Exploring the Wilderness within: An Integrative Metabolomics and Transcriptomics Study on Near-Wild and Colonized Aedes aegypti
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Insect Colonies
2.2. Mosquito Rearing
2.3. Metabolomic Profiling
2.4. Library Prep and Transcriptome Sequencing
2.5. Differential Gene Expression and Enrichment Analyses
3. Results
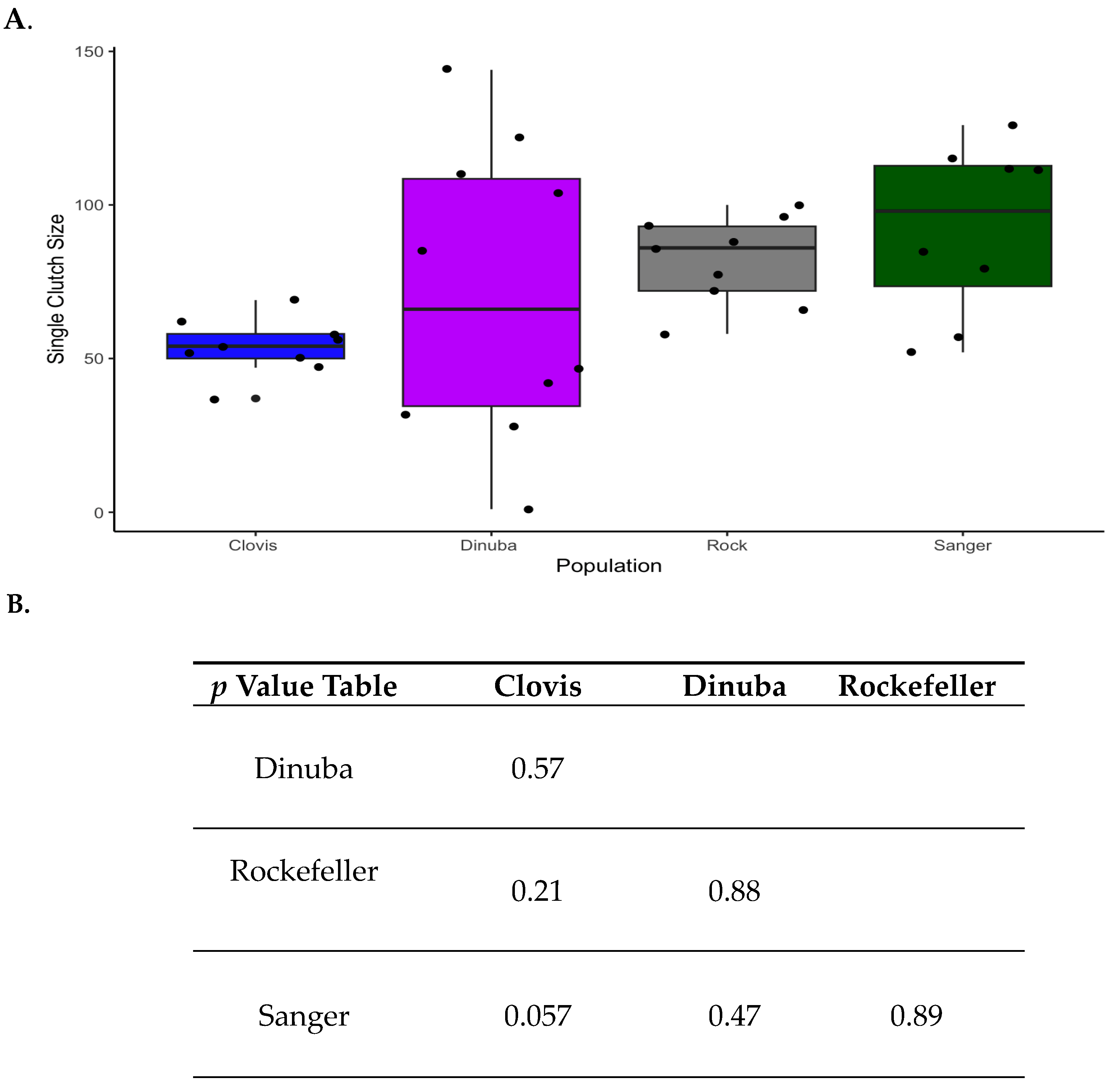
3.1. Resistant Lines and Longevity: Nutrient-Stress Effects on Lifespan
3.2. Metabolomics Panels and Transcriptome Profiles Classify Populations in Principal Component Analysis
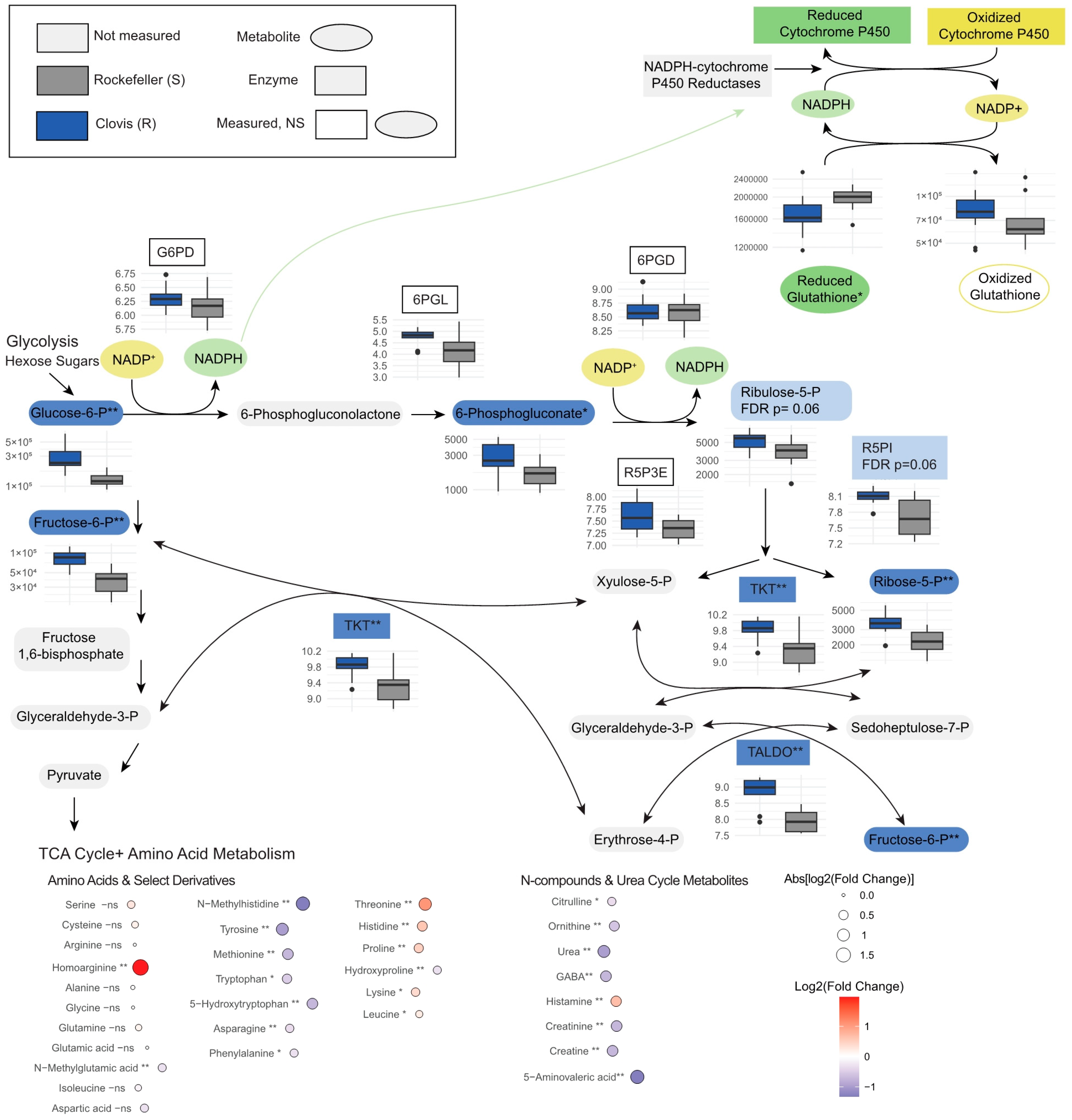
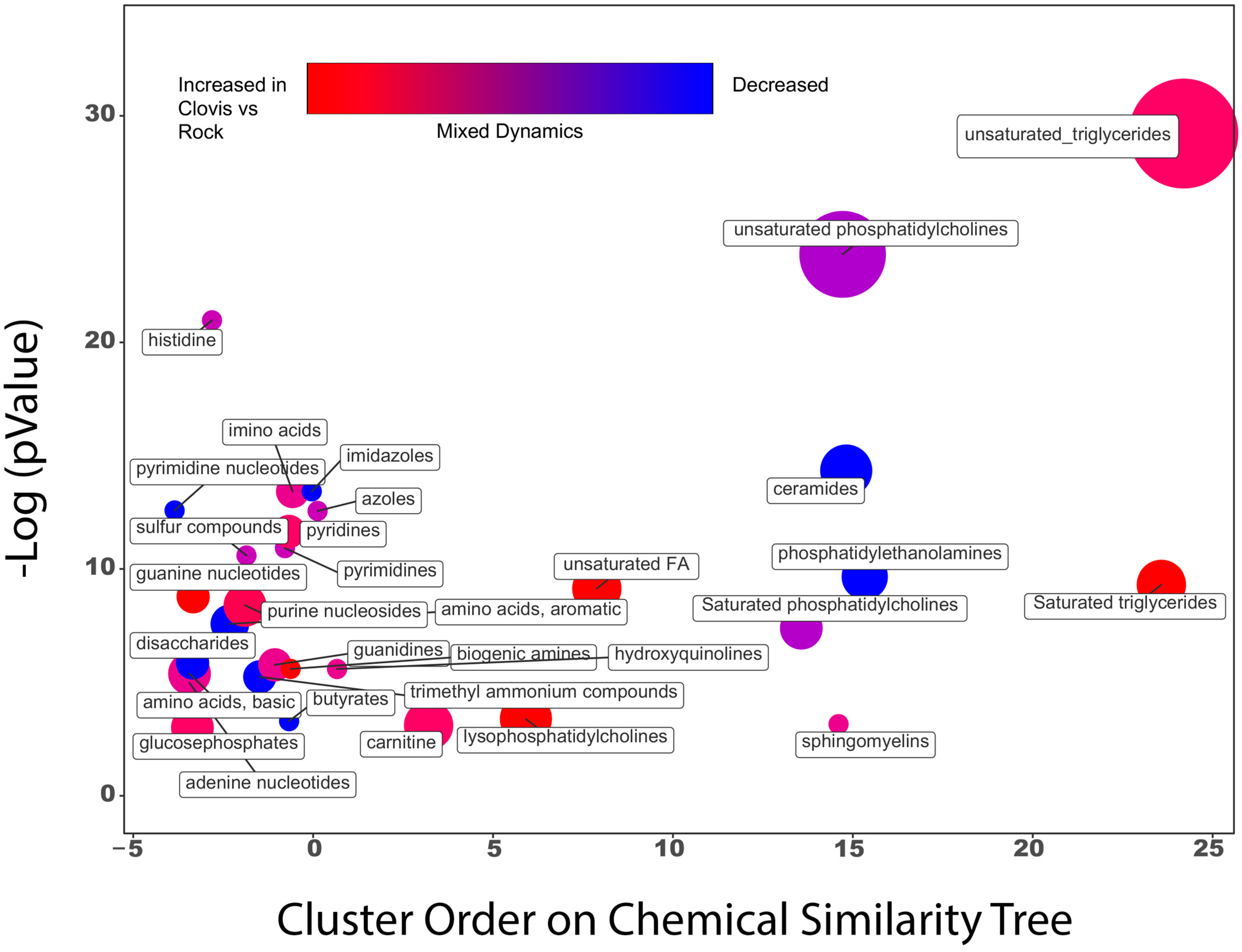
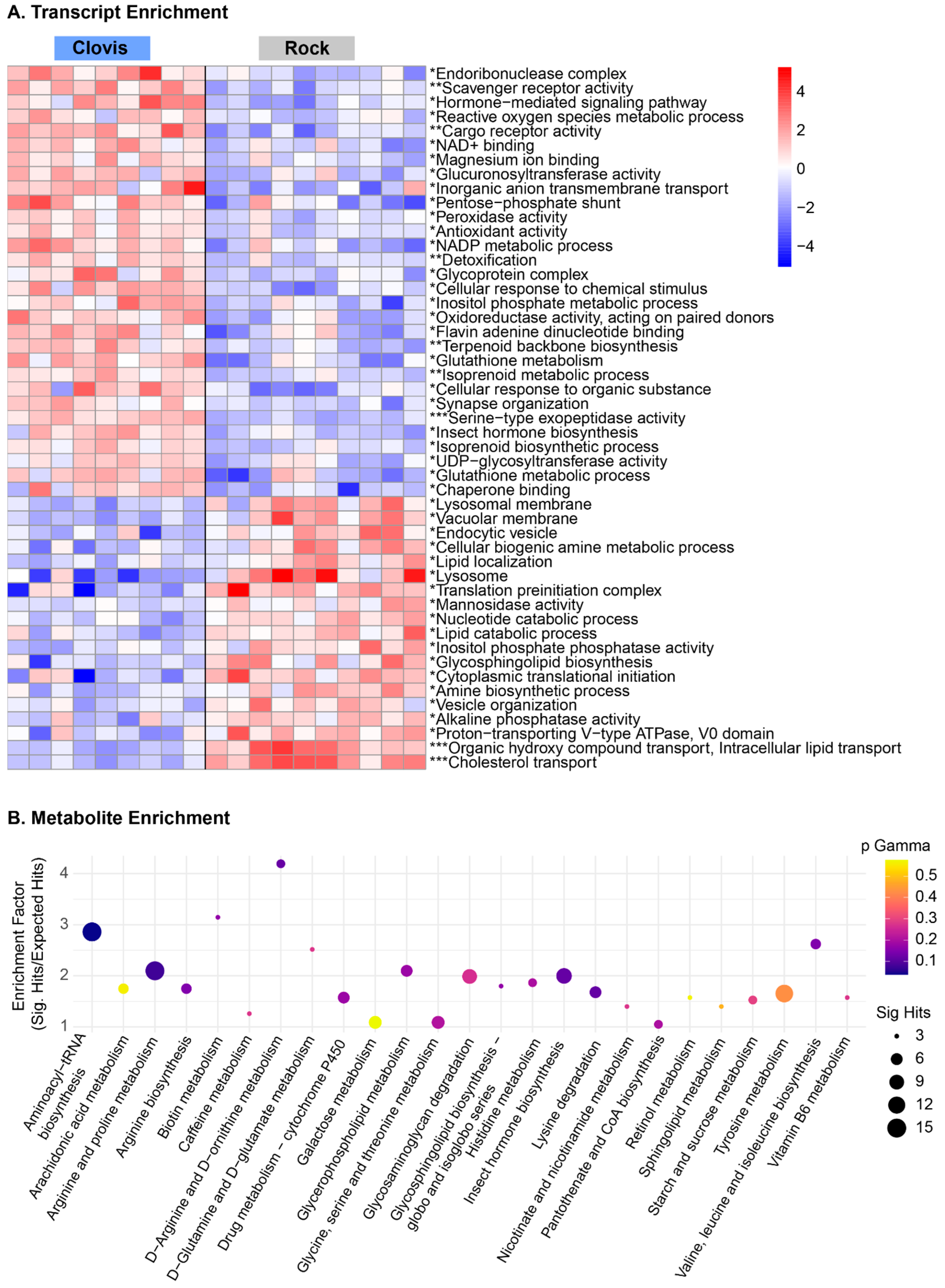
3.3. Metabolomic Profiles Reveal Enrichment in Pentose Phosphate Pathway Metabolites, Glutathione Metabolism and Lysolipids in Wild Ae. aegypti Relative to Rockefeller
3.4. Genes Associated with Detoxification Are Overexpressed in Clovis, While Immune and Catabolic Processes Are up in Rock
3.5. Metabolites Clarify Pathway Level Gene Expression Differences in Essential Metabolic Processes and Nervous System Organization
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- Kuno, G. Early History of Laboratory Breeding of Aedes aegypti (Diptera: Culicidae) Focusing on the Origins and Use of Selected Strains. J. Med. Entomol. 2010, 47, 957–971. [Google Scholar] [CrossRef] [PubMed]
- David, J.P.; Faucon, F.; Chandor-Proust, A.; Poupardin, R.; Riaz, M.A.; Bonin, A.; Navratil, V.; Reynaud, S. Comparative Analysis of Response to Selection with Three Insecticides in the Dengue Mosquito Aedes Aegypti Using MRNA Sequencing. BMC Genom. 2014, 15, 174. [Google Scholar] [CrossRef] [PubMed]
- Faucon, F.; Gaude, T.; Dusfour, I.; Navratil, V.; Corbel, V.; Juntarajumnong, W.; Girod, R.; Poupardin, R.; Boyer, F.; Reynaud, S.; et al. In the Hunt for Genomic Markers of Metabolic Resistance to Pyrethroids in the Mosquito Aedes Aegypti: An Integrated next-Generation Sequencing Approach. PLoS Negl. Trop. Dis. 2017, 11, e0005526. [Google Scholar] [CrossRef] [PubMed]
- Riaz, M.A.; Chandor-Proust, A.; Dauphin-Villemant, C.; Poupardin, R.; Jones, C.M.; Strode, C.; Régent-Kloeckner, M.; David, J.P.; Reynaud, S. Molecular Mechanisms Associated with Increased Tolerance to the Neonicotinoid Insecticide Imidacloprid in the Dengue Vector Aedes Aegypti. Aquat. Toxicol. 2013, 126, 326–337. [Google Scholar] [CrossRef] [PubMed]
- Strode, C.; Wondji, C.S.; David, J.-P.; Hawkes, N.J.; Lumjuan, N.; Nelson, D.R.; Drane, D.R.; Karunaratne, S.H.P.P.; Hemingway, J.; Black, W.C.; et al. Genomic analysis of detoxification genes in the mosquito Aedes aegypti. Insect Biochem. Mol. Biol. 2008, 38, 113–123. [Google Scholar] [CrossRef] [PubMed]
- Poupardin, R.; Riaz, M.A.; Jones, C.M.; Chandor-Proust, A.; Reynaud, S.; David, J.-P. Do Pollutants Affect Insecticide-Driven Gene Selection in Mosquitoes? Experimental Evidence from Transcriptomics. Aquat. Toxicol. 2012, 114–115, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Saavedra-Rodriguez, K.; Campbell, C.L.; Lenhart, A.; Penilla, P.; Lozano-Fuentes, S.; Black, W.C. 4th Exome-Wide Association of Deltamethrin Resistance in Aedes Aegypti from Mexico. Insect Mol. Biol. 2019, 28, 591–604. [Google Scholar] [CrossRef]
- Metzger, M.E.; Hardstone Yoshimizu, M.; Padgett, K.A.; Hu, R.; Kramer, V.L. Detection and Establishment of Aedes Aegypti and Aedes Albopictus (Diptera: Culicidae) Mosquitoes in California, 2011–2015. J. Med. Entomol. 2017, 54, 533–543. [Google Scholar] [CrossRef] [PubMed]
- Pless, E.; Gloria-Soria, A.; Evans, B.R.; Kramer, V.; Bolling, B.G.; Tabachnick, W.J.; Powell, J.R. Multiple Introductions of the Dengue Vector, Aedes Aegypti, into California. PLoS Negl. Trop. Dis. 2017, 11, e0005718. [Google Scholar] [CrossRef]
- Cornel, A.J.; Holeman, J.; Nieman, C.C.; Lee, Y.; Smith, C.; Amorino, M.; Brisco, K.K.; Barrera, R.; Lanzaro, G.C.; Mulligan, F.S., III. Surveillance, insecticide resistance and control of an invasive Aedes aegypti (Diptera: Culicidae) population in California. F1000Reserach 2016, 5, 194. [Google Scholar] [CrossRef]
- Mack, L.K.; Kelly, E.T.; Lee, Y.; Brisco, K.K.; Shen, K.V.; Zahid, A.; van Schoor, T.; Cornel, A.J.; Attardo, G.M. Frequency of Sodium Channel Genotypes and Association with Pyrethrum Knockdown Time in Populations of Californian Aedes Aegypti. Parasit. Vectors 2021, 14, 141. [Google Scholar] [CrossRef] [PubMed]
- Crawford, J.E.; Clarke, D.W.; Criswell, V.; Desnoyer, M.; Cornel, D.; Deegan, B.; Gong, K.; Hopkins, K.C.; Howell, P.; Hyde, J.S.; et al. Efficient Production of Male Wolbachia-Infected Aedes Aegypti Mosquitoes Enables Large-Scale Suppression of Wild Populations. Nat. Biotechnol. 2020, 38, 482–492. [Google Scholar] [CrossRef] [PubMed]
- Gloria-Soria, A.; Brown, J.E.; Kramer, V.; Hardstone Yoshimizu, M.; Powell, J.R. Origin of the Dengue Fever Mosquito, Aedes Aegypti, in California. PLoS Negl. Trop. Dis. 2014, 8, e3029. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.; Chong, J.; Zhou, G.; de Lima Morais, D.A.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.-É.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the Gap between Raw Spectra and Functional Insights. Nucleic Acids Res. 2021, 49, W388–W396. [Google Scholar] [CrossRef] [PubMed]
- Barupal, D.K.; Fiehn, O. Chemical Similarity Enrichment Analysis (ChemRICH) as Alternative to Biochemical Pathway Mapping for Metabolomic Datasets. Sci. Rep. 2017, 7, 14567. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.X.; Son, E.W.; Yao, R. IDEP: An Integrated Web Application for Differential Expression and Pathway Analysis of RNA-Seq Data. BMC Bioinform. 2018, 19, 534. [Google Scholar] [CrossRef] [PubMed]
- Furge, K.; Dykema, K. PGSEA: Parametric Gene Set Enrichment Analysis. In R Package Version 1480; R Core Team: Vienna, Austria, 2012. [Google Scholar]
- Liebman, K.A.; Billeter, S.A.; Yoshimizu, M.H.; Yang, F.; Metzger, M.E.; Schildhauer, S.; Payne, R.; Pakingan, M.J.; Hu, R.; Kramer, V.; et al. Identification of Molecular Determinants of Resistance to Pyrethroid Insecticides in Aedes Aegypti (Diptera: Culicidae) Populations in California, USA. J. Med. Entomol. 2019, 56, 1353–1358. [Google Scholar] [CrossRef]
- Kelly, E.T.; Mack, L.K.; Campos, M.; Grippin, C.; Chen, T.-Y.; Romero-Weaver, A.L.; Kosinski, K.J.; Brisco, K.K.; Collier, T.C.; Buckner, E.A.; et al. Evidence of Local Extinction and Reintroduction of Aedes Aegypti in Exeter, California. Front. Trop. Dis. 2021, 2, 703873. [Google Scholar] [CrossRef]
- Hardstone, M.C.; Huang, X.; Harrington, L.C.; Scott, J.G. Differences in Development, Glycogen, and Lipid Content Associated with Cytochrome P450-Mediated Permethrin Resistance in Culex Pipiens Quinquefasciatus (Diptera: Culicidae). J. Med. Entomol. 2010, 47, 188–198. [Google Scholar] [CrossRef]
- Dawaliby, R.; Trubbia, C.; Delporte, C.; Noyon, C.; Ruysschaert, J.-M.; Van Antwerpen, P.; Govaerts, C. Phosphatidylethanolamine Is a Key Regulator of Membrane Fluidity in Eukaryotic Cells. J. Biol. Chem. 2016, 291, 3658–3667. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.-X.; Zhu, M.-F.; Wang, N.; Huang, Y.-J.; Zhang, M.-J.; Zhang, C.; Ali, S.A.; Zhou, W.-W.; Zhang, C.; Mao, C.; et al. Neutral Ceramidase Is Required for the Reproduction of Brown Planthopper, Nilaparvata Lugens (Stål). Front. Physiol. 2021, 12, 629532. [Google Scholar] [CrossRef]
- Roszczyc-Owsiejczuk, K.; Zabielski, P. Sphingolipids as a Culprit of Mitochondrial Dysfunction in Insulin Resistance and Type 2 Diabetes. Front. Endocrinol. 2021, 12, 635175. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.T.; Ramesh, T.; Toh, X.R.; Nguyen, L.N. Emerging Roles of Lysophospholipids in Health and Disease. Prog. Lipid Res. 2020, 80, 101068. [Google Scholar] [CrossRef] [PubMed]
- Stanley, D.; Kim, Y. Why Most Insects Have Very Low Proportions of C20 Polyunsaturated Fatty Acids: The Oxidative Stress Hypothesis. Arch. Insect Biochem. Physiol. 2020, 103, e21622. [Google Scholar] [CrossRef] [PubMed]
- Hansen, I.A.; Boudko, D.Y.; Shiao, S.-H.; Voronov, D.A.; Meleshkevitch, E.A.; Drake, L.L.; Aguirre, S.E.; Fox, J.M.; Attardo, G.M.; Raikhel, A.S. AaCAT1 of the Yellow Fever Mosquito, Aedes Aegypti: A Novel Histidine-Specific Amino Acid Transporter from the SLC7 Family. J. Biol. Chem. 2011, 286, 10803–10813. [Google Scholar] [CrossRef] [PubMed]
- Matthews, B.J.; McBride, C.S.; DeGennaro, M.; Despo, O.; Vosshall, L.B. The Neurotranscriptome of the Aedes Aegypti Mosquito. BMC Genom. 2016, 17, 32. [Google Scholar] [CrossRef] [PubMed]
- Han, Q.; Beerntsen, B.T.; Li, J. The Tryptophan Oxidation Pathway in Mosquitoes with Emphasis on Xanthurenic Acid Biosynthesis. J. Insect Physiol. 2007, 53, 254–263. [Google Scholar] [CrossRef] [PubMed]
- Lima, V.L.A.; Dias, F.; Nunes, R.D.; Pereira, L.O.; Santos, T.S.R.; Chiarini, L.B.; Ramos, T.D.; Silva-Mendes, B.J.; Perales, J.; Valente, R.H.; et al. The Antioxidant Role of Xanthurenic Acid in the Aedes Aegypti Midgut during Digestion of a Blood Meal. PLoS ONE 2012, 7, e38349. [Google Scholar] [CrossRef]
- Zhu, Y.; Zhang, R.; Zhang, B.; Zhao, T.; Wang, P.; Liang, G.; Cheng, G. Blood Meal Acquisition Enhances Arbovirus Replication in Mosquitoes through Activation of the GABAergic System. Nat. Commun. 2017, 8, 1262. [Google Scholar] [CrossRef]
- Derilus, D.; Impoinvil, L.M.; Muturi, E.J.; McAllister, J.; Kenney, J.; Massey, S.E.; Hemme, R.; Kothera, L.; Lenhart, A. Comparative Transcriptomic Analysis of Insecticide-Resistant Aedes Aegypti from Puerto Rico Reveals Insecticide-Specific Patterns of Gene Expression. Genes 2023, 14, 1626. [Google Scholar] [CrossRef] [PubMed]
- Mack, L.K.; Attardo, G.M. Time-Series Analysis of Transcriptomic Changes Due to Permethrin Exposure Reveals That Aedes Aegypti Undergoes Detoxification Metabolism over 24 h. Sci. Rep. 2023, 13, 16564. [Google Scholar] [CrossRef] [PubMed]
- Soudi, S.; Crepeau, M.; Collier, T.C.; Lee, Y.; Cornel, A.J.; Lanzaro, G.C. Genomic Signatures of Local Adaptation in Recent Invasive Aedes Aegypti Populations in California. BMC Genomics 2023, 24, 311. [Google Scholar] [CrossRef] [PubMed]
- Carney, T.D.; Hebalkar, R.Y.; Edeleva, E.; Çiçek, I.Ö.; Shcherbata, H.R. Signaling through the Dystrophin Glycoprotein Complex Affects the Stress-Dependent Transcriptome in Drosophila. Dis. Model. Mech. 2023, 16, dmm049862. [Google Scholar] [CrossRef] [PubMed]
- Grammont, M.; Irvine, K.D. Fringe and Notch Specify Polar Cell Fate during Drosophila Oogenesis. Development 2001, 128, 2243–2253. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Sexton, T.R.; Venard, C.; Giedt, M.; Guo, Q.; Chen, Q.; Harrison, D.A. Pleiotropy of the Drosophila JAK Pathway Cytokine Unpaired 3 in Development and Aging. Dev. Biol. 2014, 395, 218–231. [Google Scholar] [CrossRef] [PubMed]
- Mayoral, J.G.; Nouzova, M.; Navare, A.; Noriega, F.G. NADP+-Dependent Farnesol Dehydrogenase, a Corpora Allata Enzyme Involved in Juvenile Hormone Synthesis. Proc. Natl. Acad. Sci. USA 2009, 106, 21091–21096. [Google Scholar] [CrossRef] [PubMed]
- Ekoka, E.; Maharaj, S.; Nardini, L.; Dahan-Moss, Y.; Koekemoer, L.L. 20-Hydroxyecdysone (20E) Signaling as a Promising Target for the Chemical Control of Malaria Vectors. Parasit. Vectors 2021, 14, 86. [Google Scholar] [CrossRef] [PubMed]
- Gruntenko, N.E.; Rauschenbach, I.Y. Interplay of JH, 20E and Biogenic Amines under Normal and Stress Conditions and Its Effect on Reproduction. J. Insect Physiol. 2008, 54, 902–908. [Google Scholar] [CrossRef]
- Freeman, J.C.; Smith, L.B.; Silva, J.J.; Fan, Y.; Sun, H.; Scott, J.G. Fitness Studies of Insecticide Resistant Strains: Lessons Learned and Future Directions. Pest Manag. Sci. 2021, 77, 3847–3856. [Google Scholar] [CrossRef]
- Brito, L.P.; Linss, J.G.B.; Lima-Camara, T.N.; Belinato, T.A.; Peixoto, A.A.; Lima, J.B.P.; Valle, D.; Martins, A.J. Assessing the Effects of Aedes Aegypti Kdr Mutations on Pyrethroid Resistance and Its Fitness Cost. PLoS ONE 2013, 8, e60878. [Google Scholar] [CrossRef] [PubMed]
- Smith, L.B.; Silva, J.J.; Chen, C.; Harrington, L.C.; Scott, J.G. Fitness Costs of Individual and Combined Pyrethroid Resistance Mechanisms, Kdr and CYP-Mediated Detoxification, in Aedes aegypti. PLoS Negl. Trop. Dis. 2021, 15, e0009271. [Google Scholar] [CrossRef] [PubMed]
- Partridge, L.; Gems, D.; Withers, D.J. Sex and Death: What Is the Connection? Cell 2005, 120, 461–472. [Google Scholar] [CrossRef] [PubMed]
- Grandison, R.C.; Piper, M.D.W.; Partridge, L. Amino-Acid Imbalance Explains Extension of Lifespan by Dietary Restriction in Drosophila. Nature 2009, 462, 1061–1064. [Google Scholar] [CrossRef] [PubMed]
- Hammerschmidt, P.; Brüning, J.C. Contribution of Specific Ceramides to Obesity-Associated Metabolic Diseases. Cell. Mol. Life Sci. 2022, 79, 395. [Google Scholar] [CrossRef] [PubMed]
- Baumann, A.A.; Benoit, J.B.; Michalkova, V.; Mireji, P.; Attardo, G.M.; Moulton, J.K.; Wilson, T.G.; Aksoy, S. Juvenile Hormone and Insulin Suppress Lipolysis between Periods of Lactation during Tsetse Fly Pregnancy. Mol. Cell. Endocrinol. 2013, 372, 30–41. [Google Scholar] [CrossRef]
- Zhao, H.; Wang, T. PE Homeostasis Rebalanced through Mitochondria-ER Lipid Exchange Prevents Retinal Degeneration in Drosophila. PLoS Genet. 2020, 16, e1009070. [Google Scholar] [CrossRef]

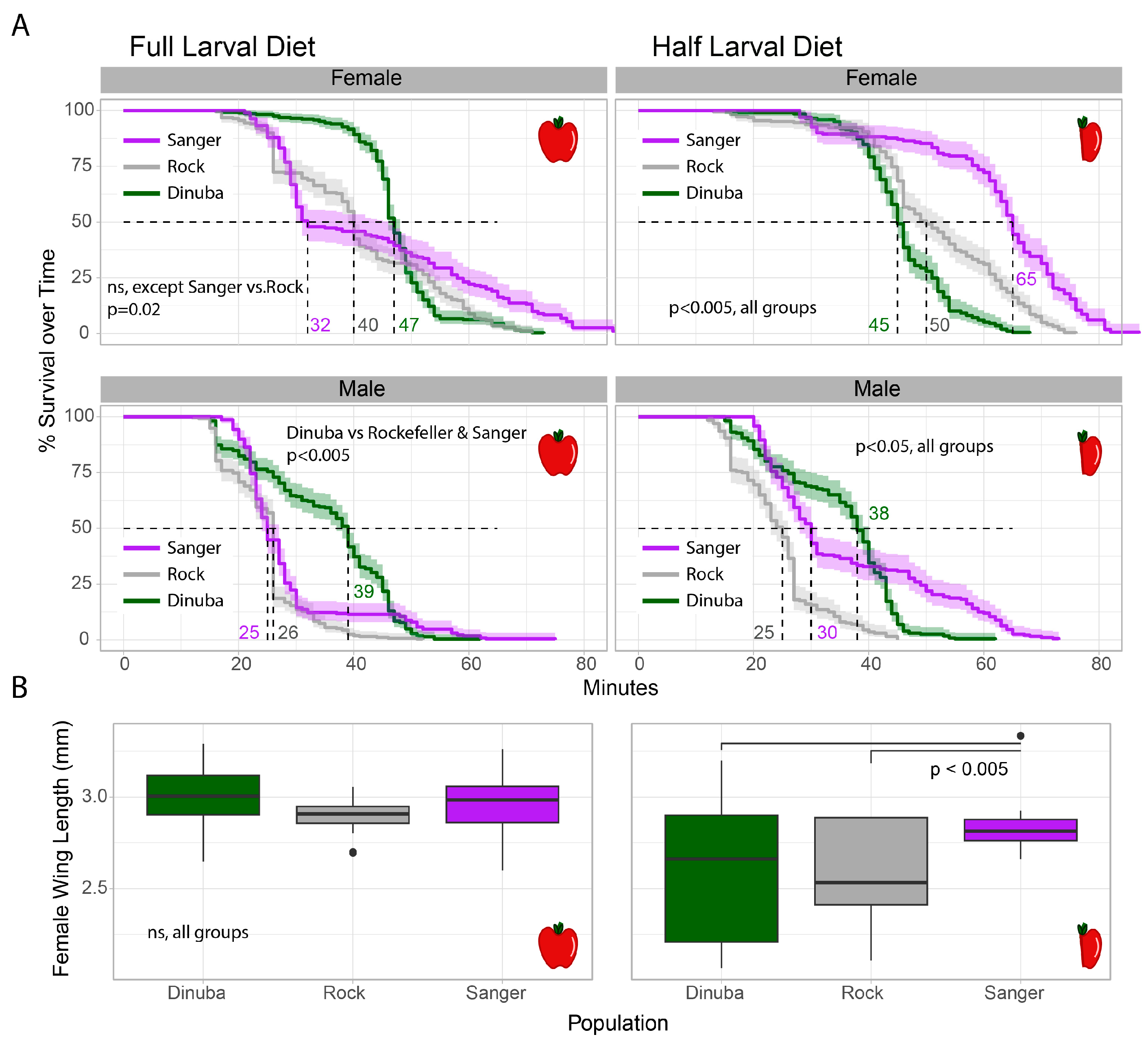

| Day | Full Diet (mg) | Half Diet (mg) |
|---|---|---|
| 1 | 372 mg (2 pinches) | 186 mg (1 pinch) |
| 2 | 46.5 mg (1 drop) | No food |
| 3 | 93 mg (2 drops) | 46.5 mg (1 drop) |
| 4 | 186 mg (4 drops) | 93 mg (2 drops) |
| 5 | 186 mg (4 drops) | 93 mg (2 drops) |
| 6 | 372 mg (8 drops) | 186 mg (4 drops) |
| 7 | 279 mg (6 drops) | 139.5 mg (3 drops) |
| 8 | 186 mg (4 drops) | 93 mg (2 drops) |
| GO Molecular Function | |||
|---|---|---|---|
| Direction | Adj. Pvalue | Genes (n) | Pathways |
| Up in Rock | 3.8 × 10−4 | 21 | Peptidase activity |
| 2.6 × 10−3 | 25 | Transition metal ion binding | |
| 6.6 × 10−3 | 34 | Hydrolase activity | |
| Up in Clovis | 6.5 × 10−5 | 13 | Tetrapyrrole binding/iron binding/monooxygenase activity |
| 2.4 × 10−3 | 2 | Farnesol dehydrogenase activity | |
| 3.1 × 10−3 | 28 | Transition metal ion binding/oxidoreductase activity | |
| GO Biological Processes | |||
| Up in Rock | 2.3 × 10−3 | 21 | Proteolysis |
| 2.3 × 10−3 | 3 | Sterol transport, intracellular lipid transport | |
| 2.6 × 10−3 | 3 | Defense response to bacterium | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kelly, E.T.; Mack, L.K.; Attardo, G.M. Exploring the Wilderness within: An Integrative Metabolomics and Transcriptomics Study on Near-Wild and Colonized Aedes aegypti. Insects 2024, 15, 507. https://doi.org/10.3390/insects15070507
Kelly ET, Mack LK, Attardo GM. Exploring the Wilderness within: An Integrative Metabolomics and Transcriptomics Study on Near-Wild and Colonized Aedes aegypti. Insects. 2024; 15(7):507. https://doi.org/10.3390/insects15070507
Chicago/Turabian StyleKelly, Erin Taylor, Lindsey K. Mack, and Geoffrey M. Attardo. 2024. "Exploring the Wilderness within: An Integrative Metabolomics and Transcriptomics Study on Near-Wild and Colonized Aedes aegypti" Insects 15, no. 7: 507. https://doi.org/10.3390/insects15070507
APA StyleKelly, E. T., Mack, L. K., & Attardo, G. M. (2024). Exploring the Wilderness within: An Integrative Metabolomics and Transcriptomics Study on Near-Wild and Colonized Aedes aegypti. Insects, 15(7), 507. https://doi.org/10.3390/insects15070507







