The Role of TcCYP6K1 and TcCYP9F2 Influences Trehalose Metabolism under High-CO2 Stress in Tribolium castaneum (Coleoptera)
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Insect Source and Feeding Method
2.2. Bioinformatics Analysis
2.3. Collection of Tissue and Developmental Expression Samples
2.4. RNA Extraction and cDNA Synthesis
2.5. Cloning of TcCYP6K1 and TcCYP9F2 Genes
2.6. Synthesis and Injection of Double-Stranded RNA (dsDNA)
2.7. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
2.8. Determination of Carbohydrate Content and Trehalase Activity
2.9. Determination of ATP Content
2.10. Data Analysis
3. Results
3.1. Sequence Analysis of Two P450 Genes in T. castaneum
3.2. Temporal and Spatial Expression Pattern of Two P450 Genes in T. castaneum
3.3. Detection of Silencing Efficiency and Survival Rate after CO2 Stress
3.4. Effect on Carbohydrate Metabolism after Silencing and under CO2 Stress
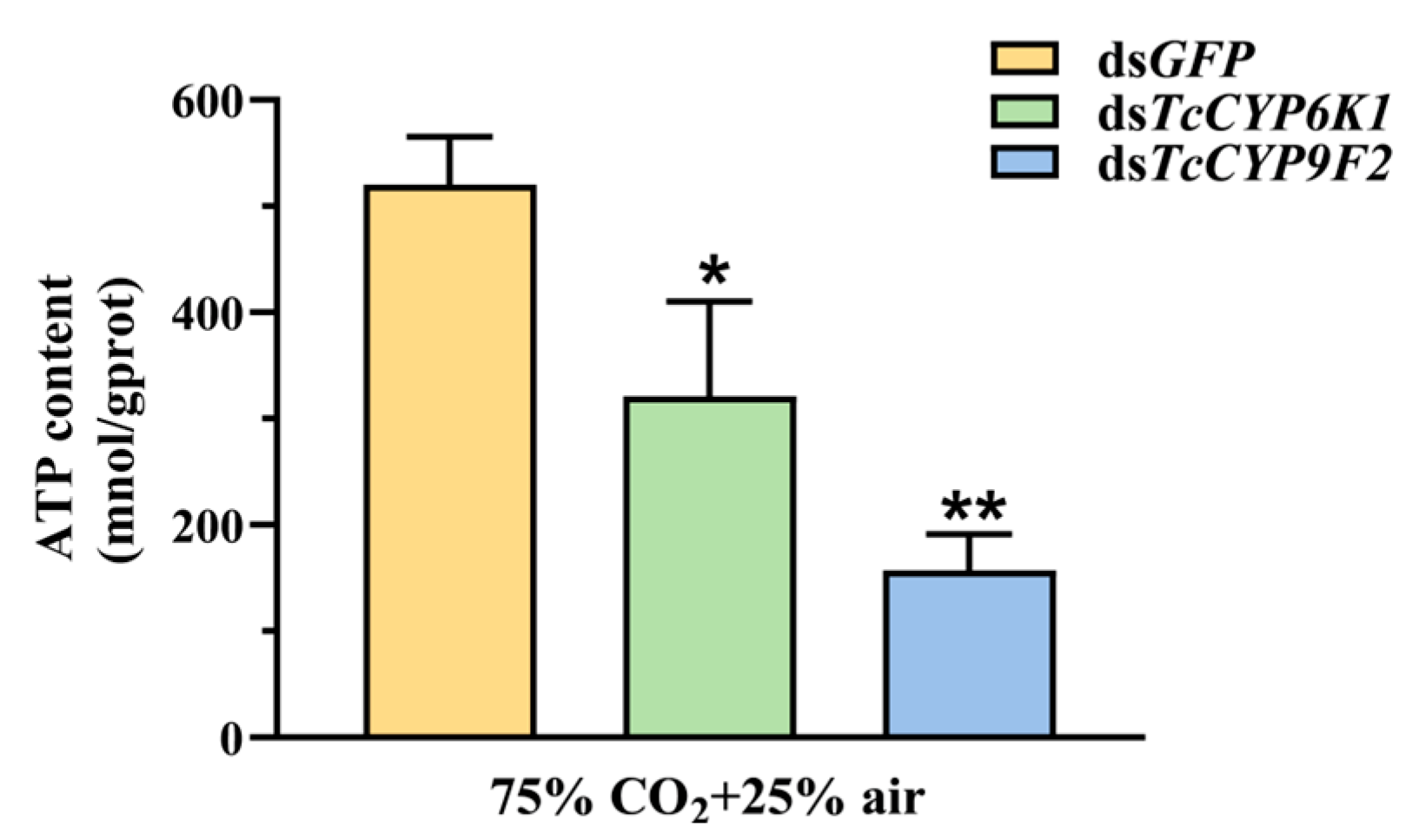
3.5. Effect on ATP Content after Silencing and under CO2 Stress
3.6. Effect on Trehalose Metabolism Pathway after Silencing and under CO2 Stress
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ingegno, B.L.; Tavella, L. Ozone gas treatment against three main pests of stored products by combination of different application parameters. J. Stored Prod. Res. 2022, 95, 101902. [Google Scholar] [CrossRef]
- Islam, W. Eco-friendly approaches for the management of red flour beetle: Tribolium castaneum (Herbst). Sci. Lett. 2017, 5, 105–114. [Google Scholar]
- Nika, E.P.; Kavallieratos, N.G.; Papanikolaou, N.E.; Malesios, C. Interactions of Oryzaephilus surinamensis (L.) (Coleoptera: Silvanidae) with two key stored-product pests under variable abiotic conditions. Entomol. Gen. 2022, 42, 471–478. [Google Scholar] [CrossRef]
- Xu, Y.A. Progress and prospect of control technology for stored grain pests—Fumigation insecticides. Sci. Technol. Cereals Oils Foods 2022, 30, 95–104. [Google Scholar]
- Haddi, K.; Nauen, R.; Benelli, G.; Guedes, R.N.C. Global perspectives on insecticide resistance in agriculture and public health. Entomol. Gen. 2023, 43, 495–500. [Google Scholar] [CrossRef]
- Caballero-Gallardo, K.; Olivero-Verbel, J.; Stashenko, E.E. Repellency and toxicity of essential oils from Cymbopogon martinii, Cymbopogon flexuosus and Lippia origanoides cultivated in Colombia against Tribolium castaneum. J. Stored Prod. Res. 2012, 50, 62–65. [Google Scholar] [CrossRef]
- Hubhachen, Z.; Jiang, H.; Schlipalius, D.; Park, Y.; Guedes, R.N.; Oppert, B.; Opit, G.; Phillips, T.W. A CAPS marker for determination of strong phosphine resistance in Tribolium castaneum from Brazil. J. Pest Sci. 2020, 93, 127–134. [Google Scholar] [CrossRef]
- Xu, J.Y.; Lv, J.H.; Bai, C.Q.; Guo, C.; Bai, J.L. Advances in Research and Application of Modified Atmosphere in the Control of Stored Grain Pests. J. Chin. Cereals Oils Assoc. 2023, 38, 21–28. [Google Scholar]
- Cui, S.; Wang, L.; Qiu, J.; Liu, Z.; Geng, X. Comparative metabolomics analysis of Callosobruchus chinensis larvae under hypoxia, hypoxia/hypercapnia and normoxia. Pest Manag. Sci. 2017, 73, 1267–1276. [Google Scholar] [CrossRef]
- Cao, Y.; Yan, Y.; Yang, W.; Xiong, Z.; Wang, L.; Li, C. Influence of carbon dioxide controlled atmosphere on Oryzaephilus surinamensis Linne and its utilization of energy substances. J. Zhejiang Univ. 2015, 41, 631–640. [Google Scholar]
- Ahn, J.E.; Zhou, X.; Dowd, S.E.; Chapkin, R.S.; Zhu-Salzman, K. Insight into hypoxia tolerance in cowpea bruchid: Metabolic repression and heat shock protein regulation via hypoxia-inducible factor 1. PLoS ONE 2013, 8, e57267. [Google Scholar] [CrossRef] [PubMed]
- Cui, S.F.; Wang, L.; Qiu, J.P.; Geng, X.Q.; Liu, Z.C. Effects of hypoxia/hypercapnia on the metablism of Callosobruchus chinensis (L.) larvae. J. Stored Prod. Res. 2019, 83, 322–330. [Google Scholar] [CrossRef]
- Zhou, M.; Zhang, X.; Pan, B.; Zhu, J.; Qian, X.; Li, X.; Xu, K.; Tang, B.; Li, C. The endogenous metabolic response of Tribolium castaneum under a high concentration of CO2. Agriculture 2022, 12, 979. [Google Scholar] [CrossRef]
- Beall, C.M. Two routes to functional adaptation: Tibetan and Andean high-altitude natives. Proc. Natl. Acad. Sci. USA 2007, 104 (Suppl. S1), 8655–8660. [Google Scholar] [CrossRef] [PubMed]
- Helenius, I.T.; Krupinski, T.; Turnbull, D.W.; Gruenbaum, Y.; Silverman, N.; Johnson, E.A.; Sporn, P.H.S.; Sznajder, J.I.; Beitel, G.J. Elevated CO2 suppresses specific Drosophila innate immune responses and resistance to bacterial infection. Proc. Natl. Acad. Sci. USA 2009, 106, 18710–18715. [Google Scholar] [CrossRef] [PubMed]
- Harrison, J.F.; Haddad, G.G. Effects of oxygen on growth and size: Synthesis of molecular, organismal, and evolutionary studies with Drosophila melanogaster. Annu. Rev. Physiol. 2011, 73, 95–113. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.Q.; Guan, L.W.; Pan, B.Y.; Xu, H.T.; Luo, Y.J.; Zhou, M.; Zhang, J.Y.; Wang, S.G.; Tang, B. Mechanism of HIF1-α-mediated regulation of Tribolium castaneum metabolism under high CO2 concentration elucidated. J. Stored Prod. Res. 2022, 99, 102030. [Google Scholar] [CrossRef]
- Liu, D.; Tian, K.; Yuan, Y.; Li, M.; Zheng, M.; Qiu, X. Prokaryotic functional expression and activity comparison of three CYP9A genes from the polyphagous pest Helicoverpa armigera Bull. Entomol. Res. 2018, 108, 77–83. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Liang, Q.M.; Zhou, W.W.; Jiang, Y.D.; Zhu, Q.Z.; Yu, H.; Zhang, C.X.; Gurr, G.M.; Zhu, Z.R. RNA interference of NADPH–cytochrome P450 reductase of the rice brown planthopper, Nilaparvata lugens, increases susceptibility to insecticides. Pest Manag. Sci. 2015, 71, 32–39. [Google Scholar] [CrossRef]
- Ullah, F.; Gul, H.; Tariq, K.; Hafeez, M.; Desneux, N.; Song, D. Silencing of Cytochrome P450 genes CYP6CY14 and CYP6DC1 in Aphis gossypii by RNA interference enhances susceptibility to clothianidin. Entomol. Gen. 2023, 43, 669–678. [Google Scholar] [CrossRef]
- Xu, D.; Liao, H.; Li, L.; Wu, M.; Xie, W.; Wu, Q.; Zhang, Y.; Zhou, X.; Wang, S. The CYP392D8 gene is not directly associated with abamectin resistance, a case study in two highly resistant Tetranychus urticae strains. Entomol. Gen. 2023, 43, 679–687. [Google Scholar] [CrossRef]
- Liang, X.; Xiao, D.; He, Y.; Yao, J.; Zhu, G.; Zhu, K.Y. Insecticide-mediated up-regulation of cytochrome P450 genes in the red flour beetle (Tribolium castaneum). Int. J. Mol. Sci. 2015, 16, 2078–2098. [Google Scholar] [CrossRef] [PubMed]
- López, M.F.; Cano-Ramírez, C.; Cesar-Ayala, A.K.; Ruiz, E.A.; Zúñiga, G. Diversity and expression of P450 genes from Dendroctonus valens LeConte (Curculionidae: Scolytinae) in response to different kairomones. Insect Biochem. Mol. Biol. 2013, 43, 417–432. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Yang, H.; Wang, Y.; Wei, L.; Lyu, J.; Shan, Z.; Zhang, X.X.; Fan, D. RNA Interference Reveals the Impacts of CYP6CY7 on Imidacloprid Resistance in Aphis glycines. Insects 2024, 15, 188. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, L.I.; Rybczynski, R.; Warren, J.T. Control and biochemical nature of the ecdysteroidogenic pathway. Annu. Rev. Entomol. 2002, 47, 883–916. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Moural, T.W.; Shah, K.; Palli, S.R. Integrated analysis of cytochrome P450 gene superfamily in the red flour beetle, Tribolium castaneum. BMC Genom. 2013, 14, 174. [Google Scholar] [CrossRef] [PubMed]
- Feyereisen, R. Evolution of insect P450. Biochem. Soc. Trans. 2006, 34, 1252–1255. [Google Scholar] [CrossRef]
- Stevens, J.L.; Snyder, M.J.; Koener, J.F.; Feyereisen, R. Inducible P450s of the CYP9 family from larval Manduca sexta midgut. Insect Biochem. Mol. Biol. 2000, 30, 559–568. [Google Scholar] [CrossRef]
- Zhu, F.; Moural, T.W.; Nelson, D.R.; Palli, S.R. A specialist herbivore pest adaptation to xenobiotics through up-regulation of multiple Cytochrome P450s. Sci. Rep. 2016, 6, 20421. [Google Scholar] [CrossRef]
- Li, W.; Berenbaum, M.R.; Schuler, M.A. Molecular analysis of multiple CYP6B genes from polyphagous Papilio species. Insect Biochem. Mol. Biol. 2001, 31, 999–1011. [Google Scholar] [CrossRef]
- Niu, G.; Rupasinghe, S.G.; Zangerl, A.R.; Siegel, J.P.; Schuler, M.A.; Berenbaum, M.R. A substrate-specific cytochrome P450 monooxygenase, CYP6AB11, from the polyphagous navel orangeworm (Amyelois transitella). Insect Biochem. Mol. Biol. 2011, 41, 244–253. [Google Scholar] [CrossRef] [PubMed]
- Yunta, C.; Ooi, J.M.; Oladepo, F.; Grafanaki, S.; Pergantis, S.A.; Tsakireli, D.; Ismail, H.M.; Paine, M.J. Chlorfenapyr metabolism by mosquito P450s associated with pyrethroid resistance identifies potential activation markers. Sci. Rep. 2023, 13, 14124. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.B.; Cai, W.J.; Wang, J.W.; Hong, G.J.; Tao, X.Y.; Wang, L.J.; Huang, Y.P.; Chen, X.Y. Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nat. Biotechnol. 2007, 25, 1307–1313. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Guo, X.; Liu, S.; Li, S.; Zhang, J.; Xue, S.; Tang, Q.; Zhang, K.; Li, R. Cytochrome P450 gene CYP6BQ8 mediates terpinen-4-ol susceptibility in the red flour beetle, Tribolium castaneum (Herbst) (Coleoptera: Tenebrionidae). Bull. Entomol. Res. 2023, 113, 271–281. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yu, R.; Tang, J.; Du, L.; Wang, Y.; Wang, J.; Liu, L.; Gao, S.; Li, B. Three cytochrome P450 CYP4 family genes regulated by the CncC signaling pathway mediate phytochemical susceptibility in the red flour beetle, Tribolium castaneum. Pest. Manag. Sci. 2022, 78, 3508–3518. [Google Scholar] [CrossRef]
- Ma, D.K.; Rothe, M.; Zheng, S.; Bhatla, N.; Pender, C.L.; Menzel, R.; Horvitz, H.R. Cytochrome P450 drives a HIF-regulated behavioral response to reoxygenation by C. elegans. Science 2013, 341, 554–558. [Google Scholar] [CrossRef] [PubMed]
- Pender, C.L.; Horvitz, H.R. Hypoxia-inducible factor cell non-autonomously regulates C. elegans stress responses and behavior via a nuclear receptor. eLife 2018, 7, e36828. [Google Scholar] [CrossRef]
- Zhou, M.; Pan, B.Y.; Guan, L.W.; Wang, Y.Y.; Xu, K.K.; Wang, S.G.; Tang, B.; Li, C. Comparative transcriptomic and metabolomics analysis of modified atmosphere responses in Tribolium castaneum (Coleoptera: Tenebrionidae). Int. J. Insect Sci. 2024, 24, 8. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.F.; Zhou, M.; Wang, Y.Y.; Jiang, X.Y.; Zhang, P.; Xu, K.K.; Tang, B.; Li, C. Characterization of the TcCYPE2 Gene and Its Role in Regulating Trehalose Metabolism in Response to High CO2 Stress. Agronomy 2023, 13, 2263. [Google Scholar] [CrossRef]
- Gupta, K.; Dhawan, R.; Kajla, M.; Misra, T.; Kumar, S.; Gupta, L. The evolutionary divergence of STAT transcription factor in different Anopheles species. Gene 2017, 596, 89–97. [Google Scholar] [CrossRef]
- Kajla, M.; Kakani, P.; Choudhury, T.P.; Gupta, K.; Gupta, L.; Kumar, S. Characterization and expression analysis of gene encoding heme peroxidase HPX15 in major Indian malaria vector Anopheles stephensi (Diptera: Culicidae). Acta Trop. 2016, 158, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Ge, X.Z.; Shi, Y.X.; Wang, S.S.; Liu, Z.H.; Cai, W.J.; Zhou, M.; Wang, S.G.; Tang, B. Sequence analysis of Harmonia axyridis pyruvate kinase gene and its regulation of trehalose metabolism. Sci. Agric. Sin. 2021, 54, 5021–5031. [Google Scholar]
- Yu, W.D.; Pan, B.Y.; Qiu, L.Y.; Huang, Z.; Zhou, T.; Ye, L.; Tang, B.; Wang, S.G. The structure characteristics and biological functions on regulating trehalose metabolism of two NlTret1s in Nilaparvata lugens. Sci. Agric. Sin. 2020, 53, 4802–4812. [Google Scholar]
- Zhang, L.; Qiu, L.Y.; Yang, H.L.; Wang, H.J.; Zhou, M.; Wang, S.G.; Tang, B. Study on the Effect of Wing Bud Chitin Metabolism and Its Developmental Network Genes in the Brown Planthopper, Nilaparvata lugens, by Knockdown of TRE Gene. Front. Physiol. 2017, 8, 750. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Liu, X.; Luo, Z.; Cai, X.; Li, Z.; Bian, L.; Xiu, C.; Chen, Z.; Li, Q.; Fu, N. Transcriptome-Wide Identification of Cytochrome P450s in Tea Black Tussock Moth (Dasychira baibarana) and Candidate Genes Involved in Type-II Sex Pheromone Biosynthesis. Insects 2024, 15, 139. [Google Scholar] [CrossRef] [PubMed]
- Seliskar, M.; Rozman, D. Mammalian cytochromes P450—Importance of tissue specificity. Biochim. Biophys. Acta 2007, 1770, 458–466. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Kang, X.; Wu, H.; Silver, K.; Zhang, J.; Ma, E.; Zhu, K.Y. Transcriptome-wide survey, gene expression profiling and exogenous chemical-induced transcriptional responses of cytochrome P450 superfamily genes in migratory locust (Locusta migratoria). Insect Biochem. Mol. Biol. 2018, 100, 66–77. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Dong, J.; Wu, H.; Zhang, H.; Zhang, J.; Ma, E. Knockdown of cytochrome P450 CYP6 family genes increases susceptibility to carbamates and pyrethroids in the migratory locust, Locusta migratoria. Chemosphere 2019, 223, 48–57. [Google Scholar] [CrossRef]
- Zhang, Y.C.; Gao, S.S.; Xue, S.; An, S.H.; Zhang, K.P. Disruption of the cytochrome P450 CYP6BQ7 gene reduces tolerance to plant toxicants in the red flour beetle, Tribolium castaneum. Int. J. Biol. Macromol. 2021, 172, 263–269. [Google Scholar] [CrossRef]
- Zhou, Z.X.; Dou, W.; Li, C.R.; Wang, J.J. CYP314A1-dependent 20-hydroxyecdysone biosynthesis is involved in regulating the development of pupal diapause and energy metabolism in the Chinese citrus fruit fly, Bactrocera minax. Pest. Manag. Sci. 2022, 78, 3384. [Google Scholar] [CrossRef] [PubMed]
- Kim, I.Y.; Choi, B.; Park, W.R.; Kim, Y.J.; Kim, B.E.; Mun, S.; Choi, H.S.; Kim, D.K. Nuclear receptor HR96 up-regulates cytochrome P450 for insecticide detoxification in Tribolium castaneum. Pest Manag. Sci. 2022, 78, 230–239. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.J.; Kim, M.R.; Kim, S.I.; Jeon, J.K. Microplate assay measurement of cytochrome P450-carbon monoxide complexes. J. Biochem. Mol. Biol. 2003, 36, 332–335. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wu, H.; Zhang, X.; Ma, W.; Zhu, W.; Silver, K.; Ma, E.; Zhang, J.; Zhu, K.Y. Metabolism of selected model substrates and insecticides by recombinant CYP6FD encoded by its gene predominately expressed in the brain of Locusta migratoria. Pestic. Biochem. Physiol. 2019, 159, 154–162. [Google Scholar] [CrossRef] [PubMed]
- Dow, J.A.; Davies, S.A. The Malpighian tubule: Rapid insights from post-genomic biology. J. Insect Physiol. 2006, 52, 365–378. [Google Scholar] [CrossRef] [PubMed]
- Dong, W.; Zhang, X.; Kong, Y.; Zhao, Z.; Mahmoud, A.; Wu, L.; Moussian, B.; Zhang, J. CYP311A1 in the anterior midgut is involved in lipid distribution and microvillus integrity in Drosophila melanogaster. Cell. Mol. Life Sci. 2022, 79, 261. [Google Scholar] [CrossRef]
- Xiong, W.; Gao, S.; Mao, J.; Wei, L.; Xie, J.; Liu, J.; Bi, J.; Song, X.; Li, B. CYP4BN6 and CYP6BQ11 mediate insecticide susceptibility and their expression is regulated by Latrophilin in Tribolium castaneum. Pest. Manag. Sci. 2019, 75, 2744–2755. [Google Scholar] [CrossRef]
- Wang, H.; Shi, Y.; Wang, L.; Liu, S.; Wu, S.; Yang, Y.; Feyereisen, R.; Wu, Y. CYP6AE gene cluster knockout in Helicoverpa armigera reveals role in detoxification of phytochemicals and insecticides. Nat. Commun. 2018, 9, 4820. [Google Scholar] [CrossRef]
- Yu, L.; Tang, W.; He, W.; Ma, X.; Vasseur, L.; Baxter, S.W.; Yang, G.; Huang, S.; Song, F.; You, M. Characterization and expression of the cytochrome P450 gene family in diamondback moth, Plutella xylostella (L.). Sci. Rep. 2015, 5, 8952. [Google Scholar] [CrossRef]
- Kajla, M.; Choudhury, T.P.; Kakani, P.; Gupta, K.; Dhawan, R.; Gupta, L.; Kumar, S. Silencing of Anopheles stephensi heme peroxidase HPX15 activates diverse immune pathways to regulate the growth of midgut bacteria. Front. Microbiol. 2016, 7, 217893. [Google Scholar] [CrossRef]
- Dhawan, R.; Gupta, K.; Kajla, M.; Kakani, P.; Choudhury, T.P.; Kumar, S.; Kumar, V.; Gupta, L. Apolipophorin-III acts as a positive regulator of Plasmodium development in Anopheles stephensi. Front. Physiol. 2017, 8, 229747. [Google Scholar] [CrossRef] [PubMed]
- Kajla, M.; Kakani, P.; Choudhury, T.P.; Kumar, V.; Gupta, K.; Dhawan, R.; Gupta, L.; Kumar, S. Anopheles stephensi heme peroxidase HPX15 suppresses midgut immunity to support Plasmodium development. Front. Immunol. 2017, 8, 243511. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Liu, K.; Liu, H.; Yin, S.; Guo, X.; Zhang, Y.; Zhang, K.; Li, R. Functional analysis of a cytochrome P450 gene CYP9Z6 responding to terpinen-4-ol in the red flour beetle, Tribolium castaneum. Pest. Biochem. Physiol. 2022, 183, 105065. [Google Scholar] [CrossRef] [PubMed]
- Bodin, P.; Burnstock, G. Purinergic signalling: ATP release. Neurochem. Res. 2001, 26, 959–969. [Google Scholar] [CrossRef] [PubMed]
- Tolmasky, D.S.; Rabossi, A.; Quesada-Allué, L.A. Synthesis and mobilization of glycogen during metamorphosis of the medfly Ceratitis capitata. Arch. Biochem. Biophys. 2001, 392, 38–47. [Google Scholar] [CrossRef] [PubMed]
- Tang, B.; Xu, Q.; Zou, Q.; Fang, Q.; Wang, S.; Ye, G. Sequencing and characterization of glycogen synthase and glycogen phosphorylase genes from Spodoptera exigua and analysis of their function in starvation and excessive sugar intake. Arch. Insect Biochem. Physiol. 2012, 80, 42–62. [Google Scholar] [CrossRef] [PubMed]
- Becker, A.; Schlöder, P.; Steele, J.E.; Wegener, G. The regulation of trehalose metabolism in insects. Experientia 1996, 52, 433–439. [Google Scholar] [CrossRef] [PubMed]
- Elbein, A.D.; Pan, Y.T.; Pastuszak, I.; Carroll, D. New insights on trehalose: A multifunctional molecule. Glycobiology 2003, 13, 17R–27R. [Google Scholar] [CrossRef] [PubMed]
- Thompson, S.N. Trehalose—The insect ‘blood’ sugar. In Advances in Insect Physiology; Elsevier: Amsterdam, The Netherlands, 2003; Supplement C, Volume 31, pp. 205–285. [Google Scholar]
- Tang, B.; Chen, J.; Yao, Q.; Pan, Z.; Xu, W.; Wang, S.; Zhang, W. Characterization of a trehalose-6-phosphate synthase gene from Spodoptera exigua and its function identification through RNA interference. J. Insect Physiol. 2010, 56, 813–821. [Google Scholar] [CrossRef]
- Mitsumasu, K.; Kanamori, Y.; Fujita, M.; Iwata, K.I.; Tanaka, D.; Kikuta, S.; Watanabe, M.; Cornette, R.; Okuda, T.; Kikawada, T. Enzymatic control of anhydrobiosis-related accumulation of trehalose in the sleeping chironomid, Polypedilum vanderplanki. FEBS J. 2010, 277, 4215–4228. [Google Scholar] [CrossRef]
- Strøm, A.R.; Kaasen, I. Trehalose metabolism in Escherichia coli: Stress protection and stress regulation of gene expression. Mol. Microbiol. 1993, 8, 205–210. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Ma, E.; Behar, K.L.; Xu, T.; Haddad, G.G. Role of trehalose phosphate synthase in anoxia tolerance and development in Drosophila melanogaster. J. Biol. Chem. 2002, 277, 3274–3279. [Google Scholar] [CrossRef] [PubMed]





| Gene | Forward Primer (5′-3′) | Reverse Primer (5′-3′) | Primer Use |
|---|---|---|---|
| dsTcCYP9F2 | CCTACAAATACTGGACCGA | CAGGAAGTACCCCAACAA | Amplification of dsRNA |
| dsTcCYP6K1 | AAGAAGCGGAGTGGTGGA | GCGAAGTAATTGTTATGGAGG | |
| dsGFP | AAGGGCGAGGAGCTGTTCACCG | CAGCAGGACCATGTGATCGCGC | |
| T7 promoter | GGATCCTAATACGACTCACTATAGG | ||
| TcRPL13a | ACCATATGACCGCAGGAAAC | GGTGAATGGAGCCACTTGTT | qRT-PCR analysis |
| TcCYP9F2 | ACCGGCTACCAAGAATCCAA | GTGACCTTTCCGTTGCAGTT | |
| TcCYP6K1 | AACCCCTTACGTTGGCATCT | GCCAGTTGTCGTTCTTTGCA | |
| TRE1-1 | AACCAAACACTCACTCATTCC | AATCCAATAAGTGTCCCAGTAG | |
| TRE1-2 | GAAGTATCGGTTGGCTCG | GAGTGGGGTTGATTGTGC | |
| TRE1-3 | CTTGAACGCCTTCCTCTG | CCATCCTCGTGGTCATAAA | |
| TRE1-4 | CTACCTAAACCGCTCCCA | TGTCCAGCCAGTACCTCAG | |
| TRE2 | TGTTGTGCGTTTGTGCTC | GGACGGCTTATTGTTGTTTA | |
| TPS | GATTCGCTACATTTACGGG | GAACGGAGACACTATGAGGAC | |
| GS | ATTGGAGGAGTCTAGGAGTGTAC | CCGAATCGCTTTCATCAT | |
| GP | CCGATGGCTCCTTATGTG | GTATGCGTTTGACGTGGAT | |
| PFK | CTACGAAAATGTCCGAAGG | GTTGCGGTCAAAAGGTGT | |
| HCK1 | GAGGTATGTCTGCGAATGC | TGGAAATGTGGGTGGAAC | |
| PK | CAACCGACGAAAAGTATGC | TTCACCCCTTTACTACTCCC | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guan, L.; Wang, X.; Wan, S.; Wang, Y.; Zhang, X.; Wang, S.; Li, C.; Tang, B. The Role of TcCYP6K1 and TcCYP9F2 Influences Trehalose Metabolism under High-CO2 Stress in Tribolium castaneum (Coleoptera). Insects 2024, 15, 502. https://doi.org/10.3390/insects15070502
Guan L, Wang X, Wan S, Wang Y, Zhang X, Wang S, Li C, Tang B. The Role of TcCYP6K1 and TcCYP9F2 Influences Trehalose Metabolism under High-CO2 Stress in Tribolium castaneum (Coleoptera). Insects. 2024; 15(7):502. https://doi.org/10.3390/insects15070502
Chicago/Turabian StyleGuan, Liwen, Xianzhong Wang, Sijing Wan, Yuanyuan Wang, Xinyu Zhang, Shigui Wang, Can Li, and Bin Tang. 2024. "The Role of TcCYP6K1 and TcCYP9F2 Influences Trehalose Metabolism under High-CO2 Stress in Tribolium castaneum (Coleoptera)" Insects 15, no. 7: 502. https://doi.org/10.3390/insects15070502
APA StyleGuan, L., Wang, X., Wan, S., Wang, Y., Zhang, X., Wang, S., Li, C., & Tang, B. (2024). The Role of TcCYP6K1 and TcCYP9F2 Influences Trehalose Metabolism under High-CO2 Stress in Tribolium castaneum (Coleoptera). Insects, 15(7), 502. https://doi.org/10.3390/insects15070502






