Impact of Ae-GRD on Ivermectin Resistance and Its Regulation by miR-71-5p in Aedes aegypti
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Mosquito Breeding
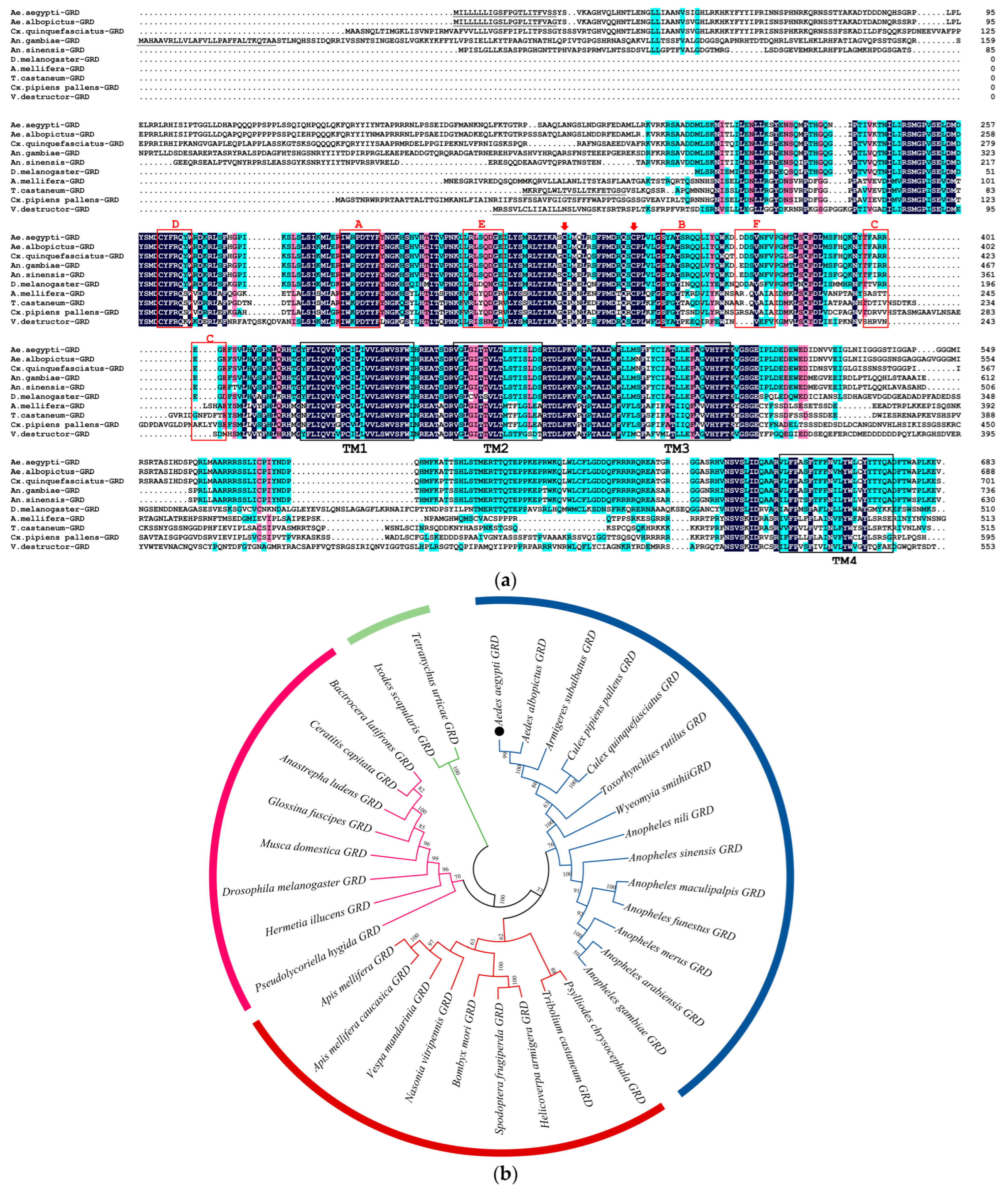
2.2. Identification of Ae-GRD
2.3. Bioinformatics Analysis and Potential miRNAs Prediction of Ae-GRD
2.4. In Vitro Synthesis and Microinjection of dsRNAs
2.5. Dual-Fluorescent Vector Construction
2.6. HEK-293T Cell Line Culture and Dual-Luciferase Activity Assay
2.7. Inhibition/Overexpression of miR-71-5p in Ae. aegypti
2.8. Real-Time qRT-PCR Analysis of Ae-GRD and miR-71-5p
2.9. Bioassays
2.10. GABA Content Determination
2.11. Statistical Analysis
3. Results
3.1. Cloning and Sequence Analysis of Ae-GRD
3.2. RNAi Efficiency of Injected dsRNA-Ae-GRD against Ae. aegypti, with Changes in Ivermectin Susceptibility
3.3. Changes in GABA Content after RNAi
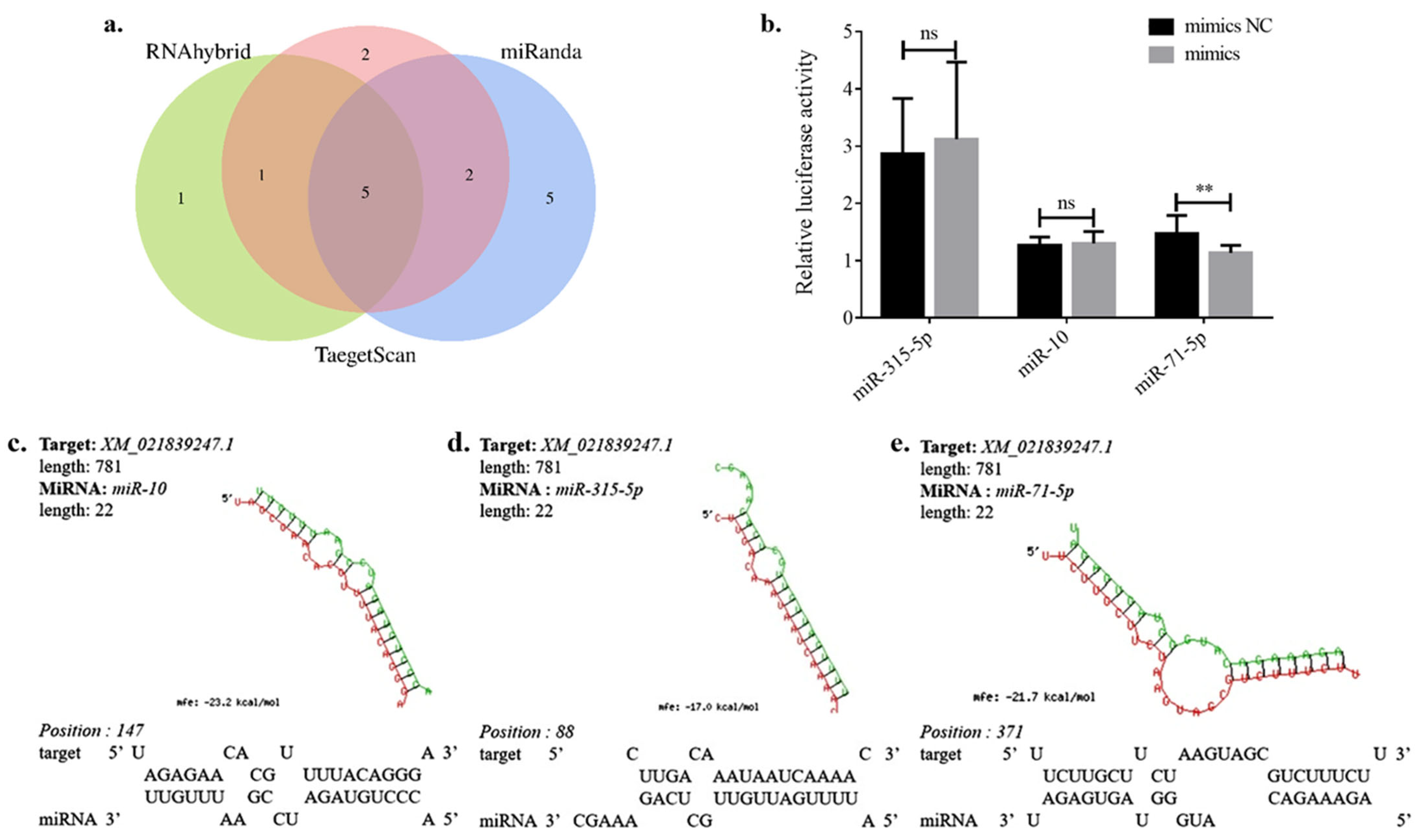
3.4. Prediction of miRNAs Targeting Ae-GRD
3.5. Dual-Luciferase Validation
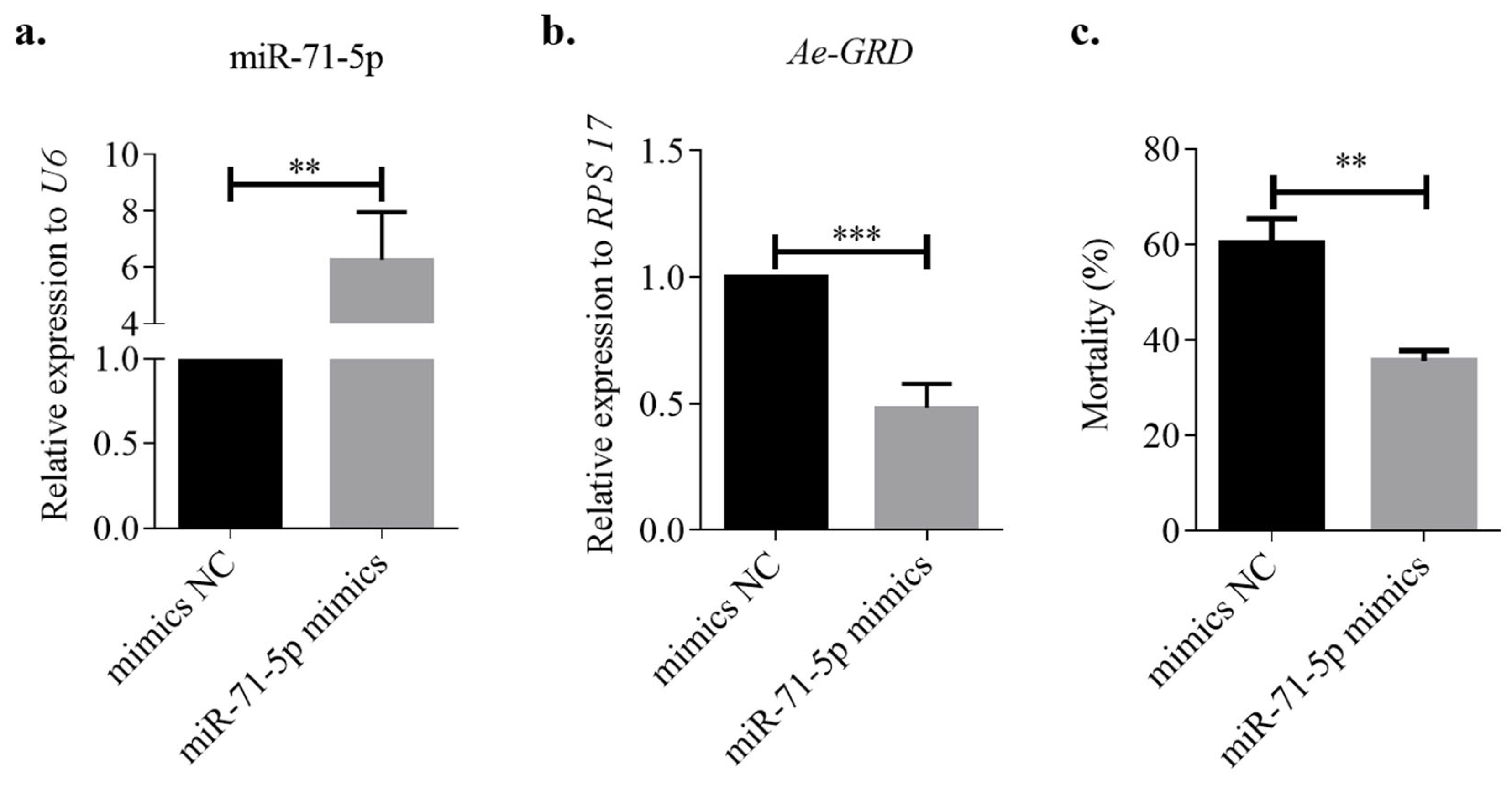
3.6. miR-71-5p Regulates Ivermectin Resistance in Ae. aegypti
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- LaBeaud, A.D.; Bashir, F.; King, C.H. Measuring the burden of arboviral diseases: The spectrum of morbidity and mortality from four prevalent infections. Popul. Health Metr. 2011, 9, 1. [Google Scholar] [CrossRef] [PubMed]
- De Swart, M.M.; Balvers, C.; Verhulst, N.O.; Koenraadt, C.J. Effects of host blood on mosquito reproduction. Trends Parasitol. 2023, 39, 575–587. [Google Scholar] [CrossRef] [PubMed]
- Sparks, T.C.; Bryant, R.J. Innovation in insecticide discovery: Approaches to the discovery of new classes of insecticides. Pest Manag. Sci. 2022, 78, 3226–3247. [Google Scholar] [CrossRef] [PubMed]
- Casida, J.E.; Durkin, K.A. Neuroactive insecticides: Targets, selectivity, resistance, and secondary effects. Annu. Rev. Entomol. 2013, 58, 99–117. [Google Scholar] [CrossRef] [PubMed]
- Smart, T.G.; Stephenson, F.A. A half century of γ-aminobutyric acid. Brain Neurosci Adv. 2019, 3, 2398212819858249. [Google Scholar] [CrossRef] [PubMed]
- Gou, Z.H.; Wang, X.; Wang, W. Evolution of neurotransmitter gamma-aminobutyric acid, glutamate and their receptors. Zool. Res. 2012, 33, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Cossart, R. Operational hub cells: A morpho-physiologically diverse class of GABAergic neurons united by a common function. Curr. Opin. Neurobiol. 2014, 26, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Miller, P.S.; Smart, T.G. Binding, activation and modulation of Cys-loop receptors. Trends Pharmacol. Sci. 2010, 31, 161–174. [Google Scholar] [CrossRef] [PubMed]
- Fujii, Y.; Ito, Y.; Harada, K.H.; Hitomi, T.; Koizumi, A.; Haraguchi, K. Comparative survey of levels of chlorinated cyclodiene pesticides in breast milk from some cities of China, Korea and Japan. Chemosphere 2012, 89, 452–457. [Google Scholar] [CrossRef]
- Wang, Q.H.; Wang, H.C.; Zhang, Y.X.; Chen, J.; Archana, U.; Biswajit, B.; Hang, J.Y.; Wu, S.Y.; Liao, C.H.; Han, Q. Functional analysis reveals ionotropic GABA receptor subunit RDL is a target site of ivermectin and fluralaner in the yellow fever mosquito, Aedes aegypti. Pest Manag. Sci. 2022, 78, 4173–4182. [Google Scholar] [CrossRef]
- Hashim, O.; Charvet, C.L.; Toubate, B.; Ahmed, A.A.E.; Lamassiaude, N.; Neveu, C.; Dimier-Poisson, I.; Debierre-Grockiego, F.; Dupuy, C. Molecular and Functional Characterization of GABA Receptor Subunits GRD and LCCH3 from Human Louse Pediculus Humanus Humanus. Mol. Pharmacol. 2021, 102, 116–127. [Google Scholar] [CrossRef] [PubMed]
- Gisselmann, G.; P Plonka, J.; Pusch, H.; Hatt, H. Drosophila melanogaster GRD and LCCH3 subunits form heteromultimeric GABA-gated cation channels. Br. J. Pharmacol. 2004, 142, 409–413. [Google Scholar] [CrossRef] [PubMed]
- Henry, C.; Cens, T.; Charnet, P.; Cohen-Solal, C.; Collet, C.; Van-Dijk, J.; Guiramand, J.; De Jésus-Ferreira, M.; Menard, C.; Mokrane, N. Heterogeneous expression of GABA receptor-like subunits LCCH3 and GRD reveals functional diversity of GABA receptors in the honeybee Apis mellifera. Br. J. Pharmacol. 2020, 177, 3924–3940. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.T.; Sheng, C.W.; Jones, A.K.; Jiang, J.; Tang, T.; Han, Z.J.; Zhao, C.Q. Functional characteristics of the lepidopteran ionotropic GABA receptor 8916 subunit interacting with the LCCH3 or the RDL subunit. J. Agric. Food. Chem. 2021, 69, 11582–11591. [Google Scholar] [CrossRef] [PubMed]
- Casida, J.E.; Durkin, K.A. Novel GABA receptor pesticide targets. Pestic. Biochem. Physiol. 2015, 121, 22–30. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Wu, Q.; Xu, Q.; He, L. From the Cover: Functional Analysis Reveals Glutamate and Gamma-Aminobutyric Acid-Gated Chloride Channels as Targets of Avermectins in the Carmine Spider Mite. Toxicol. Sci. 2017, 155, 258–269. [Google Scholar] [CrossRef] [PubMed]
- Chiara, D.C.; Dostalova, Z.; Jayakar, S.S.; Zhou, X.J.; Miller, K.W.; Cohen, J.B. Mapping general anesthetic binding site (s) in human α1β3 γ-aminobutyric acid type A receptors with [3H] TDBzl-etomidate, a photoreactive etomidate analogue. Biochemistry 2012, 51, 836–847. [Google Scholar] [CrossRef]
- Chen, I.S.; Kubo, Y. Ivermectin and its target molecules: Shared and unique modulation mechanisms of ion channels and receptors by ivermectin. J. Physiol. 2018, 596, 1833–1845. [Google Scholar] [CrossRef]
- Shan, Q.; Haddrill, J.L.; Lynch, J.W. Ivermectin, an unconventional agonist of the glycine receptor chloride channel. J. Biol. Chem. 2001, 276, 12556–12564. [Google Scholar] [CrossRef]
- Cooper, A.M.; Silver, K.; Zhang, J.Z.; Yoonseong, P.; Zhu, K.Y. Molecular mechanisms influencing efficiency of RNA interference in insects. Pest Manag. Sci. 2019, 75, 18–28. [Google Scholar] [CrossRef]
- Mor, E.; Shomron, N. Species-specific micro RNA regulation influences phenotypic variability: Perspectives on species-specific microRNA regulation. Bioessays 2013, 35, 881–888. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhao, B.; Roy, S.; Saha, T.T.; Kokoza, V.A.; Li, M.; Raikhel, A.S. microRNA-309 targets the Homeobox gene SIX4 and controls ovarian development in the mosquito Aedes aegypti. Proc. Natl. Acad. Sci. USA 2016, 113, E4828–E4836. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.C.; Guo, Q.; Wang, W.J.; Hu, S.L.; Fang, F.J.; Lv, Y.; Yu, J.; Zou, F.F.; Lei, Z.T.; Ma, K. Identification of differentially expressed microRNAs in Culex pipiens and their potential roles in pyrethroid resistance. Insect Biochem. Mol. Biol. 2014, 55, 39–50. [Google Scholar] [CrossRef] [PubMed]
- Li, X.X.; Hu, S.L.; Zhang, H.B.; Wang, H.; Zhou, Y.; Sun, Y.; Ma, L.; Shen, B. MiR-279-3p regulates deltamethrin resistance through CYP325BB1 in Culex pipiens pallens. Parasites Vectors 2021, 14, 528. [Google Scholar] [CrossRef] [PubMed]
- Matthews, B.J.; McBride, C.S.; DeGennaro, M.; Despo, O.; Vosshall, L.B. The neurotranscriptome of the Aedes aegypti mosquito. BMC Genom. 2016, 17, 32. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Kajla, M.; Kakani, P.; Choudhury, T.P.; Gupta, K.; Gupta, L.; Kumar, S. Characterization and expression analysis of gene encoding heme peroxidase HPX15 in major Indian malaria vector Anopheles stephensi (Diptera: Culicidae). Acta Trop. 2016, 158, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Gupta, K.; Dhawan, R.; Kajla, M.; Misra, T.; Kumar, S.; Gupta, L. The evolutionary divergence of STAT transcription factor in different Anopheles species. Gene 2017, 596, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Whyard, S.; Erdelyan, C.N.; Partridge, A.L.; Singh, A.D.; Beebe, N.W.; Capina, R. Silencing the buzz: A new approach to population suppression of mosquitoes by feeding larvae double-stranded RNAs. Parasites Vectors 2015, 8, 96. [Google Scholar] [CrossRef]
- Chen, J.; Wu, Y.C.; Chen, J.K.; Zhu, X.J.; Merkler, D.; Liao, C.H.; Han, Q. Elongases of Long-Chain Fatty Acids ELO2 and ELO9 Are Involved in Cuticle Formation and Function in Fecundity in the Yellow Fever Mosquito, Aedes aegypti. Insects 2023, 14, 189. [Google Scholar] [CrossRef] [PubMed]
- Kajla, M.; Choudhury, T.P.; Kakani, P.; Gupta, K.; Dhawan, R.; Gupta, L.; Kumar, S. Silencing of Anopheles stephensi heme peroxidase HPX15 activates diverse immune pathways to regulate the growth of midgut bacteria. Front. Microbiol. 2016, 7, 217893. [Google Scholar] [CrossRef] [PubMed]
- Dhawan, R.; Gupta, K.; Kajla, M.; Kakani, P.; Choudhury, T.P.; Kumar, S.; Kumar, V.; Gupta, L. Apolipophorin-III Acts as a Positive Regulator of Plasmodium Development in Anopheles stephensi. Front. Physiol. 2017, 8, 229747. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, J.M.; Hartmann, D.; Bartošová-Sojková, P.; Debat, H.; Moos, M.; Šimek, P.; Fara, J.; Palus, M.; Kučera, M.; Hajdušek, O.; et al. Blood-feeding adaptations and virome assessment of the poultry red mite Dermanyssus gallinae guided by RNA-seq. Commun. Biol. 2023, 6, 517. [Google Scholar] [CrossRef] [PubMed]
- Kobylinski, K.C.; Ubalee, R.; Ponlawat, A.; Nitatsukprasert, C.; Phasomkulsolsil, S.; Wattanakul, T.; Tarning, J.; Na-Bangchang, K.; McCardle, P.W.; Davidson, S.A.; et al. Ivermectin susceptibility and sporontocidal effect in Greater Mekong Subregion Anopheles. Malar. J. 2017, 16, 280. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, S.; Kitai, H.; Suzuki, H.I. Network regulation of microRNA biogenesis and target interaction. Cells 2023, 12, 306. [Google Scholar] [CrossRef] [PubMed]
- Lewis, B.P.; Shih, I.; Jones-Rhoades, M.W.; Bartel, D.P.; Burge, C.B. Prediction of mammalian microRNA targets. Cell 2003, 115, 787–798. [Google Scholar] [CrossRef] [PubMed]
- Buckingham, D.S.; Ihara, M.; Sattelle, B.D.; Matsuda, K. Mechanisms of action, resistance and toxicity of insecticides targeting GABA receptors. Curr. Med. Chem. 2017, 24, 2935–2945. [Google Scholar] [CrossRef] [PubMed]
- Raymond, V.; Sattelle, D.B. Novel animal-health drug targets from ligand-gated chloride channels. Nat. Rev. Drug Discov. 2002, 1, 427–436. [Google Scholar] [CrossRef]
- Meng, X.K.; Yang, X.M.; Zhang, N.; Jiang, H.; Ge, H.C.; Chen, M.X.; Qian, K.; Wang, J.J. Knockdown of the GABA receptor RDL genes decreases abamectin susceptibility in the rice stem borer, Chilo suppressalis. Pestic. Biochem. Physiol. 2019, 153, 171–175. [Google Scholar] [CrossRef]
- Huang, J.N.; Wang, T.B.; Qiu, Y.M.; Hassanyar, A.K.; Zhang, Z.N.; Sun, Q.L.; Ni, X.M.; Yu, K.J.; Guo, Y.K.; Yang, C.S.; et al. Differential Brain Expression Patterns of microRNAs Related to Olfactory Performance in Honey Bees (Apis mellifera). Genes 2023, 14, 1000. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.H.; Ge, X.; Li, Z.Q.; Wang, Y.Q.; Song, Q.S.; Stanley, D.W.; Tan, A.J.; Huang, Y.P. MicroRNA-281 regulates the expression of ecdysone receptor (EcR) isoform B in the silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 2013, 43, 692–700. [Google Scholar] [CrossRef] [PubMed]
- Song, J.S.; Li, W.W.; Zhao, H.H.; Zhou, S.T. Clustered miR-2, miR-13a, miR-13b and miR-71 coordinately target Notch gene to regulate oogenesis of the migratory locust Locusta migratoria. Insect Biochem. Mol. Biol. 2019, 106, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.L.; Wang, Y.L.; Jiang, F.; Song, T.Q.; Wang, H.M.; Liu, Q.; Zhang, J.; Zhang, J.Z.; Kang, L. miR-71 and miR-263 jointly regulate target genes chitin synthase and chitinase to control locust molting. PLoS Genet. 2016, 12, e1006257. [Google Scholar] [CrossRef]
- Harraz, M.M.; Eacker, S.M.; Wang, X.; Dawson, T.M.; Dawson, V.L. MicroRNA-223 is neuroprotective by targeting glutamate receptors. Proc. Natl. Acad. Sci. USA 2012, 109, 18962–18967. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, L.; Yin, Y.; Wang, Q.; Zhao, P.; Han, Q.; Liao, C. Impact of Ae-GRD on Ivermectin Resistance and Its Regulation by miR-71-5p in Aedes aegypti. Insects 2024, 15, 453. https://doi.org/10.3390/insects15060453
Yu L, Yin Y, Wang Q, Zhao P, Han Q, Liao C. Impact of Ae-GRD on Ivermectin Resistance and Its Regulation by miR-71-5p in Aedes aegypti. Insects. 2024; 15(6):453. https://doi.org/10.3390/insects15060453
Chicago/Turabian StyleYu, Lingling, Yanan Yin, Qiuhui Wang, Peizhen Zhao, Qian Han, and Chenghong Liao. 2024. "Impact of Ae-GRD on Ivermectin Resistance and Its Regulation by miR-71-5p in Aedes aegypti" Insects 15, no. 6: 453. https://doi.org/10.3390/insects15060453
APA StyleYu, L., Yin, Y., Wang, Q., Zhao, P., Han, Q., & Liao, C. (2024). Impact of Ae-GRD on Ivermectin Resistance and Its Regulation by miR-71-5p in Aedes aegypti. Insects, 15(6), 453. https://doi.org/10.3390/insects15060453





