The Biological Roles of microRNAs in Drosophila Development
Abstract
:Simple Summary
Abstract
1. Introduction
2. miRNA Biogenesis and Mechanism in Drosophila
3. Regulation of miRNA Expression by Hormones in Drosophila
4. Biological Roles of miRNAs in Drosophila Development
4.1. Developmental Growth and Apoptosis
4.1.1. miRNAs Involved in Developmental Growth
4.1.2. miRNAs Involved in Programmed Cell Death
4.2. miRNAs in the Development of the Nervous System
4.2.1. miR-92a/b, Bantam, and miR-124 in Neuroblasts (NBs)
4.2.2. Let-7 and miR-iab8 in the Mushroom Body (MB)
4.2.3. miR-34, miR-124, and miR-276a/b in Synaptogenesis and Dendritic Formation
4.3. miRNAs in Wing Development
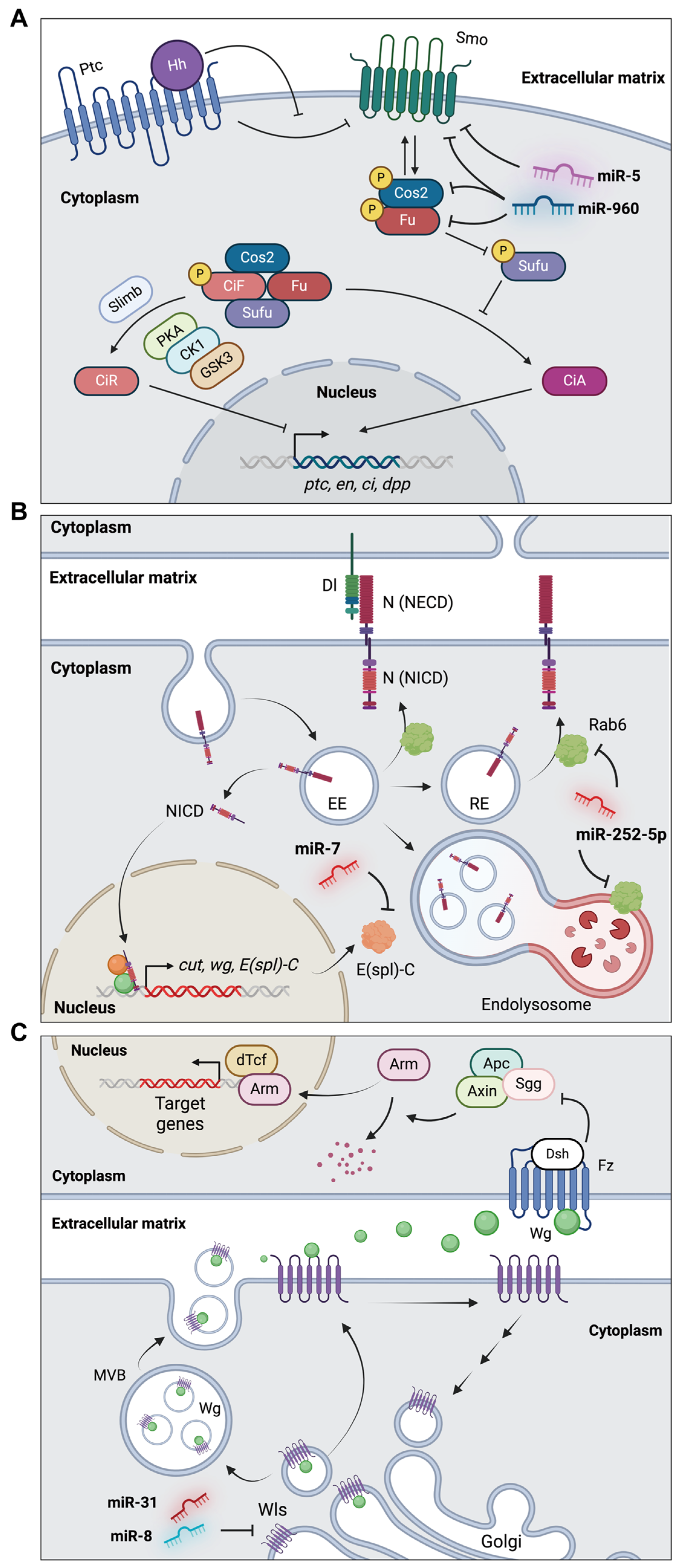
4.3.1. miR-960 and miR-5 in the Hh Signaling Pathway of Wing Development
4.3.2. miR-7 and miR-252 in the Notch Signaling Pathway of Wing Development
4.3.3. miR-31a/b and miR-8 in the Wg Signaling Pathway of Wing Development
4.4. miRNAs in Eye Development
5. Discussion and Future Research Directions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- He, L.; Hannon, G.J. MicroRNAs: Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004, 5, 522–531. [Google Scholar] [CrossRef]
- Chyb, S.; Gompel, N. Atlas of Drosophila Morphology: Wild-Type and Classical Mutants; Academic Press: London, UK, 2013. [Google Scholar]
- Wightman, B.; Ha, I.; Ruvkun, G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 1993, 75, 855–862. [Google Scholar] [CrossRef]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Lagos-Quintana, M.; Rauhut, R.; Lendeckel, W.; Tuschl, T. Identification of novel genes coding for small expressed RNAs. Science 2001, 294, 853–858. [Google Scholar] [CrossRef]
- Aravin, A.A.; Lagos-Quintana, M.; Yalcin, A.; Zavolan, M.; Marks, D.; Snyder, B.; Gaasterland, T.; Meyer, J.; Tuschl, T. The small RNA profile during Drosophila melanogaster development. Dev. Cell 2003, 5, 337–350. [Google Scholar] [CrossRef]
- Ruby, J.G.; Stark, A.; Johnston, W.K.; Kellis, M.; Bartel, D.P.; Lai, E.C. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 2007, 17, 1850–1864. [Google Scholar] [CrossRef]
- Stark, A.; Kheradpour, P.; Parts, L.; Brennecke, J.; Hodges, E.; Hannon, G.J.; Kellis, M. Systematic discovery and characterization of fly microRNAs using 12 Drosophila genomes. Genome Res. 2007, 17, 1865–1879. [Google Scholar] [CrossRef]
- Griffiths-Jones, S.; Grocock, R.J.; van Dongen, S.; Bateman, A.; Enright, A.J. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006, 34, D140–D144. [Google Scholar] [CrossRef]
- Chawla, G.; Sokol, N.S. MicroRNAs in Drosophila development. Int. Rev. Cell Mol. Biol. 2011, 286, 1–65. [Google Scholar] [CrossRef]
- Quesnelle, D.C.; Bendena, W.G.; Chin-Sang, I.D. A Compilation of the Diverse miRNA Functions in Caenorhabditis elegans and Drosophila melanogaster Development. Int. J. Mol. Sci. 2023, 24, 6963. [Google Scholar] [CrossRef]
- Lee, Y.; Kim, M.; Han, J.; Yeom, K.H.; Lee, S.; Baek, S.H.; Kim, V.N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004, 23, 4051–4060. [Google Scholar] [CrossRef]
- Cai, X.; Hagedorn, C.H.; Cullen, B.R. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 2004, 10, 1957–1966. [Google Scholar] [CrossRef]
- Borchert, G.M.; Lanier, W.; Davidson, B.L. RNA polymerase III transcribes human microRNAs. Nat. Struct. Mol. Biol. 2006, 13, 1097–1101. [Google Scholar] [CrossRef]
- Lee, Y.; Jeon, K.; Lee, J.T.; Kim, S.; Kim, V.N. MicroRNA maturation: Stepwise processing and subcellular localization. EMBO J. 2002, 21, 4663–4670. [Google Scholar] [CrossRef]
- Han, J.; Lee, Y.; Yeom, K.H.; Kim, Y.K.; Jin, H.; Kim, V.N. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004, 18, 3016–3027. [Google Scholar] [CrossRef]
- Gregory, R.I.; Yan, K.P.; Amuthan, G.; Chendrimada, T.; Doratotaj, B.; Cooch, N.; Shiekhattar, R. The Microprocessor complex mediates the genesis of microRNAs. Nature 2004, 432, 235–240. [Google Scholar] [CrossRef]
- Denli, A.M.; Tops, B.B.; Plasterk, R.H.; Ketting, R.F.; Hannon, G.J. Processing of primary microRNAs by the Microprocessor complex. Nature 2004, 432, 231–235. [Google Scholar] [CrossRef]
- Morlando, M.; Ballarino, M.; Gromak, N.; Pagano, F.; Bozzoni, I.; Proudfoot, N.J. Primary microRNA transcripts are processed co-transcriptionally. Nat. Struct. Mol. Biol. 2008, 15, 902–909. [Google Scholar] [CrossRef]
- Okamura, K.; Hagen, J.W.; Duan, H.; Tyler, D.M.; Lai, E.C. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell 2007, 130, 89–100. [Google Scholar] [CrossRef]
- Ruby, J.G.; Jan, C.H.; Bartel, D.P. Intronic microRNA precursors that bypass Drosha processing. Nature 2007, 448, 83–86. [Google Scholar] [CrossRef]
- Shibata, S.; Sasaki, M.; Miki, T.; Shimamoto, A.; Furuichi, Y.; Katahira, J.; Yoneda, Y. Exportin-5 orthologues are functionally divergent among species. Nucleic Acids Res. 2006, 34, 4711–4721. [Google Scholar] [CrossRef]
- Yi, R.; Qin, Y.; Macara, I.G.; Cullen, B.R. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003, 17, 3011–3016. [Google Scholar] [CrossRef]
- Lund, E.; Guttinger, S.; Calado, A.; Dahlberg, J.E.; Kutay, U. Nuclear export of microRNA precursors. Science 2004, 303, 95–98. [Google Scholar] [CrossRef]
- Hutvagner, G.; McLachlan, J.; Pasquinelli, A.E.; Balint, E.; Tuschl, T.; Zamore, P.D. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 2001, 293, 834–838. [Google Scholar] [CrossRef]
- Knight, S.W.; Bass, B.L. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science 2001, 293, 2269–2271. [Google Scholar] [CrossRef]
- Grishok, A.; Pasquinelli, A.E.; Conte, D.; Li, N.; Parrish, S.; Ha, I.; Baillie, D.L.; Fire, A.; Ruvkun, G.; Mello, C.C. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 2001, 106, 23–34. [Google Scholar] [CrossRef]
- Lee, Y.S.; Nakahara, K.; Pham, J.W.; Kim, K.; He, Z.; Sontheimer, E.J.; Carthew, R.W. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell 2004, 117, 69–81. [Google Scholar] [CrossRef]
- Macrae, I.J.; Zhou, K.; Li, F.; Repic, A.; Brooks, A.N.; Cande, W.Z.; Adams, P.D.; Doudna, J.A. Structural basis for double-stranded RNA processing by Dicer. Science 2006, 311, 195–198. [Google Scholar] [CrossRef]
- Zhang, H.; Kolb, F.A.; Jaskiewicz, L.; Westhof, E.; Filipowicz, W. Single processing center models for human Dicer and bacterial RNase III. Cell 2004, 118, 57–68. [Google Scholar] [CrossRef]
- Kawamata, T.; Tomari, Y. Making RISC. Trends Biochem. Sci. 2010, 35, 368–376. [Google Scholar] [CrossRef]
- Forstemann, K.; Horwich, M.D.; Wee, L.; Tomari, Y.; Zamore, P.D. Drosophila microRNAs are sorted into functionally distinct argonaute complexes after production by dicer-1. Cell 2007, 130, 287–297. [Google Scholar] [CrossRef]
- Okamura, K.; Ishizuka, A.; Siomi, H.; Siomi, M.C. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev. 2004, 18, 1655–1666. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, D.S.; Hutvagner, G.; Du, T.; Xu, Z.; Aronin, N.; Zamore, P.D. Asymmetry in the assembly of the RNAi enzyme complex. Cell 2003, 115, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Khvorova, A.; Reynolds, A.; Jayasena, S.D. Functional siRNAs and miRNAs exhibit strand bias. Cell 2003, 115, 209–216. [Google Scholar] [CrossRef] [PubMed]
- Hammond, S.M. Dicing and slicing: The core machinery of the RNA interference pathway. FEBS Lett. 2005, 579, 5822–5829. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Li, S.; Liu, S. Juvenile Hormone Studies in Drosophila melanogaster. Front. Physiol. 2021, 12, 785320. [Google Scholar] [CrossRef]
- Jindra, M.; Uhlirova, M.; Charles, J.P.; Smykal, V.; Hill, R.J. Genetic Evidence for Function of the bHLH-PAS Protein Gce/Met As a Juvenile Hormone Receptor. PLoS Genet. 2015, 11, e1005394. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Mead, E.A.; Zhu, J. Heterodimer of two bHLH-PAS proteins mediates juvenile hormone-induced gene expression. Proc. Natl. Acad. Sci. USA 2011, 108, 638–643. [Google Scholar] [CrossRef]
- He, Q.; Wen, D.; Jia, Q.; Cui, C.; Wang, J.; Palli, S.R.; Li, S. Heat shock protein 83 (Hsp83) facilitates methoprene-tolerant (Met) nuclear import to modulate juvenile hormone signaling. J. Biol. Chem. 2014, 289, 27874–27885. [Google Scholar] [CrossRef]
- He, Q.; Zhang, Y.; Zhang, X.; Xu, D.; Dong, W.; Li, S.; Wu, R. Nucleoporin Nup358 facilitates nuclear import of Methoprene-tolerant (Met) in an importin β- and Hsp83-dependent manner. Insect Biochem. Mol. Biol. 2017, 81, 10–18. [Google Scholar] [CrossRef]
- Yamanaka, N.; Rewitz, K.F.; O’Connor, M.B. Ecdysone control of developmental transitions: Lessons from Drosophila research. Annu. Rev. Entomol. 2013, 58, 497–516. [Google Scholar] [CrossRef] [PubMed]
- Truman, J.W.; Riddiford, L.M. Endocrine insights into the evolution of metamorphosis in insects. Annu. Rev. Entomol. 2002, 47, 467–500. [Google Scholar] [CrossRef] [PubMed]
- Yao, T.P.; Forman, B.M.; Jiang, Z.; Cherbas, L.; Chen, J.D.; McKeown, M.; Cherbas, P.; Evans, R.M. Functional ecdysone receptor is the product of EcR and Ultraspiracle genes. Nature 1993, 366, 476–479. [Google Scholar] [CrossRef] [PubMed]
- Yao, T.P.; Segraves, W.A.; Oro, A.E.; McKeown, M.; Evans, R.M. Drosophila ultraspiracle modulates ecdysone receptor function via heterodimer formation. Cell 1992, 71, 63–72. [Google Scholar] [CrossRef] [PubMed]
- DiBello, P.R.; Withers, D.A.; Bayer, C.A.; Fristrom, J.W.; Guild, G.M. The Drosophila Broad-Complex encodes a family of related proteins containing zinc fingers. Genetics 1991, 129, 385–397. [Google Scholar] [CrossRef] [PubMed]
- Segraves, W.A.; Hogness, D.S. The E75 ecdysone-inducible gene responsible for the 75B early puff in Drosophila encodes two new members of the steroid receptor superfamily. Genes Dev. 1990, 4, 204–219. [Google Scholar] [CrossRef] [PubMed]
- Burtis, K.C.; Thummel, C.S.; Jones, C.W.; Karim, F.D.; Hogness, D.S. The Drosophila 74EF early puff contains E74, a complex ecdysone-inducible gene that encodes two ets-related proteins. Cell 1990, 61, 85–99. [Google Scholar] [CrossRef] [PubMed]
- Riddiford, L.M.; Cherbas, P.; Truman, J.W. Ecdysone receptors and their biological actions. Vitam. Horm. 2000, 60, 1–73. [Google Scholar] [CrossRef]
- Lim, D.H.; Lee, S.; Han, J.Y.; Choi, M.S.; Hong, J.S.; Seong, Y.; Kwon, Y.S.; Lee, Y.S. Ecdysone-responsive microRNA-252-5p controls the cell cycle by targeting Abi in Drosophila. FASEB J. 2018, 32, 4519–4533. [Google Scholar] [CrossRef]
- Lim, D.H.; Lee, S.; Choi, M.S.; Han, J.Y.; Seong, Y.; Na, D.; Kwon, Y.S.; Lee, Y.S. The conserved microRNA miR-8-3p coordinates the expression of V-ATPase subunits to regulate ecdysone biosynthesis for Drosophila metamorphosis. FASEB J. 2020, 34, 6449–6465. [Google Scholar] [CrossRef]
- Sempere, L.F.; Sokol, N.S.; Dubrovsky, E.B.; Berger, E.M.; Ambros, V. Temporal regulation of microRNA expression in Drosophila melanogaster mediated by hormonal signals and broad-Complex gene activity. Dev. Biol. 2003, 259, 9–18. [Google Scholar] [CrossRef]
- Sokol, N.S.; Xu, P.; Jan, Y.N.; Ambros, V. Drosophila let-7 microRNA is required for remodeling of the neuromusculature during metamorphosis. Genes Dev. 2008, 22, 1591–1596. [Google Scholar] [CrossRef]
- Jin, X.; Wu, X.; Zhou, L.; He, T.; Yin, Q.; Liu, S. 20-Hydroxyecdysone-responsive microRNAs of insects. RNA Biol. 2020, 17, 1454–1471. [Google Scholar] [CrossRef]
- He, Q.; Zhang, Y.; Dong, W. MicroRNA miR-927 targets the juvenile hormone primary response gene Kruppel homolog1 to control Drosophila developmental growth. Insect Mol. Biol. 2020, 29, 545–554. [Google Scholar] [CrossRef]
- Garbuzov, A.; Tatar, M. Hormonal regulation of Drosophila microRNA let-7 and miR-125 that target innate immunity. Fly 2010, 4, 306–311. [Google Scholar] [CrossRef]
- Chan, K.K.; Chan, T.F.; Bendena, W.; Hui, J.H.L. Noncoding RNA Regulation of Hormonal and Metabolic Systems in the Fruit Fly Drosophila. Metabolites 2023, 13, 152. [Google Scholar] [CrossRef]
- Lim, D.H.; Lee, S.; Han, J.Y.; Choi, M.S.; Hong, J.S.; Lee, Y.S. MicroRNA miR-252 targets mbt to control the developmental growth of Drosophila. Insect Mol. Biol. 2019, 28, 444–454. [Google Scholar] [CrossRef]
- Lim, D.H.; Choi, M.S.; Jeon, J.W.; Lee, Y.S. MicroRNA miR-252-5p regulates the Notch signaling pathway by targeting Rab6 in Drosophila wing development. Insect Sci. 2023, 30, 1431–1444. [Google Scholar] [CrossRef]
- Lee, S.; Kim, N.; Jang, D.; Kim, H.K.; Kim, J.; Jeon, J.W.; Lim, D.H. Ecdysone-induced microRNA miR-276a-3p controls developmental growth by targeting the insulin-like receptor in Drosophila. Insect Mol. Biol. 2023, 32, 703–715. [Google Scholar] [CrossRef]
- Li, H.; Gavis, E.R. Drosophila FMRP controls miR-276-mediated regulation of nejire mRNA for space-filling dendrite development. G3 2022, 12, jkac239. [Google Scholar] [CrossRef]
- Hyun, S.; Lee, J.H.; Jin, H.; Nam, J.; Namkoong, B.; Lee, G.; Chung, J.; Kim, V.N. Conserved MicroRNA miR-8/miR-200 and its target USH/FOG2 control growth by regulating PI3K. Cell 2009, 139, 1096–1108. [Google Scholar] [CrossRef] [PubMed]
- Kennell, J.A.; Gerin, I.; MacDougald, O.A.; Cadigan, K.M. The microRNA miR-8 is a conserved negative regulator of Wnt signaling. Proc. Natl. Acad. Sci. USA 2008, 105, 15417–15422. [Google Scholar] [CrossRef] [PubMed]
- Varghese, J.; Lim, S.F.; Cohen, S.M. Drosophila miR-14 regulates insulin production and metabolism through its target, sugarbabe. Genes Dev. 2010, 24, 2748–2753. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Vernooy, S.Y.; Guo, M.; Hay, B.A. The Drosophila microRNA Mir-14 suppresses cell death and is required for normal fat metabolism. Curr. Biol. 2003, 13, 790–795. [Google Scholar] [CrossRef] [PubMed]
- Brennecke, J.; Hipfner, D.R.; Stark, A.; Russell, R.B.; Cohen, S.M. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 2003, 113, 25–36. [Google Scholar] [CrossRef]
- Weng, R.; Cohen, S.M. Control of Drosophila Type I and Type II central brain neuroblast proliferation by bantam microRNA. Development 2015, 142, 3713–3720. [Google Scholar] [CrossRef]
- Ge, W.; Chen, Y.W.; Weng, R.; Lim, S.F.; Buescher, M.; Zhang, R.; Cohen, S.M. Overlapping functions of microRNAs in control of apoptosis during Drosophila embryogenesis. Cell Death Differ. 2012, 19, 839–846. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.C.; Chen, C.H.; Mercer, A.; Sokol, N.S. Let-7-complex microRNAs regulate the temporal identity of Drosophila mushroom body neurons via chinmo. Dev. Cell 2012, 23, 202–209. [Google Scholar] [CrossRef] [PubMed]
- Kucherenko, M.M.; Barth, J.; Fiala, A.; Shcherbata, H.R. Steroid-induced microRNA let-7 acts as a spatio-temporal code for neuronal cell fate in the developing Drosophila brain. EMBO J. 2012, 31, 4511–4523. [Google Scholar] [CrossRef]
- Busto, G.U.; Guven-Ozkan, T.; Chakraborty, M.; Davis, R.L. Developmental inhibition of miR-iab8-3p disrupts mushroom body neuron structure and adult learning ability. Dev. Biol. 2016, 419, 237–249. [Google Scholar] [CrossRef]
- Yuva-Aydemir, Y.; Xu, X.L.; Aydemir, O.; Gascon, E.; Sayin, S.; Zhou, W.; Hong, Y.; Gao, F.B. Downregulation of the Host Gene jigr1 by miR-92 Is Essential for Neuroblast Self-Renewal in Drosophila. PLoS Genet. 2015, 11, e1005264. [Google Scholar] [CrossRef]
- Weng, R.; Cohen, S.M. Drosophila miR-124 regulates neuroblast proliferation through its target anachronism. Development 2012, 139, 1427–1434. [Google Scholar] [CrossRef] [PubMed]
- Sun, K.; Westholm, J.O.; Tsurudome, K.; Hagen, J.W.; Lu, Y.; Kohwi, M.; Betel, D.; Gao, F.B.; Haghighi, A.P.; Doe, C.Q.; et al. Neurophysiological defects and neuronal gene deregulation in Drosophila mir-124 mutants. PLoS Genet. 2012, 8, e1002515. [Google Scholar] [CrossRef]
- McNeill, E.M.; Warinner, C.; Alkins, S.; Taylor, A.; Heggeness, H.; DeLuca, T.F.; Fulga, T.A.; Wall, D.P.; Griffith, L.C.; Van Vactor, D. The conserved microRNA miR-34 regulates synaptogenesis via coordination of distinct mechanisms in presynaptic and postsynaptic cells. Nat. Commun. 2020, 11, 1092. [Google Scholar] [CrossRef]
- Gao, L.; Hou, X.; Wu, L.; Zhang, F.; Zhang, Q.; Ye, X.; Yang, Y.; Lin, X. Drosophila miR-960 negatively regulates Hedgehog signaling by suppressing Smoothened, Costal-2 and Fused. Cell. Signal. 2013, 25, 1301–1309. [Google Scholar] [CrossRef]
- Wu, L.F.; Gao, L.; Hou, X.M.; Zhang, Q.H.; Li, S.; Yang, Y.F.; Lin, X.H. Drosophila miR-5 suppresses Hedgehog signaling by directly targeting Smoothened. FEBS Lett. 2012, 586, 4052–4060. [Google Scholar] [CrossRef]
- Stark, A.; Brennecke, J.; Russell, R.B.; Cohen, S.M. Identification of Drosophila MicroRNA targets. PLoS Biol. 2003, 1, E60. [Google Scholar] [CrossRef]
- Lai, E.C.; Tam, B.; Rubin, G.M. Pervasive regulation of Drosophila Notch target genes by GY-box-, Brd-box-, and K-box-class microRNAs. Genes Dev. 2005, 19, 1067–1080. [Google Scholar] [CrossRef]
- Li, X.; Carthew, R.W. A microRNA mediates EGF receptor signaling and promotes photoreceptor differentiation in the Drosophila eye. Cell 2005, 123, 1267–1277. [Google Scholar] [CrossRef]
- Jung, J.E.; Lee, J.Y.; Kim, I.R.; Park, S.M.; Kang, J.W.; Kim, Y.H.; Park, H.R.; Lee, J.H. MicroRNA-31 Regulates Expression of Wntless in Both Drosophila melanogaster and Human Oral Cancer Cells. Int. J. Mol. Sci. 2020, 21, 7232. [Google Scholar] [CrossRef] [PubMed]
- Duan, H.; de Navas, L.F.; Hu, F.; Sun, K.; Mavromatakis, Y.E.; Viets, K.; Zhou, C.; Kavaler, J.; Johnston, R.J.; Tomlinson, A.; et al. The mir-279/996 cluster represses receptor tyrosine kinase signaling to determine cell fates in the Drosophila eye. Development 2018, 145, dev159053. [Google Scholar] [CrossRef] [PubMed]
- Edgar, B.A. How flies get their size: Genetics meets physiology. Nat. Rev. Genet 2006, 7, 907–916. [Google Scholar] [CrossRef] [PubMed]
- Liguori, F.; Mascolo, E.; Verni, F. The Genetics of Diabetes: What We Can Learn from Drosophila. Int. J. Mol. Sci. 2021, 22, 11295. [Google Scholar] [CrossRef] [PubMed]
- Varghese, J.; Cohen, S.M. microRNA miR-14 acts to modulate a positive autoregulatory loop controlling steroid hormone signaling in Drosophila. Genes Dev. 2007, 21, 2277–2282. [Google Scholar] [CrossRef] [PubMed]
- Steller, H. Regulation of apoptosis in Drosophila. Cell Death Differ. 2008, 15, 1132–1138. [Google Scholar] [CrossRef]
- Denton, D.; Aung-Htut, M.T.; Kumar, S. Developmentally programmed cell death in Drosophila. Biochim. Biophys. Acta 2013, 1833, 3499–3506. [Google Scholar] [CrossRef] [PubMed]
- Bejarano, F.; Chang, C.H.; Sun, K.; Hagen, J.W.; Deng, W.M.; Lai, E.C. A comprehensive in vivo screen for anti-apoptotic miRNAs indicates broad capacities for oncogenic synergy. Dev. Biol. 2021, 475, 10–20. [Google Scholar] [CrossRef] [PubMed]
- Homem, C.C.; Knoblich, J.A. Drosophila neuroblasts: A model for stem cell biology. Development 2012, 139, 4297–4310. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.; Lee, A.; Luo, L. Development of the Drosophila mushroom bodies: Sequential generation of three distinct types of neurons from a neuroblast. Development 1999, 126, 4065–4076. [Google Scholar] [CrossRef]
- Zhu, S.; Chiang, A.S.; Lee, T. Development of the Drosophila mushroom bodies: Elaboration, remodeling and spatial organization of dendrites in the calyx. Development 2003, 130, 2603–2610. [Google Scholar] [CrossRef]
- Tabata, T.; Kornberg, T.B. Hedgehog is a signaling protein with a key role in patterning Drosophila imaginal discs. Cell 1994, 76, 89–102. [Google Scholar] [CrossRef] [PubMed]
- Basler, K.; Struhl, G. Compartment boundaries and the control of Drosophila limb pattern by hedgehog protein. Nature 1994, 368, 208–214. [Google Scholar] [CrossRef]
- Hooper, J.E.; Scott, M.P. Communicating with Hedgehogs. Nat. Rev. Mol. Cell Biol. 2005, 6, 306–317. [Google Scholar] [CrossRef]
- Huangfu, D.; Anderson, K.V. Signaling from Smo to Ci/Gli: Conservation and divergence of Hedgehog pathways from Drosophila to vertebrates. Development 2006, 133, 3–14. [Google Scholar] [CrossRef]
- Irvine, K.D.; Vogt, T.F. Dorsal-ventral signaling in limb development. Curr. Opin. Cell Biol. 1997, 9, 867–876. [Google Scholar] [CrossRef] [PubMed]
- Kopan, R.; Ilagan, M.X. The canonical Notch signaling pathway: Unfolding the activation mechanism. Cell 2009, 137, 216–233. [Google Scholar] [CrossRef]
- Bray, S.J. Notch signalling: A simple pathway becomes complex. Nat. Rev. Mol. Cell Biol. 2006, 7, 678–689. [Google Scholar] [CrossRef] [PubMed]
- Aparicio, R.; Simoes Da Silva, C.J.; Busturia, A. MicroRNA miR-7 contributes to the control of Drosophila wing growth. Dev. Dyn. 2015, 244, 21–30. [Google Scholar] [CrossRef]
- Swarup, S.; Verheyen, E.M. Wnt/Wingless signaling in Drosophila. Cold Spring Harb. Perspect. Biol. 2012, 4, a007930. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Chia, J.; Canning, C.A.; Jones, C.M.; Bard, F.A.; Virshup, D.M. WLS retrograde transport to the endoplasmic reticulum during Wnt secretion. Dev. Cell 2014, 29, 277–291. [Google Scholar] [CrossRef]
- Port, F.; Kuster, M.; Herr, P.; Furger, E.; Banziger, C.; Hausmann, G.; Basler, K. Wingless secretion promotes and requires retromer-dependent cycling of Wntless. Nat. Cell Biol. 2008, 10, 178–185. [Google Scholar] [CrossRef] [PubMed]
- Waghmare, I.; Page-McCaw, A. Wnt Signaling in Stem Cell Maintenance and Differentiation in the Drosophila Germarium. Genes 2018, 9, 127. [Google Scholar] [CrossRef]
- Goodman, R.M.; Thombre, S.; Firtina, Z.; Gray, D.; Betts, D.; Roebuck, J.; Spana, E.P.; Selva, E.M. Sprinter: A novel transmembrane protein required for Wg secretion and signaling. Development 2006, 133, 4901–4911. [Google Scholar] [CrossRef]
- DasGupta, R.; Kaykas, A.; Moon, R.T.; Perrimon, N. Functional genomic analysis of the Wnt-wingless signaling pathway. Science 2005, 308, 826–833. [Google Scholar] [CrossRef]
- DasGupta, R.; Nybakken, K.; Booker, M.; Mathey-Prevot, B.; Gonsalves, F.; Changkakoty, B.; Perrimon, N. A case study of the reproducibility of transcriptional reporter cell-based RNAi screens in Drosophila. Genome Biol. 2007, 8, R203. [Google Scholar] [CrossRef] [PubMed]
- Kumar, J.P. Building an ommatidium one cell at a time. Dev. Dyn. 2012, 241, 136–149. [Google Scholar] [CrossRef]
- Freeman, M. Reiterative use of the EGF receptor triggers differentiation of all cell types in the Drosophila eye. Cell 1996, 87, 651–660. [Google Scholar] [CrossRef] [PubMed]
- Sun, K.; Jee, D.; de Navas, L.F.; Duan, H.; Lai, E.C. Multiple In Vivo Biological Processes Are Mediated by Functionally Redundant Activities of Drosophila mir-279 and mir-996. PLoS Genet. 2015, 11, e1005245. [Google Scholar] [CrossRef]
- Chi, S.W.; Zang, J.B.; Mele, A.; Darnell, R.B. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature 2009, 460, 479–486. [Google Scholar] [CrossRef]
- Hafner, M.; Landthaler, M.; Burger, L.; Khorshid, M.; Hausser, J.; Berninger, P.; Rothballer, A.; Ascano, M., Jr.; Jungkamp, A.C.; Munschauer, M.; et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 2010, 141, 129–141. [Google Scholar] [CrossRef]
- Konig, J.; Zarnack, K.; Rot, G.; Curk, T.; Kayikci, M.; Zupan, B.; Turner, D.J.; Luscombe, N.M.; Ule, J. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat. Struct. Mol. Biol. 2010, 17, 909–915. [Google Scholar] [CrossRef]
- Van Nostrand, E.L.; Pratt, G.A.; Shishkin, A.A.; Gelboin-Burkhart, C.; Fang, M.Y.; Sundararaman, B.; Blue, S.M.; Nguyen, T.B.; Surka, C.; Elkins, K.; et al. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nat. Methods 2016, 13, 508–514. [Google Scholar] [CrossRef]
- Wessels, H.H.; Lebedeva, S.; Hirsekorn, A.; Wurmus, R.; Akalin, A.; Mukherjee, N.; Ohler, U. Global identification of functional microRNA-mRNA interactions in Drosophila. Nat. Commun. 2019, 10, 1626. [Google Scholar] [CrossRef]
- Helwak, A.; Kudla, G.; Dudnakova, T.; Tollervey, D. Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell 2013, 153, 654–665. [Google Scholar] [CrossRef]
- Moore, M.J.; Scheel, T.K.; Luna, J.M.; Park, C.Y.; Fak, J.J.; Nishiuchi, E.; Rice, C.M.; Darnell, R.B. miRNA-target chimeras reveal miRNA 3′-end pairing as a major determinant of Argonaute target specificity. Nat. Commun. 2015, 6, 8864. [Google Scholar] [CrossRef] [PubMed]
- Cai, Z.; Cao, C.; Ji, L.; Ye, R.; Wang, D.; Xia, C.; Wang, S.; Du, Z.; Hu, N.; Yu, X.; et al. RIC-seq for global in situ profiling of RNA-RNA spatial interactions. Nature 2020, 582, 432–437. [Google Scholar] [CrossRef] [PubMed]
- Martin, H.C.; Wani, S.; Steptoe, A.L.; Krishnan, K.; Nones, K.; Nourbakhsh, E.; Vlassov, A.; Grimmond, S.M.; Cloonan, N. Imperfect centered miRNA binding sites are common and can mediate repression of target mRNAs. Genome Biol. 2014, 15, R51. [Google Scholar] [CrossRef] [PubMed]




| miRNA | Target Gene Name (Symbol) | Validation Methods 1 | Tissues/Cells | References | Signaling Pathways |
|---|---|---|---|---|---|
| miR-252 | mushroom bodies tiny (mbt) | Ago1 CLIP-seq, WB, Luc rep | Fat body | [58] | - |
| Abelson interacting protein (Abi) | Ago1 CLIP-seq, WB, RT-qPCR, Luc rep | S2 cell | [50] | - | |
| Rab6 (Rab6) | Ago1 CLIP-seq, RT-qPCR, Luc rep | Wing | [59] | Notch | |
| miR-276a | Insulin-like receptor (InR) | Ago1 CLIP-seq, Luc rep | Wing/fat body | [60] | IIS |
| nejire (nej) | EGFP rep | Neuron | [61] | - | |
| miR-8 | u-shaped (ush) | RT-qPCR, WB, Luc rep | Fat body | [62] | IIS |
| CG32767, wntless (wls) | Luc rep | Wing | [63] | Wg | |
| miR-14 | sugarbabe (sug) | Luc rep, EGFP rep | Insulin-producing neurosecretory cell | [64] | IIS |
| Death related ICE-like caspase (Drice) | WB | Eye | [65] | Apoptosis | |
| Bantam | head involution defective (hid) | EGFP rep, IS | Wing | [66] | Apoptosis |
| brain tumor (brat), prospero (pros) | RT-qPCR, Luc rep | Neuroblast | [67] | - | |
| miR-6 | head involution defective (hid), reaper (rpr), grim (grim), sickle (skl) | RT-qPCR, Luc rep | Central nervous system | [68] | Apoptosis |
| miR-11 | |||||
| let-7 | Chronologically inappropriate morphogenesis (chinmo) | Luc rep, IS | Mushroom body | [69] | - |
| abrupt (ab) | IS | Mushroom body | [70] | - | |
| miR-iab8 | CG12229, Ceramide phosphoethanolamine synthase (Cpes) | Behavioral analyses | Mushroom body | [71] | - |
| miR-92a/b | Jing interacting gene regulatory 1 (jigr1) | RT-qPCR, IS, WB, Luc rep | Neuroblast | [72] | - |
| miR-124 | anachronism (ana) | RT-qPCR, Luc rep | Neuroblast | [73] | - |
| saxophone (sax), wishful thinking (wit), thickveins (tkv), Mothers against dpp (Mad), Medea (Med) | Luc rep | Central nervous system | [74] | - | |
| miR-34 | Neurexin IV (Nrx-IV), hu li tai shao (hts) | IS | Central nervous system | [75] | - |
| miR-960 | smoothened (smo), costa (cos2), fused (fu) | Luc rep, IS, EGFP rep | Wing | [76] | Hh |
| miR-5 | smoothened (smo) | Luc rep, EGFP rep, IS | Wing | [77] | Hh |
| miR-7 | Enhancer of split complex (E(spl)-C) | EGFP rep | Wing | [78,79] | Notch |
| Yan (=anterior open (aop)) | EGFP rep, IS | Eye | [80] | - | |
| miR-31a/b | wntless (wls) | RT-qPCR, WB | Wing | [81] | Wg |
| miR-279/996 | rhomboid (rho), roughoid (ru), bride of sevenless (boss) | EGFP rep, Luc rep | Eye | [82] | - |
| Sequencing Techniques | Features 1 | Reference | |
|---|---|---|---|
| RNA–Protein interaction | HITS-CLIP | - High-throughput CLIP-seq method using 254 nm UVC - Identifies mRNA fragments and miRNAs associated with Ago protein - Requires additional bioinformatic analysis to confirm miRNA–mRNA interactions | [110] |
| PAR-CLIP | - Modified from HITS-CLIP - Uses 4-thiouridine and 312–365 nm UVA/B to enhance RNA–protein crosslinking | [111] | |
| iCLIP | - Modified from HITS-CLIP - Improves efficiency using DNA circularization after reverse transcription | [112] | |
| eCLIP | - Modified from HITS-CLIP - Enhances efficiency using adapter ligation after reverse transcription | [113] | |
| RNA–RNA interaction | CLASH | - Direct ligation of miRNA and mRNA fragments - Includes Ago protein denaturation and two purification steps | [115] |
| CLEAR-CLIP | - Direct ligation of miRNA and mRNA fragments - Single purification step; does not require full denaturation of Ago | [116] | |
| RIC-seq | - Captures global RNA–RNA interactions using in situ proximity ligation with pCp-biotin - May require Ago immunoprecipitation to enrich miRNA–mRNA interactions | [117] | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jang, D.; Kim, C.J.; Shin, B.H.; Lim, D.-H. The Biological Roles of microRNAs in Drosophila Development. Insects 2024, 15, 491. https://doi.org/10.3390/insects15070491
Jang D, Kim CJ, Shin BH, Lim D-H. The Biological Roles of microRNAs in Drosophila Development. Insects. 2024; 15(7):491. https://doi.org/10.3390/insects15070491
Chicago/Turabian StyleJang, Daegyu, Chae Jeong Kim, Bo Hyun Shin, and Do-Hwan Lim. 2024. "The Biological Roles of microRNAs in Drosophila Development" Insects 15, no. 7: 491. https://doi.org/10.3390/insects15070491
APA StyleJang, D., Kim, C. J., Shin, B. H., & Lim, D.-H. (2024). The Biological Roles of microRNAs in Drosophila Development. Insects, 15(7), 491. https://doi.org/10.3390/insects15070491







