De Novo Genome Assembly at Chromosome-Scale of Hermetia illucens (Diptera Stratiomyidae) via PacBio and Omni-C Proximity Ligation Technology
Abstract
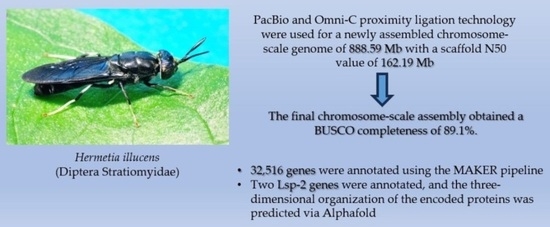
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Hermetia Illucens Mass Rearing
2.2. Genome Library Construction and Sequencing
2.3. Genome Assembly
2.4. Genome and Gene Prediction Quality Evaluation
2.5. Genome Visualization and Analysis of the Chromatin Organization
2.6. Genome Annotation
3. Results
3.1. Hermetia Illucens Rearing and Crossing
3.2. Genome Library Construction and Sequencing and Genome Assembly
3.3. Genome Quality Evaluation
3.4. Genome Visualization and Analysis of the Chromatin Organization
3.5. Genome Annotation
3.6. Lsp-2 Annotation
4. Discussion
4.1. Comparative Genome Assembly Analysis
4.2. Structural Organization of the Genome
4.3. Genome Annotation
4.4. Lsp-2
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Spranghers, T.; Noyez, A.; Schildermans, K.; Clercq, P.D. Cold Hardiness of the Black Soldier Fly (Diptera: Stratiomyidae). J. Econ. Entomol. 2017, 110, 7–1501. [Google Scholar] [CrossRef]
- Silva, G.D.P.; Hesselberg, T. A Review of the Use of Black Soldier Fly Larvae, Hermetia illucens (Diptera: Stratiomyidae), to Compost Organic Waste in Tropical Regions. Neotrop. Entomol. 2020, 49, 62–151. [Google Scholar] [CrossRef]
- Liu, C.; Wang, C.; Yao, H. Comprehensive Resource Utilization of Waste Using the Black Soldier Fly (Hermetia illucens (L.)) (Diptera: Stratiomyidae). Animals 2019, 9, 349. [Google Scholar] [CrossRef]
- Van Huis, A.; Van Itterbeeck, J.; Klunder, H.; Mertens, E.; Halloran, A.; Muir, G.; Vantomme, P. Edible Insects: Future Prospects for Food and Feed Security (No. 171); Food and Agriculture Organization of the United Nations: Rome, Italy, 2013. [Google Scholar]
- Choi, Y.C.; Choi, J.Y.; Kim, J.G.; Kim, M.S.; Kim, W.T.; Park, K.H.; Bae, S.W.; Jeong, G.S. Potential Usage of Food Waste as a Natural Fertilizer after Digestion by Hermetia illucens (Diptera: Stratiomyidae). Int. J. Indust. Entomol. 2009, 19, 4–171. [Google Scholar]
- Franco, A.; Scieuzo, C.; Salvia, R.; Petrone, A.M.; Tafi, E.; Moretta, A.; Schmitt, E.; Falabella, P. Lipids from Hermetia illucens, an Innovative and Sustainable Source. Sustainability 2021, 13, 10198. [Google Scholar] [CrossRef]
- Leong, S.Y.; Kutty, S.R.M.; Malakahmad, A.; Tan, C.K. Feasibility study of biodiesel production using lipids of Hermetia illucens larva fed with organic waste. Waste Manag. 2016, 47, 84–90. [Google Scholar] [CrossRef]
- Lee, A.F.; Bennett, J.A.; Manayil, J.C.; Wilson, K. Heterogeneous catalysis for sustainable biodiesel production via esterification and transesterification. Chem. Soc. Rev. 2014, 43, 916–7887. [Google Scholar] [CrossRef]
- Jayanegara, A.; Haryati, R.P.; Nafisah, A.; Suptijah, P.; Ridla, M.; Laconi, E.B. Derivatization of Chitin and Chitosan from Black Soldier Fly (Hermetia illucens) and Their Use as Feed Additives: An In vitro Study. Adv. Anim. Vet. Sci. 2020, 8, 472–477. [Google Scholar] [CrossRef]
- Kumar, M.N.V.R. A review of chitin and chitosan applications. React. Funct. Polym. 2000, 46, 1–27. [Google Scholar] [CrossRef]
- Elhag, O.; Zhou, D.; Song, Q.; Soomro, A.A.; Cai, M.; Zheng, L.; Yu, Z.; Zhang, J. Screening, Expression, Purification and Functional Characterization of Novel Antimicrobial Peptide Genes from Hermetia illucens (L.). PLoS ONE 2017, 12, e0169582. [Google Scholar] [CrossRef]
- Xia, J.; Ge, C.; Yao, H. Antimicrobial Peptides from Black Soldier Fly (Hermetia illucens) as Potential Antimicrobial Factors Representing an Alternative to Antibiotics in Livestock Farming. Animals 2021, 11, 1937. [Google Scholar] [CrossRef]
- Moretta, A.; Salvia, R.; Scieuzo, C.; Somma, A.D.; Vogel, H.; Pucci, P.; Sgambato, A.; Wolff, M.; Falabella, P. A bioinformatic study of antimicrobial peptides identified in the Black Soldier Fly (BSF) Hermetia illucens (Diptera: Stratiomyidae). Sci. Rep. 2020, 10, 16875. [Google Scholar] [CrossRef]
- Erickson, M.C.; Islam, M.; Sheppa, C.; Liao, J.; Doyle, M.P. Reduction of Escherichia coli O157:H7 and Salmonella enterica Serovar Enteritidis in Chicken Manure by Larvae of the Black Soldier Fly. J. Food Prot. 2016, 67, 90–685. [Google Scholar] [CrossRef]
- Liu, Q.; Tomberlin, J.K.; Brady, J.A.; Sanford, M.R.; Yu, Z. Black Soldier Fly (Diptera: Stratiomyidae) Larvae Reduce Escherichia coli in Dairy Manure. Environ. Entomol. 2008, 37, 30–1525. [Google Scholar] [CrossRef]
- Barragan-Fonseca, K.B.; Dicke, M.; Van Loon, J.J.A. Nutritional value of the black soldier fly (Hermetia illucens L.) and its suitability as animal feed—A review. J. Insects Food Feed. 2017, 3, 20–105. [Google Scholar] [CrossRef]
- Wang, Y.S.; Shelomi, M. Review of Black Soldier Fly (Hermetia illucens) as Animal Feed and Human Food. Foods 2017, 6, 91. [Google Scholar] [CrossRef]
- Generalovic, T.N.; McCarthy, S.A.; Warren, I.A.; Wood, J.M.D.; Torrance, J.; Sims, Y.; Quail, M.; Howe, K.; Pipan, M.; Durbin, R.; et al. A high-quality, chromosome-level genome assembly of the Black Soldier Fly (Hermetia illucens L.). G3 2021, 11, jkab085. [Google Scholar] [CrossRef]
- Burton, J.N.; Adey, A.; Patwardhan, R.P.; Qiu, R.; Kitzman, J.O.; Shendure, J. Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions. Nat. Biotechnol. 2013, 31, 25–1119. [Google Scholar] [CrossRef]
- Zhan, S.; Fang, G.; Cai, M.; Kou, Z.; Xu, J.; Cao, Y.; Bai, L.; Zhang, Y.; Jiang, Y.; Luo, X.; et al. Genomic landscape and genetic manipulation of the black soldier fly Hermetia illucens, a natural waste recycler. Cell Res. 2020, 30, 50–60. [Google Scholar] [CrossRef]
- Inaki, M.; Yoshikawa, S.; Thomas, J.B.; Aburatani, H.; Nose, A. Wnt4 Is a Local Repulsive Cue that Determines Synaptic Target Specificity. Curr. Biol. 2007, 17, 9–1574. [Google Scholar] [CrossRef]
- Akam, M.E.; Roberts, D.B.; Wolfe, J. Drosophila hemolymph proteins: Purification, characterization, and genetic mapping of larval serum protein 2 in D. melanogaster. Biochem. Genet. 1978, 16, 19–101. [Google Scholar] [CrossRef]
- Beneš, H.; Edmondson, R.G.; Fink, P.; Kejzlarová-Lepesant, J.; Lepesant, J.A.; Miles, J.P.; Spivey, D.W. Adult expression of the Drosophila Lsp-2 gene. Dev. Biol. 1990, 142, 46–138. [Google Scholar] [CrossRef]
- Powell, D.; Sato, J.D.; Brock, H.W.; Roberts, D.B. Regulation of synthesis of the larval serum proteins of Drosophila melanogaster. Dev. Biol. 1984, 102, 15–206. [Google Scholar] [CrossRef]
- Cheng, H.; Concepcion, G.T.; Feng, X.; Zhang, H.; Li, H. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat. Methods 2021, 18, 5–170. [Google Scholar] [CrossRef]
- Laetsch, D.R.; Blaxter, M.L. BlobTools: Interrogation of genome assemblies. F1000Research 2017, 6, 1287. [Google Scholar] [CrossRef]
- Guan, D.; McCarthy, S.A.; Wood, J.; Howe, K.; Wang, Y.; Durbin, R. Identifying and removing haplotypic duplication in primary genome assemblies. Bioinformatics 2020, 36, 8–2896. [Google Scholar] [CrossRef]
- Putnam, N.H.; O’Connell, B.L.; Stites, J.C.; Rice, B.J.; Blanchette, M.; Calef, R.; Troll, C.J.; Fields, A.; Hartley, P.D.; Sugnet, C.W.; et al. Chromosome-scale shotgun assembly using an in vitro method for long-range linkage. Genome Res. 2016, 26, 50–342. [Google Scholar] [CrossRef]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 2–3210. [Google Scholar] [CrossRef]
- Kerpedjiev, P.; Abdennur, N.; Lekschas, F.; McCallum, C.; Dinkla, K.; Strobelt, H.; Luber, J.M.; Ouellette, S.B.; Azhir, A.; Kumar, N.; et al. HiGlass: Web-based visual exploration and analysis of genome interaction maps. Genome Biol. 2018, 19, 125. [Google Scholar] [CrossRef]
- Durand, N.C.; Shamim, M.S.; Machol, I.; Rao, S.S.P.; Huntley, M.H.; Lander, E.S.; Aiden, E.L. Juicer Provides a One-Click System for Analyzing Loop-Resolution Hi-C Experiments. Cell Syst. 2016, 3, 8–95. [Google Scholar] [CrossRef]
- Oliver, J.L.; Carpena, P.; Hackenberg, M.; Bernaola-Galván, P. IsoFinder: Computational prediction of isochores in genome sequences. Nucleic Acids Res. 2004, 32, 92–287. [Google Scholar] [CrossRef]
- Stanke, M.; Morgenstern, B. AUGUSTUS: A web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res. 2005, 33, 7–465. [Google Scholar] [CrossRef]
- Hoff, K.J.; Stanke, M. WebAUGUSTUS—A web service for training AUGUSTUS and predicting genes in eukaryotes. Nucleic Acids Res. 2013, 41, 8–123. [Google Scholar] [CrossRef]
- Stanke, M.; Diekhans, M.; Baertsch, R.; Haussler, D. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics 2008, 24, 44–637. [Google Scholar] [CrossRef]
- Smit, A.F.A.; Hubley, R. RepeatModeler Open-1.0. Available online: http://www.repeatmasker.org (accessed on 30 July 2022).
- Smit, A.F.A.; Hubley, R.; Green, P. RepeatMasker Open-4.0. Available online: http://www.repeatmasker.org (accessed on 30 July 2022).
- Chan, P.P.; Lin, B.Y.; Mak, A.J.; Lowe, T.M. tRNAscan-SE 2.0: Improved detection and functional classification of transfer RNA genes. Nucleic Acids Res. 2021, 49, gkab688. [Google Scholar] [CrossRef]
- Korf, I. Gene finding in novel genomes. BMC Bioinform. 2004, 5, 59. [Google Scholar] [CrossRef]
- Vogel, H.; Müller, A.; Heckel, D.G.; Gutzeit, H.; Vilcinskas, A. Nutritional immunology: Diversification and diet-dependent expression of antimicrobial peptides in the black soldier fly Hermetia illucens. Dev. Comp. Immunol. 2018, 78, 8–141. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Cantarel, B.L.; Korf, I.; Robb, S.M.C.; Parra, G.; Ross, E.; Moore, B.; Holt, C.; Alvarado, A.S.; Yandell, M. MAKER: An easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 2008, 18, 96–188. [Google Scholar] [CrossRef]
- Boeckmann, B.; Bairoch, A.; Apweiler, R.; Blatter, M.C.; Estreicher, A.; Gasteiger, E.; Martin, M.J.; Michoud, K.; O’Donovan, C.; Phan, I.; et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003, 31, 70–365. [Google Scholar] [CrossRef]
- Consortium, T.U.; Bateman, A.; Martin, M.J.; Orchard, S.; Magrane, M.; Agivetova, R.; Ahmad, S.; Alpi, E.; Bowler-Barnett, E.H.; Britto, R.; et al. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2020, 49, 9–480. [Google Scholar]
- Marchler-Bauer, A.; Bryant, S.H. CD-Search: Protein domain annotations on the fly. Nucleic Acids Res. 2004, 32, 31–327. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 10–403. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 9–583. [Google Scholar] [CrossRef]
- Mirdita, M.; Schütze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making protein folding accessible to all. Nat. Methods 2022, 19, 82–679. [Google Scholar] [CrossRef]
- Wang, J.; Youkharibache, P.; Zhang, D.; Lanczycki, C.J.; Geer, R.C.; Madej, T.; Phan, L.; Ward, M.; Lu, S.; Marchler, G.H.; et al. iCn3D, a Web-based 3D Viewer for Sharing 1D/2D/3D Representations of Biomolecular Structures. Bioinformatics 2019, 36, 5–131. [Google Scholar] [CrossRef]
- Wang, J.; Youkharibache, P.; Marchler-Bauer, A.; Lanczycki, C.; Zhang, D.; Lu, S.; Madej, T.; Marchler, G.H.; Cheng, T.; Chong, L.C.; et al. iCn3D: From Web-Based 3D Viewer to Structural Analysis Tool in Batch Mode. Front. Mol. Biosci. 2022, 9, 831740. [Google Scholar] [CrossRef]
- Vicoso, B.; Bachtrog, D. Reversal of an ancient sex chromosome to an autosome in Drosophila. Nature 2013, 499, 332–335. [Google Scholar] [CrossRef]
- Cammarano, R.; Costantini, M.; Bernardi, G. The isochore patterns of invertebrate genomes. BMC Genomics 2009, 10, 538. [Google Scholar] [CrossRef]
- Fujita, M.K.; Edwards, S.V.; Ponting, C.P. The Anolis Lizard Genome: An Amniote Genome without Isochores. Genome Biol. Evol. 2011, 3, 84–974. [Google Scholar] [CrossRef]
- Jørgensen, F.G.; Schierup, M.H.; Clark, A.G. Heterogeneity in Regional GC Content and Differential Usage of Codons and Amino Acids in GC-Poor and GC-Rich Regions of the Genome of Apis mellifera. Mol. Biol. Evol. 2007, 24, 9–611. [Google Scholar] [CrossRef]
- Gramates, L.S.; Agapite, J.; Attrill, H.; Calvi, B.R.; Crosby, M.A.; Dos Santos, G.; Goodman, J.L.; Goutte-Gattat, D.; Jenkins, V.K.; Kaufman, T.; et al. FlyBase: A guided tour of highlighted features. Genetics 2022, 220, iyac035. [Google Scholar] [CrossRef]
- Burmester, T.; Masse, H.C., Jr.; Zakharkin, S.O.; Benes, H. The Evolution of Hexamerins and the Phylogeny of Insects. J. Mol. Evol. 1998, 47, 108–193. [Google Scholar] [CrossRef]
- Beneš, H.; Neal, K.C.; Willis, R.L.; Gadde, D.; Castleberry, A.B.; Korochkina, S.E. Overlapping Lsp-2 gene sequences target expression to both the larval and adult Drosophila fat body. Insect Mol. Biol. 1996, 5, 39–49. [Google Scholar] [CrossRef]
- Mousseron-Grall, S.; Kejzlarová-Lepesant, J.; Burmester, T.; Chihara, C.; Barray, M.; Delain, E.; Pictet, R.; Lepesant, J.A. Sequence, Structure and Evolution of the Ecdysone-Inducible Lsp-2 Gene of Drosophila Melanogaster. Eur. J. Biochem. 1997, 245, 8–191. [Google Scholar] [CrossRef]
- Xu, J.; Zhang, Y. How significant is a protein structure similarity with TM-score = 0.5? Bioinformatics 2010, 26, 95–889. [Google Scholar] [CrossRef]









| Assembly | Total Length (bp) | N50 (Mb) | L50 | Scaffold Number |
|---|---|---|---|---|
| Initial draft assembly | 1,311,107,399 | 3,793,743 | 78 | 4762 |
| Primary filtered assembly | 888,493,946 | 5,340,478 | 44 | 1185 |
| Final Assembly | 888,595,941 | 162,194,137 | 3 | 169 |
| Genome Annotation Statistics | Number |
|---|---|
| Total number of genes | 32,516 |
| Total coding region (bp) | 36,672,320 |
| Average length of genes (bp) | 1127.82 |
| Number of single-exon genes | 2091 |
| Species Name | Scaffold Number | N50 Value (Mb) | BUSCO % | ||||
|---|---|---|---|---|---|---|---|
| C | S | D | F | M | |||
| Final assembly | 169 | 162.19 | 89.1 | 87.0 | 1.2 | 1.1 | 9.8 |
| Hermetia illucens (reference genome) | 20 | 180.36 | 98.6 | 97.8 | 0.8 | 0.5 | 0.9 |
| Hermetia illucens (GCA_009835165.1) | 2806 | 1.70 | 98.9 | 91.1 | 7.8 | 0.6 | 0.5 |
| Drosophila melanogaster (GCA_000001215.4) | 1870 | 25.29 | 99.7 | 99.0 | 0.7 | 0.2 | 0.1 |
| Drosophila virilis (GCA_000005245.1) | 13,530 | 31.08 | 99.1 | 98.1 | 1.0 | 0.4 | 0.5 |
| Musca domestica (GCA_000371365.1) | 20,487 | 0.23 | 98.6 | 96.9 | 1.7 | 0.4 | 1.0 |
| Stomoxys calcitrans (GCA_001015335.1) | 12,042 | 0.50 | 98.4 | 97.7 | 0.7 | 1.0 | 0.5 |
| Glossina morsitans (GCA_001077435.1) | 24,071 | / | 98.9 | 96.6 | 2.3 | 0.6 | 0.5 |
| Aedes aegypti (GCA_002204515.1) | 2310 | 0.41 | 98.9 | 94.5 | 4.4 | 0.4 | 0.7 |
| Culex quinquefasciatus (GCA_000209185.1) | 3171 | 0.49 | 96.7 | 91.8 | 4.9 | 0.8 | 2.5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Costagli, S.; Abenaim, L.; Rosini, G.; Conti, B.; Giovannoni, R. De Novo Genome Assembly at Chromosome-Scale of Hermetia illucens (Diptera Stratiomyidae) via PacBio and Omni-C Proximity Ligation Technology. Insects 2024, 15, 133. https://doi.org/10.3390/insects15020133
Costagli S, Abenaim L, Rosini G, Conti B, Giovannoni R. De Novo Genome Assembly at Chromosome-Scale of Hermetia illucens (Diptera Stratiomyidae) via PacBio and Omni-C Proximity Ligation Technology. Insects. 2024; 15(2):133. https://doi.org/10.3390/insects15020133
Chicago/Turabian StyleCostagli, Simone, Linda Abenaim, Giulia Rosini, Barbara Conti, and Roberto Giovannoni. 2024. "De Novo Genome Assembly at Chromosome-Scale of Hermetia illucens (Diptera Stratiomyidae) via PacBio and Omni-C Proximity Ligation Technology" Insects 15, no. 2: 133. https://doi.org/10.3390/insects15020133
APA StyleCostagli, S., Abenaim, L., Rosini, G., Conti, B., & Giovannoni, R. (2024). De Novo Genome Assembly at Chromosome-Scale of Hermetia illucens (Diptera Stratiomyidae) via PacBio and Omni-C Proximity Ligation Technology. Insects, 15(2), 133. https://doi.org/10.3390/insects15020133