Simple Summary
The ubiquitin system plays an important role in defense against viruses. In this study, we cloned and characterized the polyubiquitin-C (UBC) gene in Laodelphax striatellus. Expression profile analysis showed that LsUBC was highly expressed in the salivary glands, midgut, and reproductive system. Moreover, the expression of LsUBC was upregulated during viral infection, and inhibition of LsUBC expression increased viral accumulation. These results indicated that LsUBC plays a vital role in inhibiting viral accumulation in L. striatellus.
Abstract
Many hosts utilize the ubiquitin system to defend against viral infection. As a key subunit of the ubiquitin system, the role of polyubiquitin in the viral infection of insects is unclear. Here, we identified the full-length cDNA of the polyubiquitin-C (UBC) gene in Laodelphax striatellus, the small brown planthopper (SBPH). LsUBC was expressed in various tissues and was highly expressed in salivary glands, midgut, and reproductive systems. Furthermore, the LsUBC expression profiles in the developmental stages showed that LsUBC was ubiquitously expressed in seven developmental stages and was highest expressed in female adults with SBPH. qRT-PCR analyses indicated that rice stripe virus (RSV) infection promoted the LsUBC expression. Knockdown of LsUBC mRNA via RNA interference increased RSV accumulation. These findings suggest that LsUBC inhibits RSV accumulation in L. striatellus.
1. Introduction
Ubiquitin is a family of highly conserved proteins in eukaryotic organisms and is involved in the regulation of many cellular events [1,2]. In the canonical function of ubiquitin, the C-terminal glycine residue of ubiquitin can be linked to the epsilon-amino group of a lysine residue in substrate proteins through the action of ubiquitin-protein ligase [3,4]. Genes encoding ubiquitin can be divided into monomeric and polymeric ubiquitin genes. The monomeric can be fused to other carboxyl-extension protein-coding regions. The polyubiquitin genes contain multiple tandem ubiquitin repeat regions with no spacer sequences between monomer units [5]. Polyubiquitin-C (UBC) is one of the sources of ubiquitin and is a stress-regulated polyubiquitin gene. UBC proteins play an essential role in maintaining ubiquitin levels and cellular homeostasis under stress conditions in mammals [6,7]. Unanchored ubiquitin chains are reported to connect the proteasome and autophagy to eliminate abnormal or foreign protein accumulation [8].
Numerous hosts (e.g., plants, insects, and mammals) utilize the ubiquitin system to defend against viral infection. For example, ubiquitin-like protein 5 directly interacted with rice stripe virus (RSV) P3 protein to mediate the viral P3 degradation through the 26S proteasome system in plants [9]. Also, ubiquitin-proteasome system could decrease the replication of the Turnip yellow mosaic virus by regulating the accumulation of viral protein [10]. Conversely, viruses have evolved many strategies to exploit the ubiquitin system to evade host immune responses. For example, Tomato yellow leaf curl virus C2 protein interacted with ubiquitin-40S ribosomal protein S27a (RPS27A) to compromise the degradation of JAZ1 protein and inhibit plant jasmonic acid defense [11]. In addition, both rice black-streaked dwarf virus (RBSDV) and RSV interfered with insect 26S proteasome to attenuate plant defense response and facilitated virus accumulation and transmission [12,13]. Similarly, human immunodeficiency virus type 1 (HIV-1) hijacked ubiquitin systems to evade host interferon responses in mammals [14].
The small brown planthopper (SBPH, Laodelphax striatellus) is one of the most serious pests and has caused severe losses in rice production in East Asia. SBPH also acts as a vector for transmission of rice stripe virus (RSV) in a persistent and circulative-propagative manner [15,16,17]. RSV is a typical member of the genus Tenuivirus and contains four single-stranded, negative-sense RNA segments that encode seven viral proteins. RSV particles first establish infection in the midgut epithelium, then spread to various tissues and ultimately reach the salivary glands or ovaries under the transportation of hemolymph [15,18]. Recently, several studies have reported that multiple proteins in the ubiquitin system are involved in viral infection and inhibit virus accumulation in SBPHs [12,13,19]. However, the ubiquitin UBC protein involved in RSV infection has not been identified to date.
In this study, we cloned and characterized the L. striatellus UBC (LsUBC). The expression of LsUBC was upregulated during viral infection. In addition, inhibition of LsUBC expression facilitated RSV accumulation in L. striatellus. These results indicated that LsUBC plays a negative role in mediating viral infection.
2. Materials and Methods
2.1. Virus, Plants and Insects
The RSV-infected rice plants were originally obtained from Jiangsu Province, China, and were maintained in the laboratory. The viruliferous (RSV-infected) and nonviruliferous (RSV-free) SBPHs were reared separately on rice plants inside a growth incubator at 26 °C with 80% relative humidity and a photoperiod of 14 h light and 10 h dark. To ensure a high infection rate, the offspring of viruliferous females were monitored and collected for RT-PCR assay. The infection rate of the viruliferous population used in the experiments was approximately 90%.
2.2. Identification and Phylogenetic Analysis of LsUBC
To identify the putative UBC sequence in L. striatellus, annotated UBC sequences in other insect species were collected and used as a query to compare similar sequences in the SBPH genome [20]. The candidate sequences with high E-values were extracted and confirmed by Blastp search against a non-redundant protein database at the National Center for Biotechnology Information (NCBI). The open reading frame sequences were recovered with the ORF Finder program (https://www.ncbi.nlm.nih.gov/orffinder; accessed on 27 June 2023). The sequence function annotation was analyzed using the NCBI Conserved Domain Database (https://www.ncbi.nlm.nih.gov/cdd/; accessed on 27 June 2023). A phylogenetic tree was obtained by the ML algorithm using RAxML-NG with 1000 bootstrap replications [21].
2.3. Tissue Collection
Adult SBPHs were collected and dissected using tweezers. Each type of tissue sample was obtained from 20 female or male insects. The dissected tissues were washed twice with 1xPBS and stored with the RNAiso Plus solution (Takara, Beijing, China) at –80 °C. Six tissue types were isolated (epidermis, salivary glands, midgut, ovary, testis, and fat body) and three replicates per tissue were collected for expression analysis of the LsUBC gene.
2.4. RNA Extraction and qRT-PCR
Total RNA was extracted from whole insects or tissue samples using RNAiso Plus solution. Total RNA was then transcribed into complementary DNA (cDNA) using the HiScript II Q RT SuperMix for qPCR (+gDNA wiper) (Vazyme, China) following the standard protocols. Quantitative real-time PCR (qRT-PCR) was performed using Hieff qPCR SYBR Green Master Mix (Yeasen, China). The expression level of the LsUBC gene was calculated with the 2−ΔΔCt method and a housekeeping gene (Actin) was amplified as an internal standard. Three biological replicates were carried out for each experiment, and two technical replicates were conducted for each biological replicate.
2.5. Western Blotting
SBPH samples were collected and ground with liquid nitrogen. Total protein samples were then collected from the extraction buffer and treated with 6× SDS loading buffer. After boiled for 10 min, the samples were separated by 10% SDS-PAGE gels and transferred onto polyvinylidene fluoride (PVDF) membranes. The membranes were inoculated with primary antibodies against RSV-NP (1:5000 dilution) or ACTB (1:5000 dilution) followed by probed with horseradish peroxidase (HRP)-conjugated goat anti-mouse antibody at 1:10,000 dilution. The membranes were detected using the Luminescent Image Analyzer AI680 (GE, Danderyd, Sweden).
2.6. RNA Interference (RNAi)
The dsRNAs targeting LsUBC and GFP genes were synthesized using the T7 High Yield Transcription Kit as recommended by the manufacturer (Vazyme, Nanjing, China). The PCR primers with T7 promoter sequences are available in Table S1. Third-instar nymphs were microinjected with 25 nl of dsLsUBC or dsGFP into insect hemolymph using a TransferMan 4r micromanipulator (Eppendorf, Hamburg, Germany). Following microinjection, nymphs were transferred to healthy rice seedlings until used for qRT-PCR or Western blotting analysis.
3. Results
3.1. Identification and Characterization of LsUBC
The sequence of the LsUBC gene was obtained by comparing the SBPH genome with UBC genes from other insect species. This sequence was then compared to the NCBI non-redundant protein database through Blastp search. The result showed that the amino acid sequence of LsUBC was highly conserved with the UBC sequences of other species and shared the highest identity (90%) with the Pimephales promelas UBC. Subsequently, the LsUBC gene was confirmed by PCR amplification and verified by Sanger sequencing.
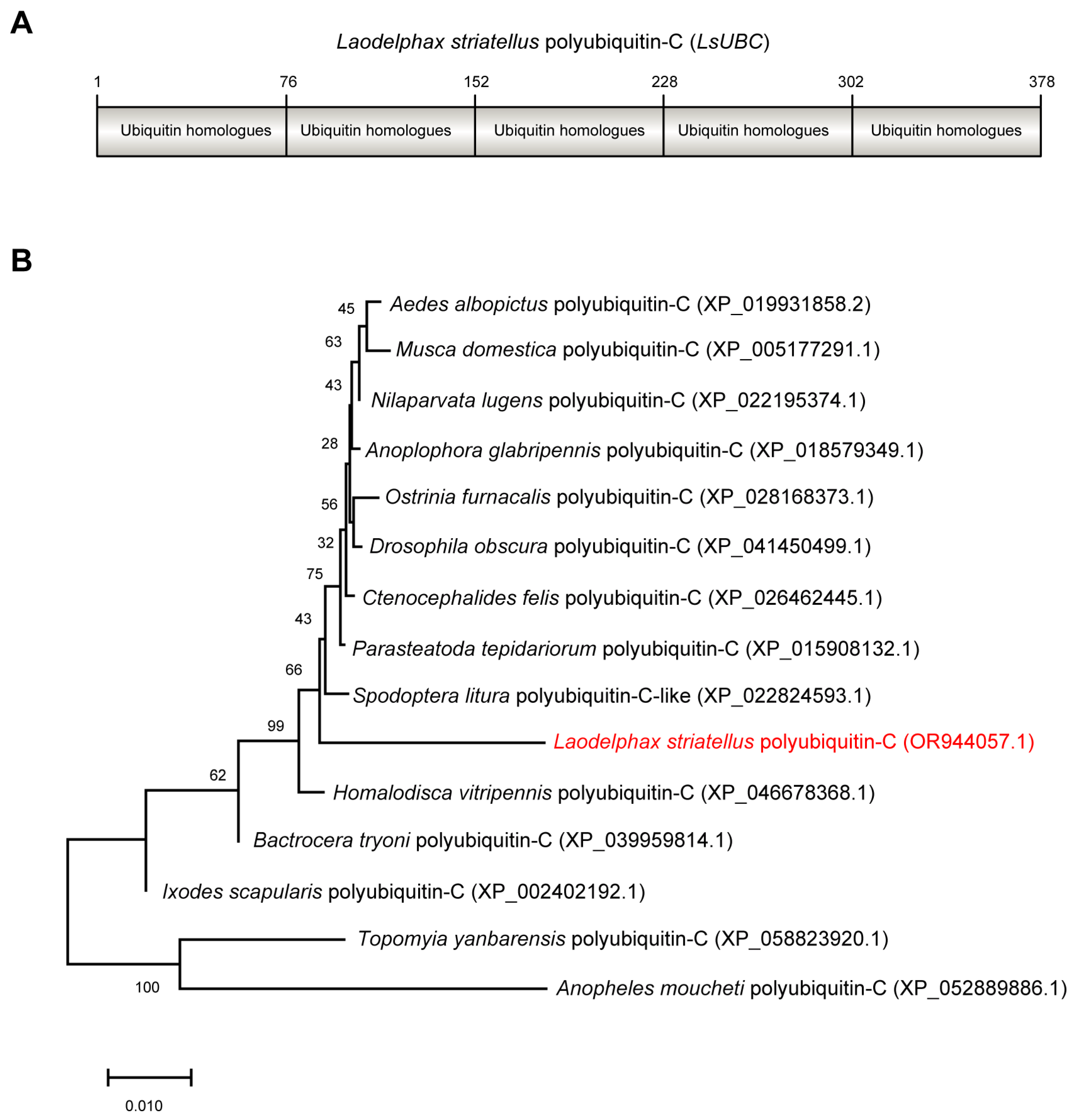
The ORF sequence of LsUBC (GenBank accession number OR944057.1) was 1137 bp in length and encoded 378 amino acids with a calculated molecular mass of 42 kDa. Conserved domain analysis by NCBI Conserved Domain Database search found that LsUBC contained five repeated ubiquitin homologues (UBQ) (Figure 1A). Phylogenetic analysis using amino acid sequences showed that LsUBC clustered with other insect UBCs deposited in the NCBI database with a high branch support (Figure 1B).
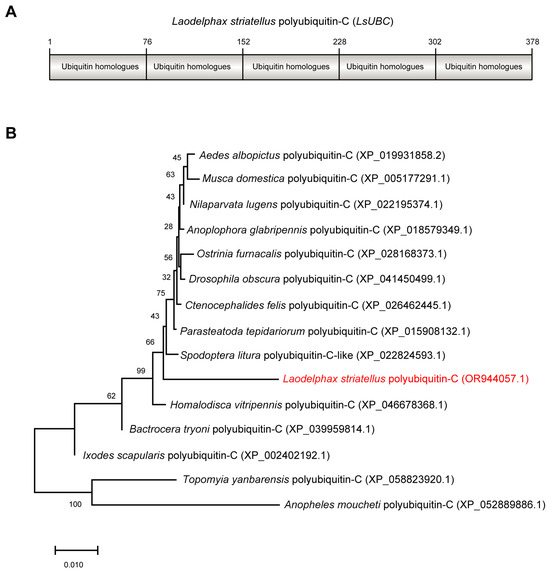
Figure 1.
Sequence characterization and phylogenetic analysis of Laodelphax striatellus polyubiquitin-C (LsUBC) gene. (A) Schematic diagrams showing the amino acid sequence of LsUBC. The position of each single monomeric unit is marked. (B) Phylogenetic analysis using the amino acid sequences of polyubiquitin-C from L. striatellus and other insect species. A phylogenetic tree was obtained by the ML algorithm using RAxML-NG with 1000 bootstrap replications. The position of the LsUBC on the phylogenetic tree is labeled in red font.
Sequence alignment analysis using UBCs from L. striatellus and other insects revealed that these UBCs were highly conserved in each insect species and contained multiple tandem repeating monomeric units (76 amino acids). In addition, several amino acids at the C-terminus of LsUBC are not conserved (Figure 2).

Figure 2.
Sequence alignment of polyubiquitin-C from L. striatellus and other insect species. The amino acid sequences of UBC from the following 16 insect species were aligned: L. striatellus; Sogatella furcifera; Nilaparvata lugens; Ostrinia furnacalis; Homalodisca vitripennis; Aedes albopictus; Drosophila obscura; Musca domestica; Topomyia yanbarensis; Anopheles moucheti; Ctenocephalides felis; Ixodes scapularis; Parasteatoda tepidariorum; Spodoptera litura; Anoplophora glabripennis; Bactrocera tryoni. Residues that are fully conserved are shaded in black, and those that are partially conserved are shown in gray.
3.2. Expression Analysis of LsUBC in SBPHs
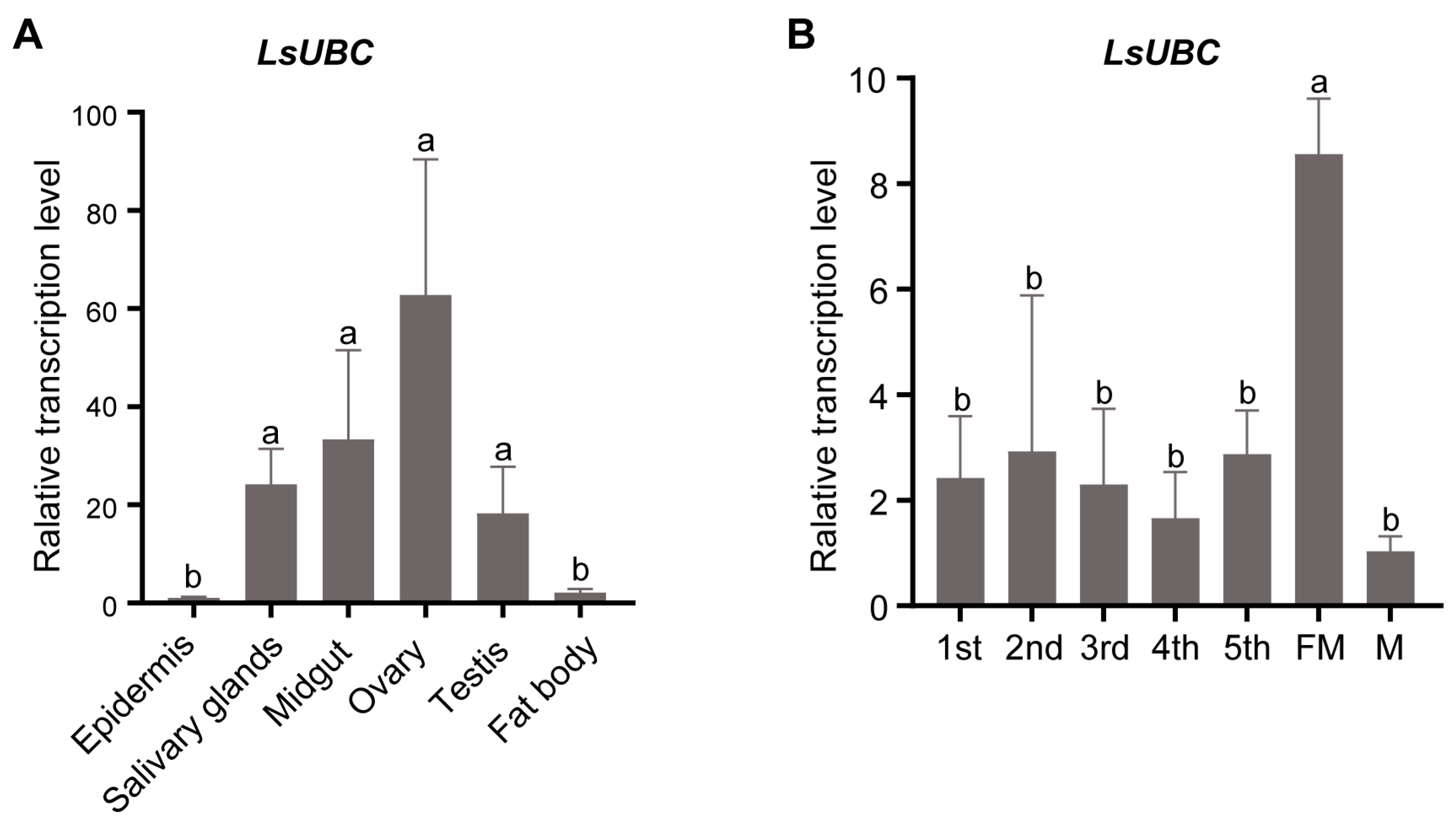
To investigate the detailed expression profiles, we performed a qRT-PCR assay to examine the expression of LsUBC in different tissues and developmental stages of SBPH. The results in Figure 3A showed that LsUBC was expressed in all six tissues. Meanwhile, LsUBC was mainly highly expressed in four tissues (salivary glands, midgut, ovaries, and testis) and most highly expressed in ovaries (Figure 3A). In addition, the LsUBC expression profiles in the seven developmental stages were also detected. The results showed that LsUBC was ubiquitously expressed in seven collected developmental stages of SBPH. The highest transcription level of LsUBC was observed in female adult samples (Figure 3B).
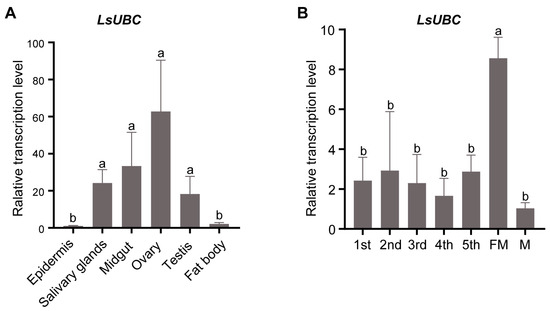
Figure 3.
Analyses of LsUBC expression in different tissues and different developmental stages of L. striatellus. (A) qRT-PCR analysis of LsUBC expression in six different tissues. Similar LsUBC expression levels were found in the epidermis and fat body, whereas high expression was detected in the salivary glands, midgut, ovaries, and testis. Tissues obtained from 50 RSV-free SBPH adults were considered to be a single replicate, and the experiment was performed in three independent replicates. (B) qRT-PCR analysis of LsUBC expression in seven developmental stages. A total of 15 RSV-free SBPHs were considered to be a single replicate, and the experiment contained three replicates. Different letters indicate significant differences in LsUBC expression levels (p < 0.05).
3.3. Rice Stripe Virus Infection Upregulates the Expression of LsUBC in SBPHs
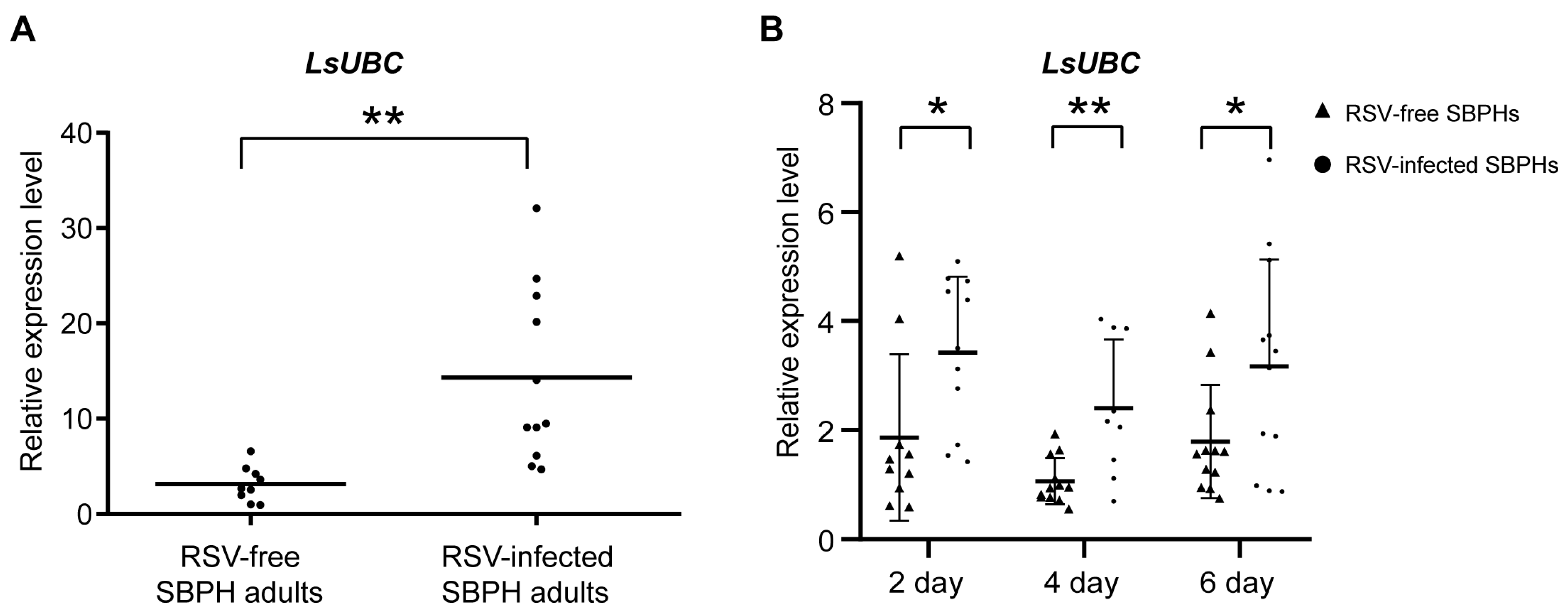
To determine whether the LsUBC was involved in viral infection, we compared the expression of LsUBC between RSV-infected and RSV-free SBPH adults. Compared with the RSV-free SBPHs, the transcript level of the LsUBC was significantly increased in the RSV-infected insect population (Figure 4A). Moreover, we conducted a time course study to detect the expression of LsUBC in SBPHs. After 3 days of feeding on RSV-infected rice plants, SBPH samples were transferred to un-infected rice seedlings and collected at 2, 4, and 6 days post RSV feeding. To eliminate the effect of the insect developmental stage on LsUBC, another group of SBPHs feeding on rice without RSV was collected as a control. Compared with the controls, the transcript level of the LsUBC was higher in RSV-acquired SBPH groups (Figure 4B). These results indicated that RSV infection increases LsUBC expression in SBPHs.
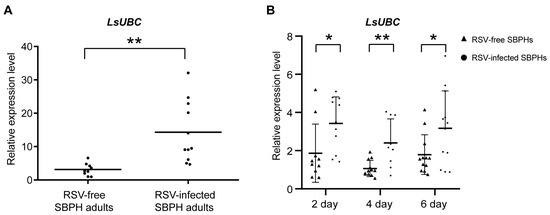
Figure 4.
Analyses of the LsUBC expression during RSV infection in L. striatellus. (A) qRT-PCR analysis of LsUBC expression in RSV-free and RSV-infected SBPH adults. (B) qRT-PCR analysis of LsUBC expression at 2, 4, and 6 days post RSV feeding, respectively. SBPHs feeding on rice without RSV were considered as a control (RSV-free SBPHs). Each dot represents a single insect sample. Experiments were performed on three independent replicates. *, p < 0.05; **, p < 0.01.
3.4. Inhibition of LsUBC Expression Increases RSV Accumulation in SBPHs
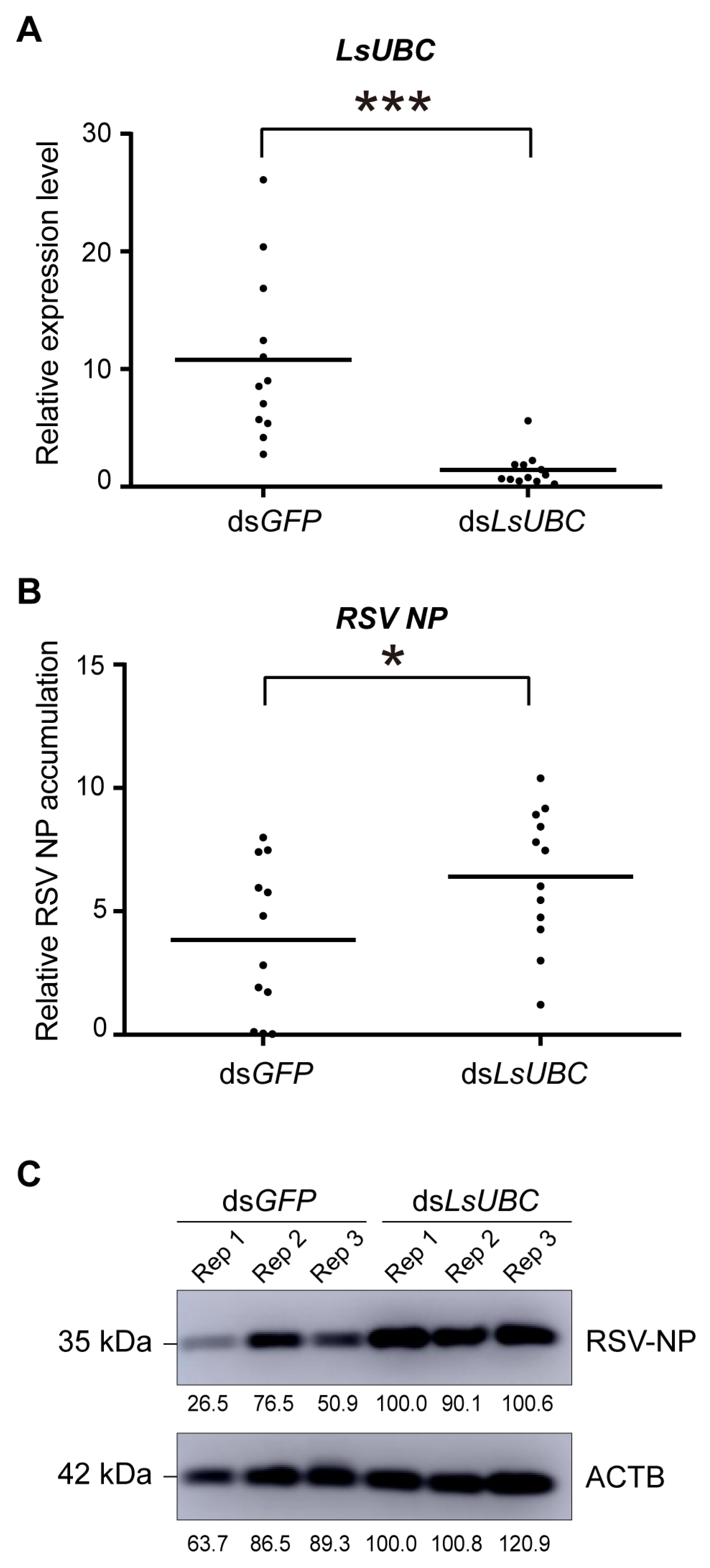
To further explore the effect of LsUBC on RSV accumulation, RSV-infected third-instar nymphs were microinjected with dsRNAs targeting LsUBC (dsLsUBC) or GFP (dsGFP). At two days post microinjection, we first examined the expression of LsUBC. Compared with the dsGFP-treated group, the transcript level of LsUBC was significantly lower in the dsLsUBC-treated group. Further, we examined the accumulation of RSV nucleocapsid protein (NP) at the RNA level and protein level. As shown in Figure 5B, the RNA level of RSV NP was significantly increased in the dsLsUBC-treated SBPHs compared with the dsGFP controls. Similarly, the amount of RSV NP in dsLsUBC-treated SBPHs also obviously increased, as determined by Western blotting assay (Figure 5C). These results suggested that inhibition of LsUBC expression facilitated RSV accumulation in SBPHs.
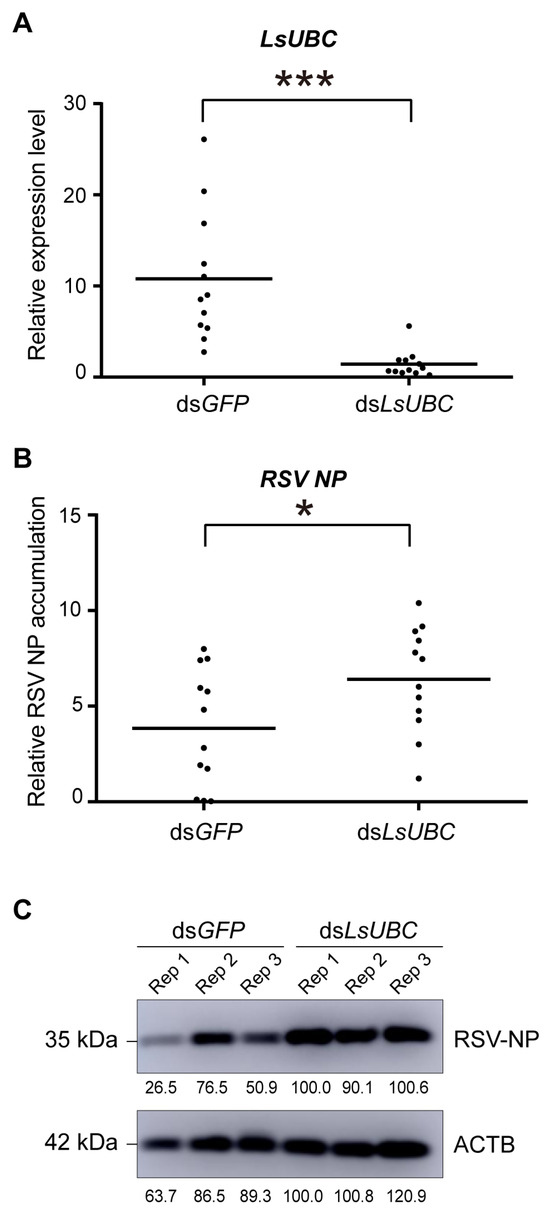
Figure 5.
Effect of LsUBC knockdown on RSV accumulation in L. striatellus. The expression of LsUBC (A) and RSV nucleocapsid protein (NP) (B) in dsGFP- and dsLsUBC-treated RSV-infected SBPHs were determined by qRT-PCR. Each dot represents a single insect sample. Experiments were performed on three independent replicates. *, p < 0.05; ***, p < 0.001. (C) The protein level accumulation of RSV NP in dsGFP- and dsLsUBC-treated RSV-infected SBPHs was detected using a Western blotting assay. ACTB was used as a loading control.
4. Discussion
As a persistently transmitted virus, RSV needs to accumulate in the insect vector. At the same time, numerous antiviral immune systems in insects are activated to inhibit excessive RSV accumulation. Among them, the ubiquitin system plays an important role in resisting viral infection and spread in SBPHs. Here, we characterized a polyubiquitin gene LsUBC and explored its effect on viral infection. The results indicate that LsUBC plays a positive role in inhibiting excessive accumulation of RSV NP in L. striatellus.
Sequence alignment of LsUBC and other insect UBCs revealed these sequences all have highly tandem repeating monomeric units. However, the number of monomeric units varies considerably among insect species. The polyubiquitin gene family was generally thought to be subjected to concerted evolution. Currently, there are two mechanisms for operating the evolution of the UBC gene, namely gene conversion and unequal crossing-over [22,23,24,25]. In C. elegans, and A. thaliana, the monomeric units within the polyubiquitin gene were highly diverged in silent substitution sites. In the evolution of the mouse UBC gene, unequal crossing-over was considered to be the main mechanism of homogenization within the rodent lineage [25]. Whether there is a similar mechanism for the evolution of insect UBC warrants further investigation.
Although expressed in all SBPH tissues, LsUBC is highly expressed in salivary glands, the midgut, and the reproductive system. This tissue distribution is closely related to the process of RSV infection. During the long-term co-evolution of insects and viruses, RSV infection activates the ubiquitin system and protects against RSV accumulation. Other subunits in the ubiquitin system have been reported to be involved in virus accumulation in insect vectors. For example, the silencing of two regulatory particles non-ATPase (RPN3 and RPN8) increased RSV accumulation and viral proteins could hijack the two RPN proteins to facilitate virus accumulation [12,13]. In Culex mosquitoes, West Nile virus infection increased the expression of cullin RING ubiquitin ligase Cullin4. Knockdown of Cullin4 activated the JAK-STAT pathway and inhibited viral replication [26]. Similarly, another Cullin3 protein has also been reported to be a proviral host factor during chikungunya virus infection in Aedes aegypti [27]. We show that the expression of LsUBC increased during RSV infection and repression of LsUBC facilitated RSV accumulation. The molecular mechanism of how LsUBC protein inhibits RSV accumulation remains to be further explored.
In conclusion, the full-length cDNA of LsUBC was obtained and sequenced in L. striatellus. The expression of LsUBC was highly expressed in salivary glands, midgut, and reproductive systems. In addition, LsUBC was highly expressed in RSV-infected SBPHs and inhibited RSV accumulation in L. striatellus. These results indicated that the expression of LsUBC correlated with RSV accumulation in SBPHs. This study provides new insight into the persistent RSV infection in SBPH.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/insects15030149/s1, Supplementary Table S1. Primers used in this study. Figures S1 and S2. Original Images for Western Blots.
Author Contributions
Conceptualization, B.-X.L. and G.L.; methodology, B.-X.L. and Y.-H.Q.; software, B.-X.L.; validation, Y.-H.Q., J.-M.L. and G.L.; formal analysis, B.-X.L.; investigation, B.-X.L.; resources, Y.-H.Q.; data curation, C.-X.Z.; writing—original draft preparation, B.-X.L.; writing—review and editing, G.L.; visualization, B.-X.L.; supervision, J.-P.C.; project administration, G.L.; funding acquisition, G.L. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (32370149) and the Zhejiang Provincial Natural Science Foundation of China (LY24C140001).
Data Availability Statement
The data presented in this study are openly available in GenBank (OR944057.1).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Swatek, K.N.; Komander, D. Ubiquitin modifications. Cell Res. 2016, 26, 399–422. [Google Scholar] [CrossRef] [PubMed]
- Pickart, C.M.; Eddins, M.J. Ubiquitin: Structures, functions, mechanisms. Biochim. Biophys. Acta 2004, 1695, 55–72. [Google Scholar] [CrossRef] [PubMed]
- Cadwell, K.; Coscoy, L. Ubiquitination on nonlysine residues by a viral E3 ubiquitin ligase. Science 2005, 309, 127–130. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Herr, R.A.; Chua, W.J.; Lybarger, L.; Wiertz, E.J.; Hansen, T.H. Ubiquitination of serine, threonine, or lysine residues on the cytoplasmic tail can induce ERAD of MHC-I by viral E3 ligase mK3. J. Cell Biol. 2007, 177, 613–624. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Ye, Y. Polyubiquitin chains: Functions, structures, and mechanisms. Cell Mol. Life Sci. 2008, 65, 2397–2406. [Google Scholar] [CrossRef] [PubMed]
- Ryu, K.Y.; Maehr, R.; Gilchrist, C.A.; Long, M.A.; Bouley, D.M.; Mueller, B.; Ploegh, H.L.; Kopito, R.R. The mouse polyubiquitin gene UbC is essential for fetal liver development, cell-cycle progression and stress tolerance. EMBO J. 2007, 26, 2693–2706. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.N.; Choi, J.; Ryu, H.W.; Ryu, K.Y. Disruption of polyubiquitin gene Ubc leads to attenuated resistance against arsenite-induced toxicity in mouse embryonic fibroblasts. Biochim. Biophys. Acta 2015, 1853, 996–1009. [Google Scholar] [CrossRef]
- Hao, R.; Nanduri, P.; Rao, Y.; Panichelli, R.S.; Ito, A.; Yoshida, M.; Yao, T.P. Proteasomes activate aggresome disassembly and clearance by producing unanchored ubiquitin chains. Mol. Cell 2013, 51, 819–828. [Google Scholar] [CrossRef]
- Chen, B.; Lin, L.; Lu, Y.; Peng, J.; Zheng, H.; Yang, Q.; Rao, S.; Wu, G.; Li, J.; Chen, Z.; et al. Ubiquitin-Like protein 5 interacts with the silencing suppressor p3 of rice stripe virus and mediates its degradation through the 26S proteasome pathway. PLoS Pathog. 2020, 16, e1008780. [Google Scholar] [CrossRef]
- Camborde, L.; Planchais, S.; Tournier, V.; Jakubiec, A.; Drugeon, G.; Lacassagne, E.; Pflieger, S.; Chenon, M.; Jupin, I. The ubiquitin-proteasome system regulates the accumulation of Turnip yellow mosaic virus RNA-dependent RNA polymerase during viral infection. Plant Cell 2010, 22, 3142–3152. [Google Scholar] [CrossRef]
- Li, P.; Liu, C.; Deng, W.H.; Yao, D.M.; Pan, L.L.; Li, Y.Q.; Liu, Y.Q.; Liang, Y.; Zhou, X.P.; Wang, X.W. Plant begomoviruses subvert ubiquitination to suppress plant defenses against insect vectors. PLoS Pathog. 2019, 15, e1007607. [Google Scholar] [CrossRef]
- Li, Y.; Zhu, L.; Gao, J.; Ma, H.; Li, C.; Song, Y.; Zhu, X.; Zhu, C. Silencing suppressors of rice black-streaked dwarf virus and rice stripe virus hijack the 26S proteasome of Laodelphax striatellus to facilitate virus accumulation and transmission. Pest. Manag. Sci. 2022, 78, 2940–2951. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Wu, J.; Fu, S.; Li, C.; Zhu, Z.R.; Zhou, X. Rice stripe tenuivirus nonstructural protein 3 hijacks the 26S proteasome of the small brown planthopper via direct interaction with regulatory particle non-ATPase subunit 3. J. Virol. 2015, 89, 4296–4310. [Google Scholar] [CrossRef] [PubMed]
- Viswanathan, K.; Fruh, K.; DeFilippis, V. Viral hijacking of the host ubiquitin system to evade interferon responses. Curr. Opin. Microbiol. 2010, 13, 517–523. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Fu, S.; Tao, X.; Zhou, X. Rice stripe virus: Exploring Molecular Weapons in the Arsenal of a Negative-Sense RNA Virus. Annu. Rev. Phytopathol. 2021, 59, 351–371. [Google Scholar] [CrossRef] [PubMed]
- Lu, G.; Li, S.; Zhou, C.; Qian, X.; Xiang, Q.; Yang, T.; Wu, J.; Zhou, X.; Zhou, Y.; Ding, X.S.; et al. Tenuivirus utilizes its glycoprotein as a helper component to overcome insect midgut barriers for its circulative and propagative transmission. PLoS Pathog. 2019, 15, e1007655. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, B.X.; Mao, Q.Z.; Zhuo, J.C.; Huang, H.J.; Lu, J.B.; Zhang, C.X.; Li, J.M.; Chen, J.P.; Lu, G. The JAK-STAT pathway promotes persistent viral infection by activating apoptosis in insect vectors. PLoS Pathog. 2023, 19, e1011266. [Google Scholar] [CrossRef]
- Wu, W.; Zheng, L.; Chen, H.; Jia, D.; Li, F.; Wei, T. Nonstructural protein NS4 of Rice Stripe Virus plays a critical role in viral spread in the body of vector insects. PLoS ONE 2014, 9, e88636. [Google Scholar] [CrossRef]
- Li, Y.; Zhou, Z.; Shen, M.; Ge, L.; Liu, F. Ubiquitin-Conjugating Enzyme E2 E Inhibits the Accumulation of Rice Stripe Virus in Laodelphax striatellus (Fallen). Viruses 2020, 12, 908. [Google Scholar] [CrossRef]
- Zhu, J.; Jiang, F.; Wang, X.; Yang, P.; Bao, Y.; Zhao, W.; Wang, W.; Lu, H.; Wang, Q.; Cui, N.; et al. Genome sequence of the small brown planthopper, Laodelphax striatellus. Gigascience 2017, 6, 1–12. [Google Scholar] [CrossRef]
- Kozlov, A.M.; Darriba, D.; Flouri, T.; Morel, B.; Stamatakis, A. RAxML-NG: A fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 2019, 35, 4453–4455. [Google Scholar] [CrossRef] [PubMed]
- Nei, M.; Rogozin, I.B.; Piontkivska, H. Purifying selection and birth-and-death evolution in the ubiquitin gene family. Proc. Natl. Acad. Sci. USA 2000, 97, 10866–10871. [Google Scholar] [CrossRef] [PubMed]
- Nenoi, M.; Mita, K.; Ichimura, S.; Kawano, A. Higher frequency of concerted evolutionary events in rodents than in man at the polyubiquitin gene VNTR locus. Genetics 1998, 148, 867–876. [Google Scholar] [CrossRef] [PubMed]
- Keeling, P.J.; Doolittle, W.F. Concerted evolution in protists: Recent homogenization of a polyubiquitin gene in Trichomonas vaginalis. J. Mol. Evol. 1995, 41, 556–562. [Google Scholar] [CrossRef] [PubMed]
- Perelygin, A.A.; Kondrashov, F.A.; Rogozin, I.B.; Brinton, M.A. Evolution of the mouse polyubiquitin-C gene. J. Mol. Evol. 2002, 55, 202–210. [Google Scholar] [CrossRef] [PubMed]
- Paradkar, P.N.; Duchemin, J.B.; Rodriguez-Andres, J.; Trinidad, L.; Walker, P.J. Cullin4 Is Pro-Viral during West Nile Virus Infection of Culex Mosquitoes. PLoS Pathog. 2015, 11, e1005143. [Google Scholar] [CrossRef] [PubMed]
- Dubey, S.K.; Mehta, D.; Chaudhary, S.; Hasan, A.; Sunil, S. An E3 Ubiquitin Ligase Scaffolding Protein Is Proviral during Chikungunya Virus Infection in Aedes aegypti. Microbiol. Spectr. 2022, 10, e0059522. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).