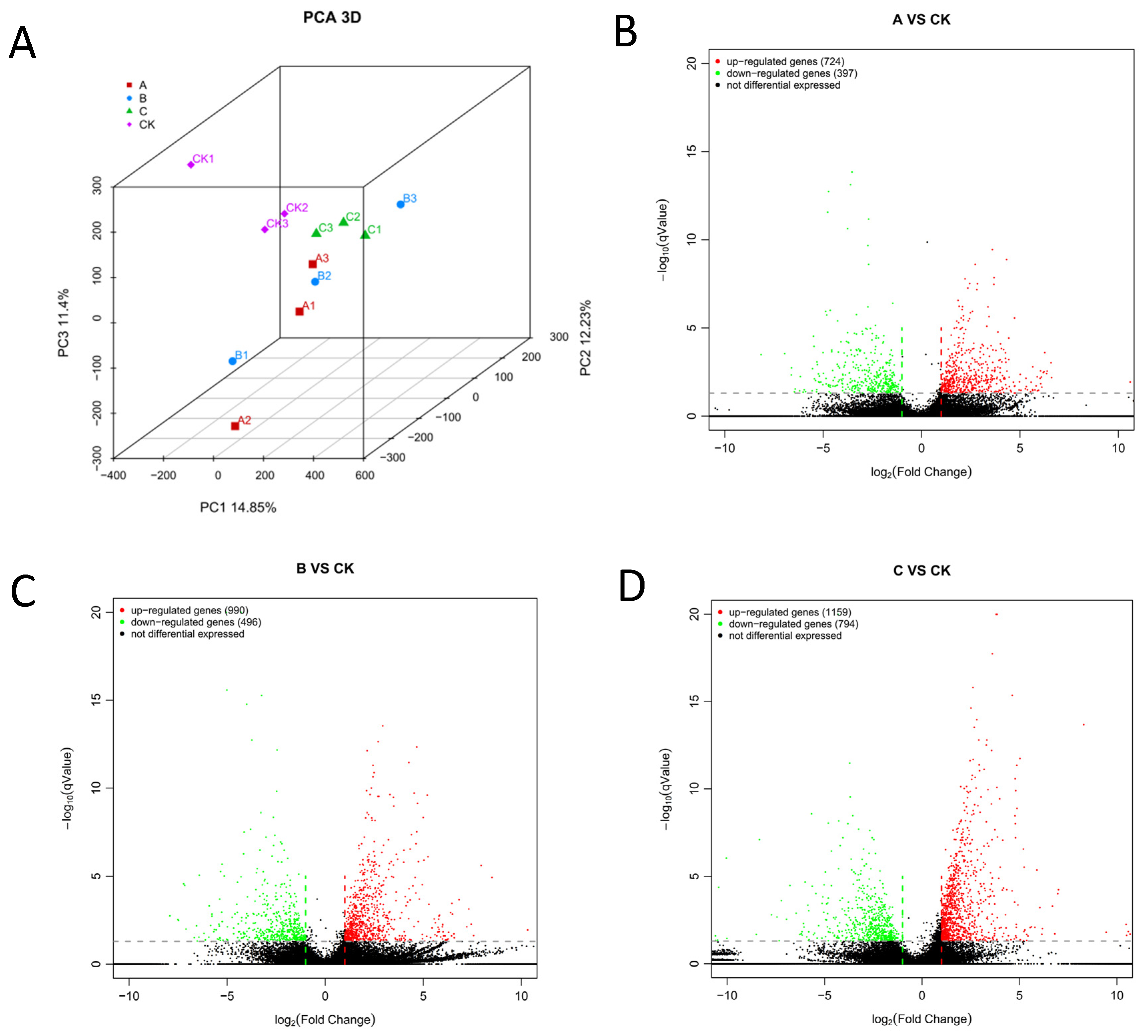
3.1.1. Quality Control of Samples and Analysis of Differences in Gene Expression
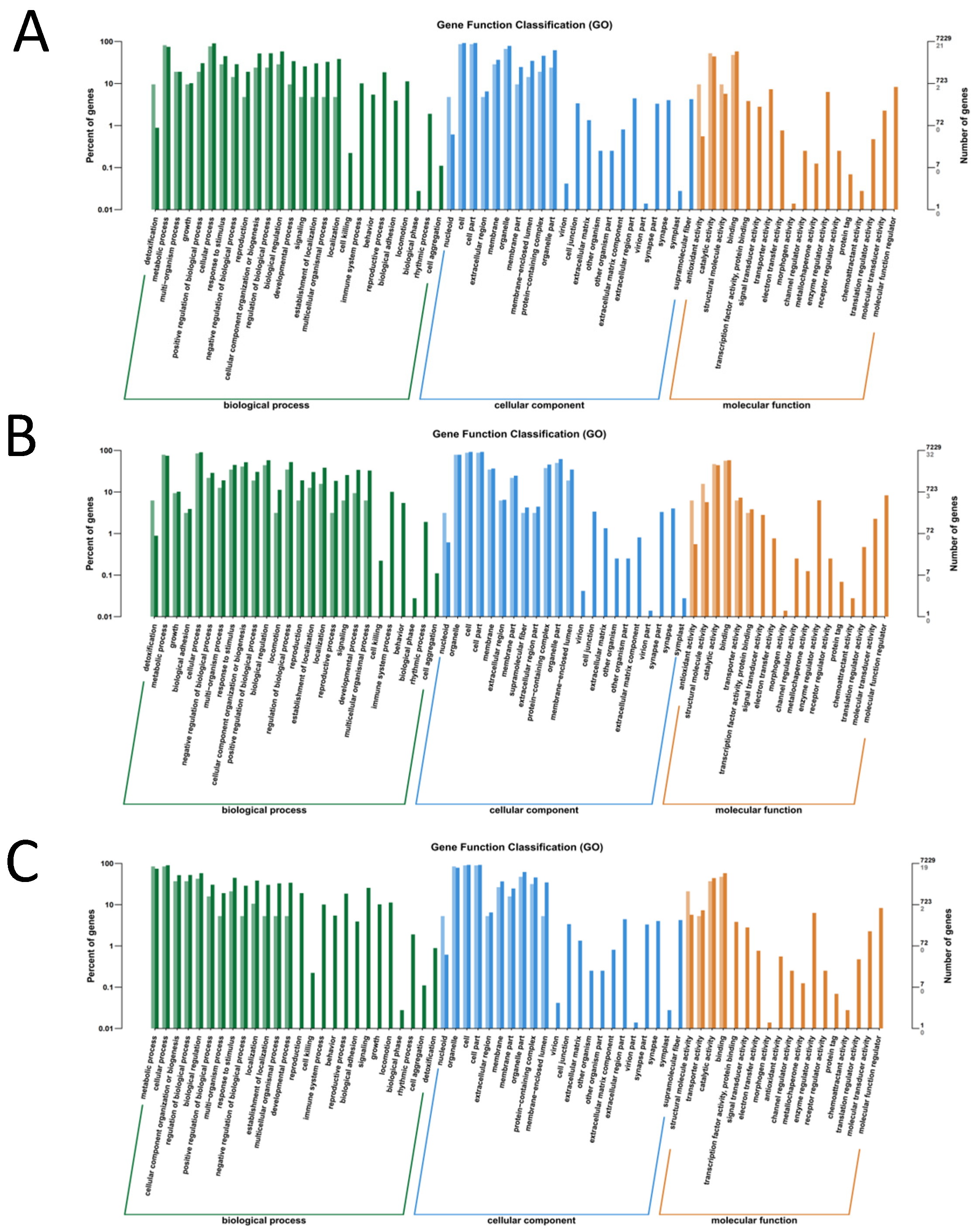
3.1.2. GO Enrichment Analysis of Differentially Expressed Genes
3.1.3. KEGG Pathway Analysis of Differentially Expressed Genes
3.2. Metabolomic Analysis of the Immune Response of L. glycinivorella after Infection with B. bassiana
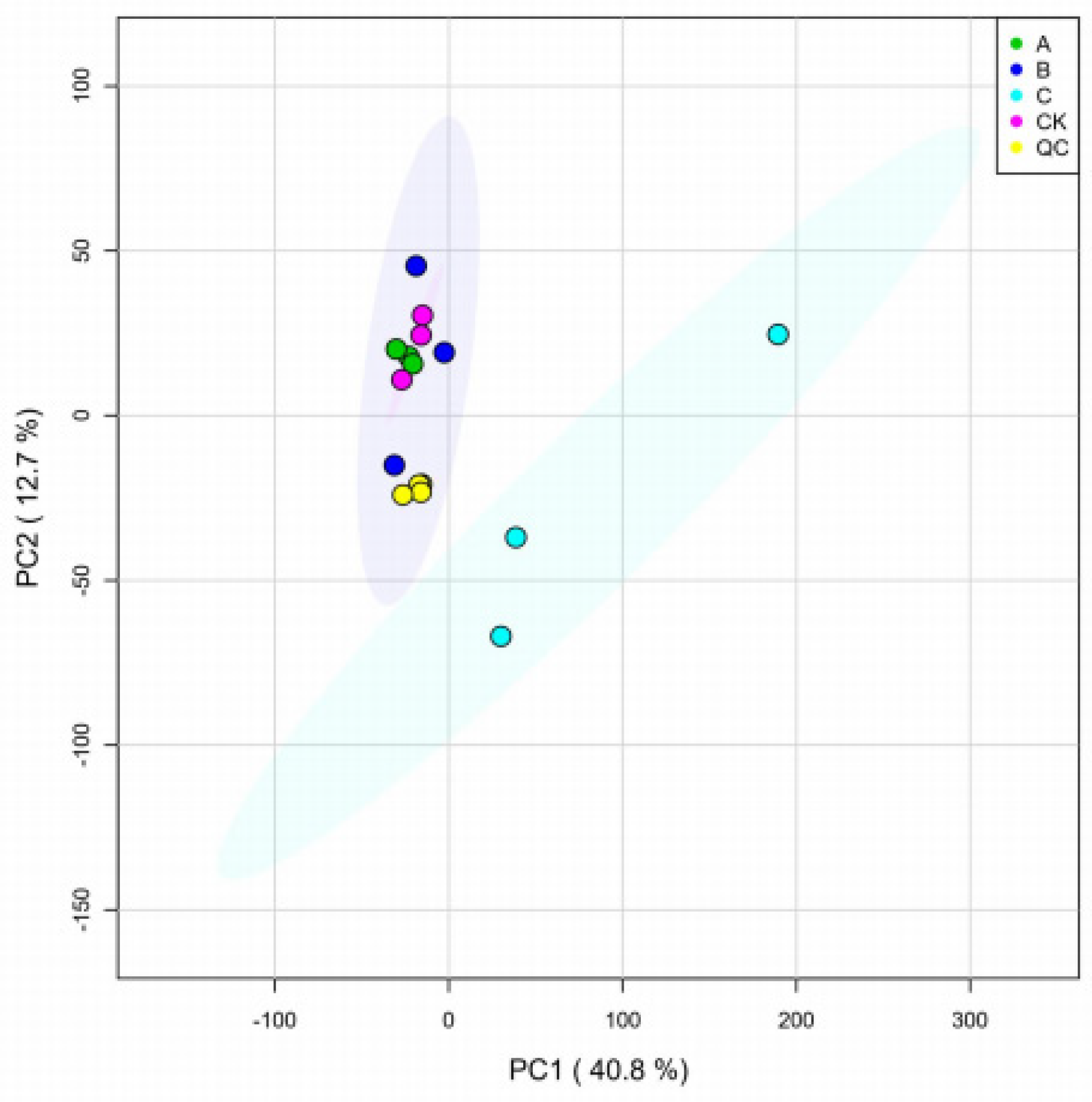
3.2.1. Quality Control of Metabolome Samples
3.2.2. Screening of Differentially Expressed Metabolites Related to Antibacterial Activity
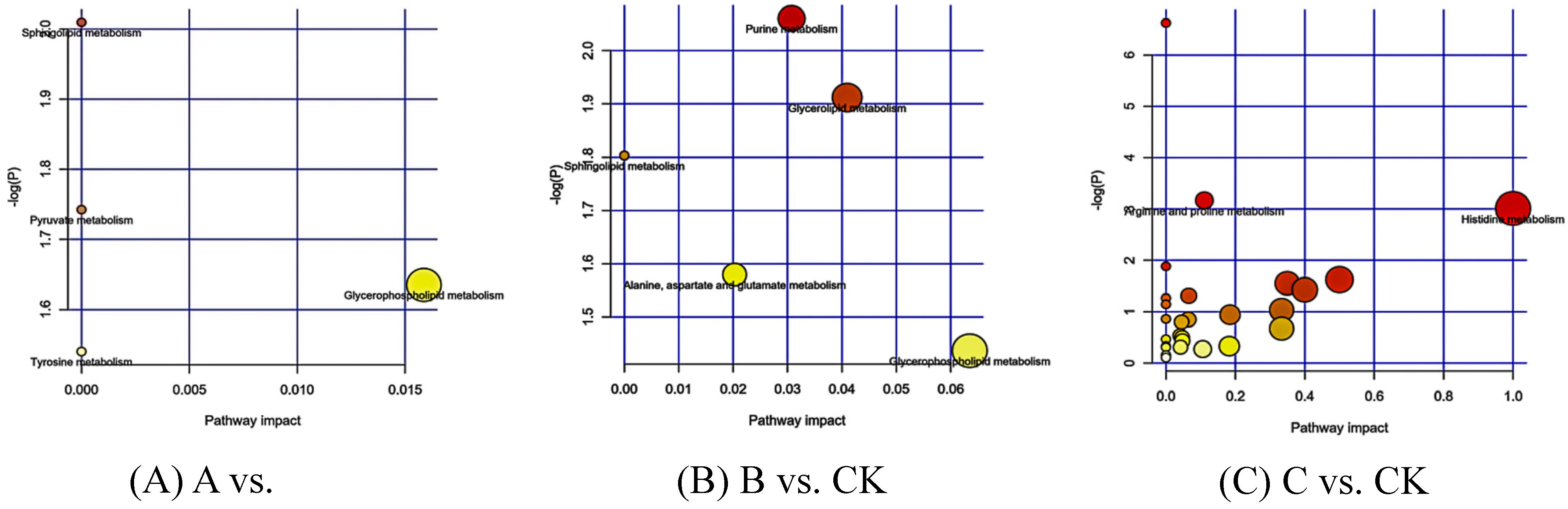
3.2.3. KEGG Enrichment of the Differential Metabolites
3.3. Transcriptome and Metabolome Analysis of Immune Response in L. glycinivorella Infected with B. bassiana
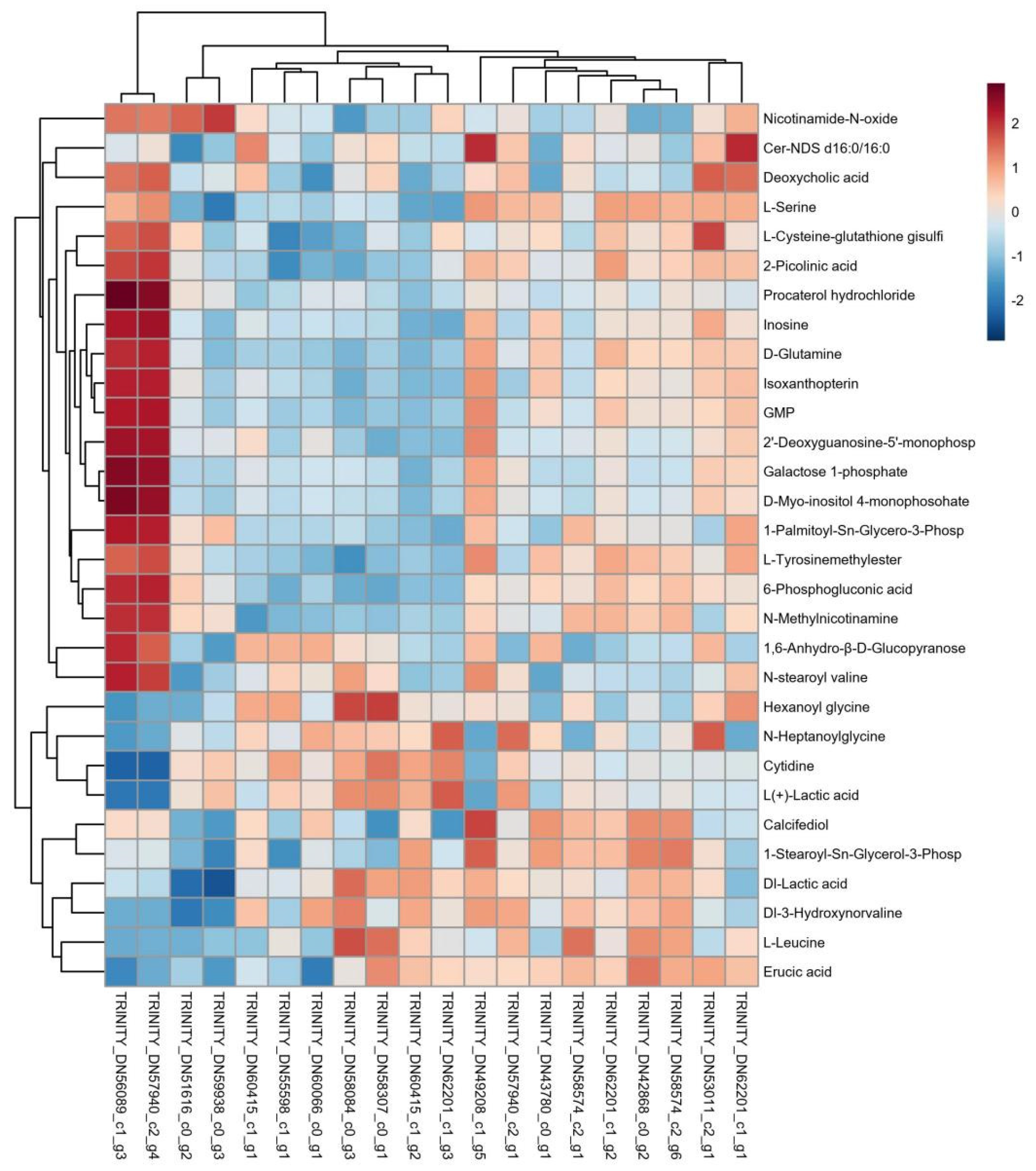
3.3.1. Cluster Analysis of Differentially Expressed Genes and Metabolites
3.3.2. Analysis of Antibacterial-Related Differential Genes and Metabolite Regulation Mechanisms
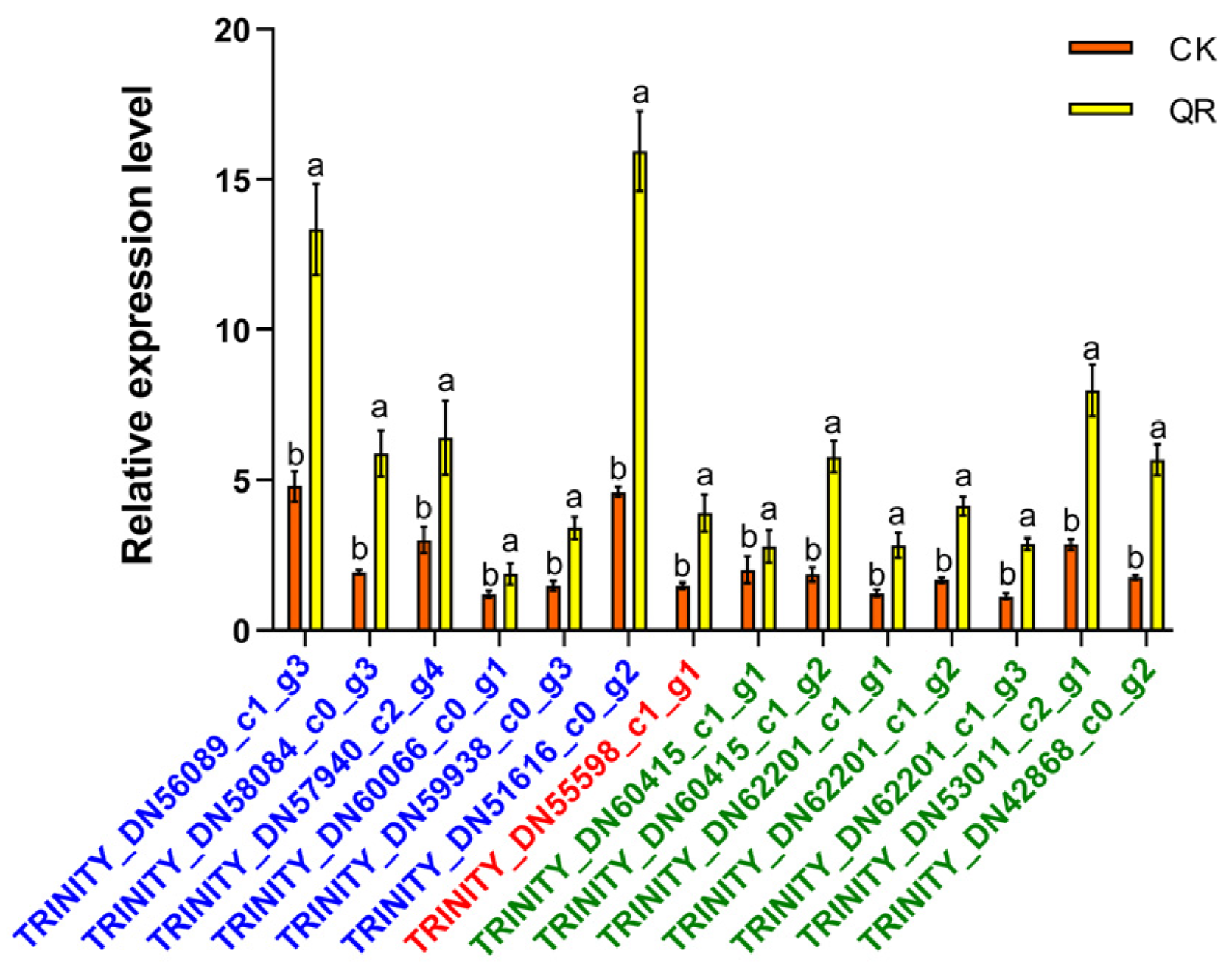
3.4. qRT-PCR Validation of Transcriptome Sequencing Data
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ran, R.; Li, T.; Liu, X.; Ni, H.; Li, W.; Meng, F. RNA interference-mediated silencing of genes involved in the immune responses of the soybean pod borer Leguminivora glycinivorella (Lepidoptera: Olethreutidae). PeerJ 2018, 6, e4931. [Google Scholar] [CrossRef] [PubMed]
- Meng, F.; Li, Y.; Zang, Z.; Li, N.; Ran, R.; Cao, Y.; Li, T.; Zhou, Q.; Li, W. Expression of the double-stranded RNA of the soybean pod borer Leguminivora glycinivorella (Lepidoptera: Tortricidae) ribosomal protein P0 gene enhances the resistance of transgenic soybean plants. Pest Manag. Sci. 2017, 73, 2447–2455. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Wang, Z.; Wang, R.; Zhang, X.; Li, M.; Xin, J.; Qin, Y.; Zhang, C.; Meng, F. Transcriptomic and proteomic analyses of the mechanisms of overwintering diapause in soybean pod borer (Leguminivora glycinivorella). Pest Manag. Sci. 2020, 76, 4248–4257. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Song, B.; Yu, C.; Zhang, J.; Zhang, J.; Bi, R.; Li, X.; Ren, X.; Zhu, Y.; Yao, D. Identifying Soybean Pod Borer (Leguminivora glycinivorella) Resistance QTLs and the Mechanism of Induced Defense Using Linkage Mapping and RNA-Seq Analysis. Int. J. Mol. Sci. 2022, 23, 10910. [Google Scholar] [CrossRef] [PubMed]
- Melotto, G.; Potter, B.D.; Koch, R.L.; Lindsey, A.R. Spatial and temporal dynamics of soybean gall midge (Resseliella maxima) parasitism by Synopeas maximum. Pest Manag. Sci. 2023, 79, 5096–5105. [Google Scholar] [CrossRef]
- Gui, J.; Xu, H.; Fei, J. Non-Destructive Detection of Soybean Pest Based on Hyperspectral Image and Attention-ResNet Meta-Learning Model. Sensors 2023, 23, 678. [Google Scholar] [CrossRef]
- Hu, D.-H.; He, J.; Zhou, Y.-W.; Feng, J.-T.; Zhang, X. Synthesis and field evaluation of the sex pheromone analogues to soybean pod borer Leguminivora glycinivorella. Molecules 2012, 17, 12140–12150. [Google Scholar] [CrossRef]
- Pareddy, D.; Chennareddy, S.; Anthony, G.; Sardesai, N.; Mall, T.; Minnicks, T.; Karpova, O.; Clark, L.; Griffin, D.; Bishop, B. Improved soybean transformation for efficient and high throughput transgenic production. Transgenic Res. 2020, 29, 267–281. [Google Scholar] [CrossRef]
- Manczinger, L. Biological control of agricultural pests by filamentous fungi. Acta Microbiol. Imm. Hung. 1999, 46, 259–267. [Google Scholar] [CrossRef][Green Version]
- Tong, Z.; Sun, M.; Zhou, Z.; Dong, X.; Hu, B.; Duan, J. The fate and effect of chlorpyrifos and lambda-cyhalothrin in soybean (Glycine max L. Merril) field. Ecotox. Environ. Safe 2021, 209, 111861. [Google Scholar] [CrossRef]
- Zhan, Y.; Chen, S.; Wang, G.; Fu, J.; Lan, Y. Biological control technology and application based on agricultural unmanned aerial vehicle (UAV) intelligent delivery of insect natural enemies (Trichogramma) carrier. Pest Manag. Sci. 2021, 77, 3259–3272. [Google Scholar] [CrossRef] [PubMed]
- Fontes, E.M.G.; Laumann, R. Special Section on Biological Control. Neotrop. Entomol. 2019, 48, 873–874. [Google Scholar] [CrossRef]
- Ab Rahman, S.F.S.; Singh, E.; Pieterse, C.M.; Schenk, P.M. Emerging microbial biocontrol strategies for plant pathogens. Plant Sci. 2018, 267, 102–111. [Google Scholar] [CrossRef]
- Martin, E.A.; Reineking, B.; Seo, B.; Steffan-Dewenter, I. Natural enemy interactions constrain pest control in complex agricultural landscapes. Proc. Natl. Acad. Sci. USA 2013, 110, 5534–5539. [Google Scholar] [CrossRef] [PubMed]
- Thies, C.; Tscharntke, T. Landscape structure and biological control in agroecosystems. Science 1999, 285, 893–895. [Google Scholar] [CrossRef]
- Fei, M.; Gols, R.; Harvey, J.A. The biology and ecology of parasitoid wasps of predatory arthropods. Annu. Rev. Entomol. 2023, 68, 109–128. [Google Scholar] [CrossRef]
- Lacey, L.A.; Shapiro-Ilan, D.I. Microbial control of insect pests in temperate orchard systems: Potential for incorporation into IPM. Annu. Rev. Entomol. 2008, 53, 121–144. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.M. Biological control with Trichogramma: Advances, successes, and potential of their use. Annu. Rev. Entomol. 1996, 41, 375–406. [Google Scholar] [CrossRef]
- Sanger, F.; Coulson, A.; Friedmann, T.; Air, G.; Barrell, B.; Brown, N.; Fiddes, J.; Hutchison Iii, C.; Slocombe, P.; Smith, M. The nucleotide sequence of bacteriophage φX174. J. Mol. Biol. 1978, 125, 225–246. [Google Scholar] [CrossRef]
- Sultan, M.; Schulz, M.H.; Richard, H.; Magen, A.; Klingenhoff, A.; Scherf, M.; Seifert, M.; Borodina, T.; Soldatov, A.; Parkhomchuk, D. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science 2008, 321, 956–960. [Google Scholar] [CrossRef]
- Margulies, M.; Egholm, M.; Altman, W.E.; Attiya, S.; Bader, J.S.; Bemben, L.A.; Berka, J.; Braverman, M.S.; Chen, Y.-J.; Chen, Z. Genome sequencing in microfabricated high-density picolitre reactors. Nature 2005, 437, 376–380. [Google Scholar] [CrossRef] [PubMed]
- Seo, T.S.; Bai, X.; Kim, D.H.; Meng, Q.; Shi, S.; Ruparel, H.; Li, Z.; Turro, N.J.; Ju, J. Four-color DNA sequencing by synthesis on a chip using photocleavable fluorescent nucleotides. Proc. Natl. Acad. Sci. USA 2005, 102, 5926–5931. [Google Scholar] [CrossRef] [PubMed]
- Mo, Y.; Jiao, Y. Advances and applications of single-cell omics technologies in plant research. Plant J. 2022, 110, 1551–1563. [Google Scholar] [CrossRef] [PubMed]
- Schrimpe-Rutledge, A.C.; Codreanu, S.G.; Sherrod, S.D.; Mclean, J.A. Untargeted metabolomics strategies—Challenges and emerging directions. J. Am. Soc. Mass. Spectrom. 2016, 27, 1897–1905. [Google Scholar] [CrossRef] [PubMed]
- Tokarz, J.; Haid, M.; Cecil, A.; Prehn, C.; Artati, A.; Möller, G.; Adamski, J. Endocrinology meets metabolomics: Achievements, pitfalls, and challenges. Trends Endocrinol. Metab. 2017, 28, 705–721. [Google Scholar] [CrossRef]
- Pang, R.; Chen, M.; Yue, L.; Xing, K.; Li, T.; Kang, K.; Liang, Z.; Yuan, L.; Zhang, W. A distinct strain of Arsenophonus symbiont decreases insecticide resistance in its insect host. PLoS Genet. 2018, 14, e1007725. [Google Scholar] [CrossRef] [PubMed]
- Meng, F.; Yang, M.; Li, Y.; Li, T.; Liu, X.; Wang, G.; Wang, Z.; Jin, X.; Li, W. Functional analysis of RNA interference-related soybean pod borer (Lepidoptera) genes based on transcriptome sequences. Front. Physiol. 2018, 9, 383. [Google Scholar] [CrossRef]
- Arseneau, J.R.; Steeves, R.; Laflamme, M. Modified low-salt CTAB extraction of high-quality DNA from contaminant-rich tissues. Mol. Ecol. Resour. 2017, 17, 686–693. [Google Scholar] [CrossRef]
- Anholt, R.R.; Dilda, C.L.; Chang, S.; Fanara, J.-J.; Kulkarni, N.H.; Ganguly, I.; Rollmann, S.M.; Kamdar, K.P.; Mackay, T.F. The genetic architecture of odor-guided behavior in Drosophila: Epistasis and the transcriptome. Nat. Genet. 2003, 35, 180–184. [Google Scholar] [CrossRef]
- Vera, J.C.; Wheat, C.W.; Fescemyer, H.W.; Frilander, M.J.; Crawford, D.L.; Hanski, I.; Marden, J.H. Rapid transcriptome characterization for a nonmodel organism using 454 pyrosequencing. Mol. Ecol. 2008, 17, 1636–1647. [Google Scholar] [CrossRef]
- Malone, J.H.; Oliver, B. Microarrays, deep sequencing and the true measure of the transcriptome. BMC Biol. 2011, 9, 34. [Google Scholar] [CrossRef]
- Zhao, P.; Xue, H.; Zhu, X.; Wang, L.; Zhang, K.; Li, D.; Ji, J.; Niu, L.; Gao, X.; Luo, J. Silencing of cytochrome P450 gene CYP321A1 effects tannin detoxification and metabolism in Spodoptera litura. Int. J. Biol. Macromol. 2022, 194, 895–902. [Google Scholar] [CrossRef] [PubMed]
- Elzaki, M.E.A.; Li, Z.-F.; Wang, J.; Xu, L.; Liu, N.; Zeng, R.-S.; Song, Y.-Y. Activiation of the nitric oxide cycle by citrulline and arginine restores susceptibility of resistant brown planthoppers to the insecticide imidacloprid. J. Hazard. Mater. 2020, 396, 122755. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Feng, W.; Ye, J.; Li, Z.; Zhou, G. Metabolomic Changes in Sogatella furcifera under Southern rice black-streaked dwarf virus Infection and Temperature Stress. Viruses 2018, 10, 344. [Google Scholar] [CrossRef] [PubMed]
- Kumar, D.; Bansal, G.; Narang, A.; Basak, T.; Abbas, T.; Dash, D. Integrating transcriptome and proteome profiling: Strategies and applications. Proteomics 2016, 16, 2533–2544. [Google Scholar] [CrossRef] [PubMed]
- Hasin, Y.; Seldin, M.; Lusis, A. Multi-omics approaches to disease. Genome Biol. 2017, 18, 83. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Chen, Q.; Tan, Y.; Shuang, S.; Dai, R.; Jiang, X.; Temuer, B. Combined transcriptome and metabolome analysis of alfalfa response to thrips infection. Genes. 2021, 12, 1967. [Google Scholar] [CrossRef] [PubMed]
- Wojda, I.; Cytryńska, M.; Zdybicka-Barabas, A.; Kordaczuk, J. Insect defense proteins and peptides. In Vertebrate and Invertebrate Respiratory Proteins, Lipoproteins and Other Body Fluid Proteins; Springer: Berlin/Heidelberg, Germany, 2020; pp. 81–121. [Google Scholar]
- Merkling, S.H.; Lambrechts, L. Taking insect immunity to the single-cell level. Trends Immunol. 2020, 41, 190–199. [Google Scholar] [CrossRef]
- Gassmann, A.J.; Carrière, Y.; Tabashnik, B.E. Fitness costs of insect resistance to Bacillus thuringiensis. Annu. Rev. Entomol. 2009, 54, 147–163. [Google Scholar] [CrossRef]
- Müller, L.; Soares, G.C.; Josende, M.E.; Monserrat, J.M.; Ventura-Lima, J. Comparison of the toxic effects of organic and inorganic arsenic in Caenorhabditis elegans using a multigenerational approach. Toxicol. Res. 2022, 11, 402–416. [Google Scholar] [CrossRef]
- Bk, S.K.; Moural, T.; Zhu, F. Functional and structural diversity of insect glutathione S-transferases in xenobiotic adaptation. Int. J. Biol. Sci. 2022, 18, 5713. [Google Scholar]
- Gao, H.; Lin, X.; Yang, B.; Liu, Z. The roles of GSTs in fipronil resistance in Nilaparvata lugens: Over-expression and expression induction. Pestic. Biochem. Physiol. 2021, 177, 104880. [Google Scholar] [CrossRef] [PubMed]
- Pavlidi, N.; Vontas, J.; Van Leeuwen, T. The role of glutathione S-transferases (GSTs) in insecticide resistance in crop pests and disease vectors. Curr. Opin. Insect Sci. 2018, 27, 97–102. [Google Scholar] [CrossRef]
- Li, Y.; Sun, H.; Tian, Z.; Su, X.; Li, Y.; Ye, X.; Zhou, Y.; Zheng, S.; Liu, J.; Zhang, Y. The determination of Plutella xylostella (L.) GSTs (PxGSTs) involved in the detoxification metabolism of Tolfenpyrad. Pest Manag. Sci. 2020, 76, 4036–4045. [Google Scholar] [CrossRef] [PubMed]
- Tower, J. Heat shock proteins and Drosophila aging. Exp. Gerontol. 2011, 46, 355–362. [Google Scholar] [CrossRef] [PubMed]
- Punyavathi; Manjunatha, H. Comprehensive analysis of differentially expressed proteins in the male and female Bombyx mori larval instars exposed to thermal stress. Arch. Insect Biochem. Physiol. 2020, 105, e21719. [Google Scholar]
- Li, J.; Moghaddam, S.H.H.; Du, X.; Zhong, B.-X.; Chen, Y.-Y. Comparative analysis on the expression of inducible HSPs in the silkworm, Bombyx mori. Mol. Biol. Rep. 2012, 39, 3915–3923. [Google Scholar] [CrossRef]
- Barman, M.; Samanta, S.; Ahmed, B.; Dey, S.; Chakraborty, S.; Deeksha, M.; Dutta, S.; Samanta, A.; Tarafdar, J.; Roy, D. Transcription dynamics of heat-shock proteins (Hsps) and endosymbiont titres in response to thermal stress in whitefly, Bemisia tabaci (Asia-I). Front. Physiol. 2023, 13, 2762. [Google Scholar] [CrossRef]
- Berenbaum, M.R.; Bush, D.S.; Liao, L.-H. Cytochrome P450-mediated mycotoxin metabolism by plant-feeding insects. Curr. Opin. Insect Sci. 2021, 43, 85–91. [Google Scholar] [CrossRef]
- Scott, J.G.; Wen, Z. Cytochromes P450 of insects: The tip of the iceberg. Pest Manag. Sci. 2001, 57, 958–967. [Google Scholar] [CrossRef]
- Le Goff, G.; Hilliou, F. Resistance evolution in Drosophila: The case of CYP6G1. Pest Manag. Sci. 2017, 73, 493–499. [Google Scholar] [CrossRef] [PubMed]
- Nauen, R.; Bass, C.; Feyereisen, R.; Vontas, J. The role of cytochrome P450s in insect toxicology and resistance. Annu. Rev. Entomol. 2022, 67, 105–124. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fei, H.; Cui, J.; Zhu, S.; Xia, Y.; Xing, Y.; Gao, Y.; Shi, S. Integrative Analyses of Transcriptomics and Metabolomics in Immune Response of Leguminivora glycinivorella Mats to Beauveria bassiana Infection. Insects 2024, 15, 126. https://doi.org/10.3390/insects15020126
Fei H, Cui J, Zhu S, Xia Y, Xing Y, Gao Y, Shi S. Integrative Analyses of Transcriptomics and Metabolomics in Immune Response of Leguminivora glycinivorella Mats to Beauveria bassiana Infection. Insects. 2024; 15(2):126. https://doi.org/10.3390/insects15020126
Chicago/Turabian StyleFei, Hongqiang, Juan Cui, Shiyu Zhu, Ye Xia, Yichang Xing, Yu Gao, and Shusen Shi. 2024. "Integrative Analyses of Transcriptomics and Metabolomics in Immune Response of Leguminivora glycinivorella Mats to Beauveria bassiana Infection" Insects 15, no. 2: 126. https://doi.org/10.3390/insects15020126
APA StyleFei, H., Cui, J., Zhu, S., Xia, Y., Xing, Y., Gao, Y., & Shi, S. (2024). Integrative Analyses of Transcriptomics and Metabolomics in Immune Response of Leguminivora glycinivorella Mats to Beauveria bassiana Infection. Insects, 15(2), 126. https://doi.org/10.3390/insects15020126





