Microbiomes of Two Pest Fly Species of Pennsylvania Mushroom Houses
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection Sites and Sampling
2.2. Sample Processing for Bacterial 16S rRNA Sequencing
2.3. Analysis of Bacterial Communities of Flies
2.4. Post-Illumina Sequence Confirmation and Phylogenetic Placement of Wolbachia Sequences
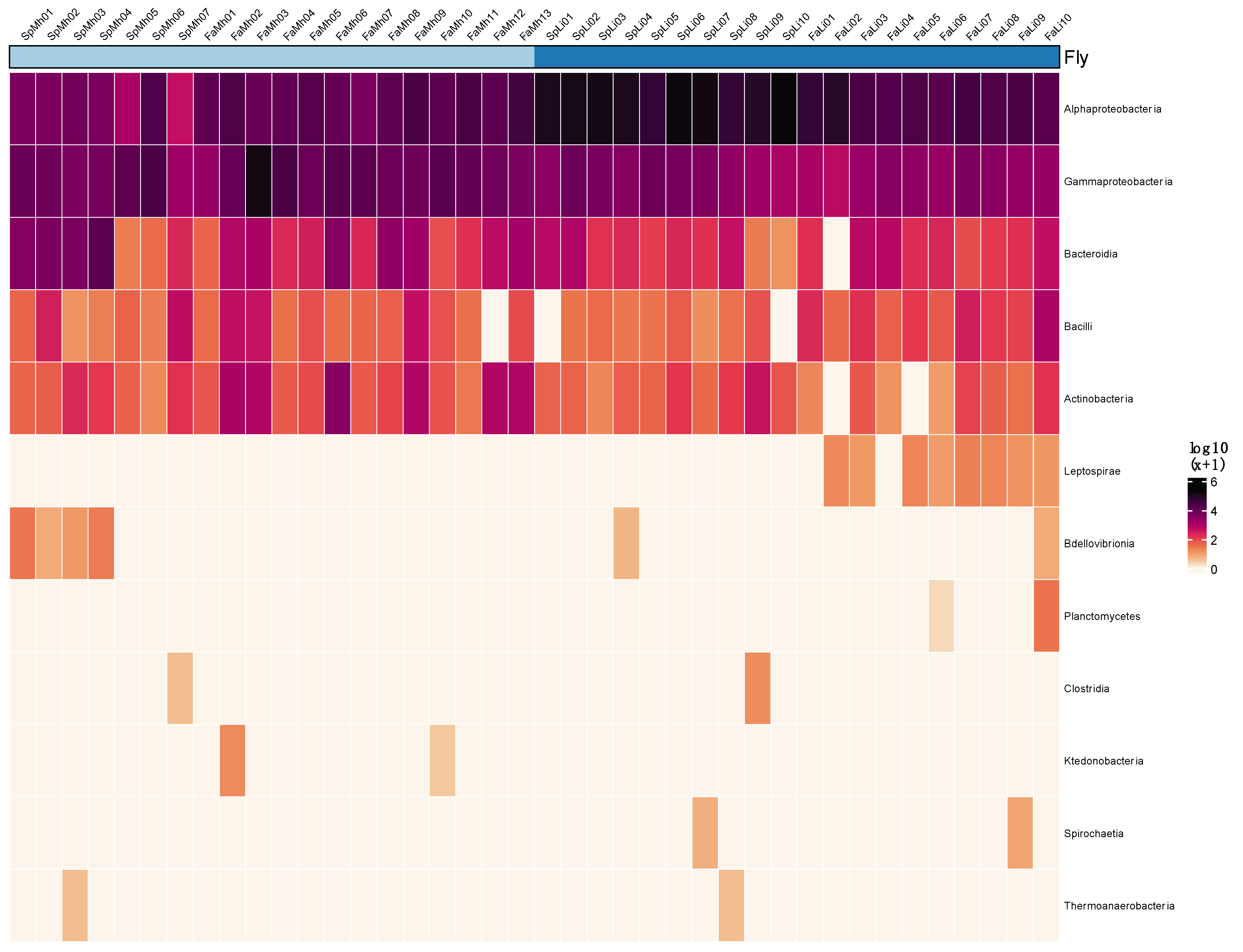
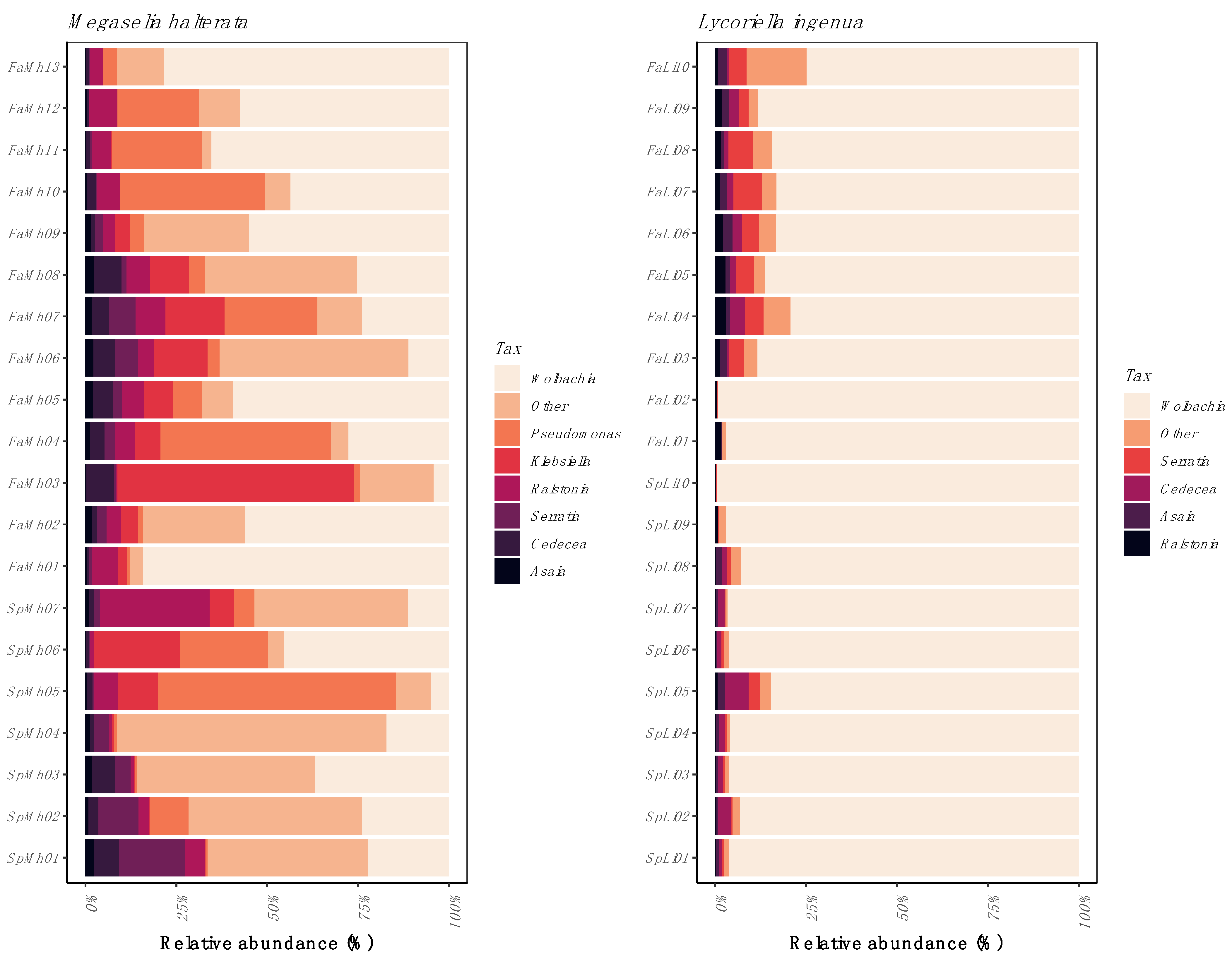
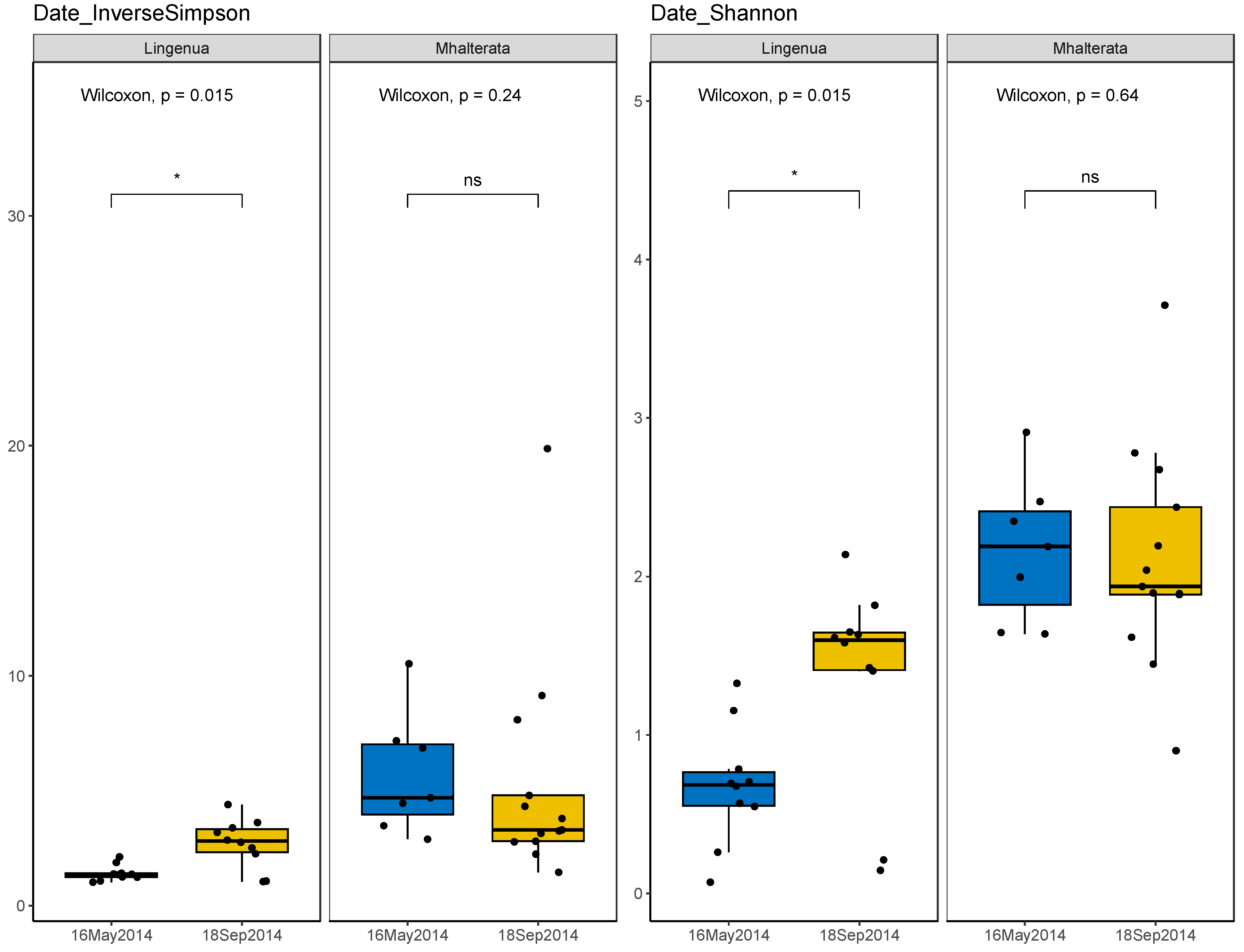
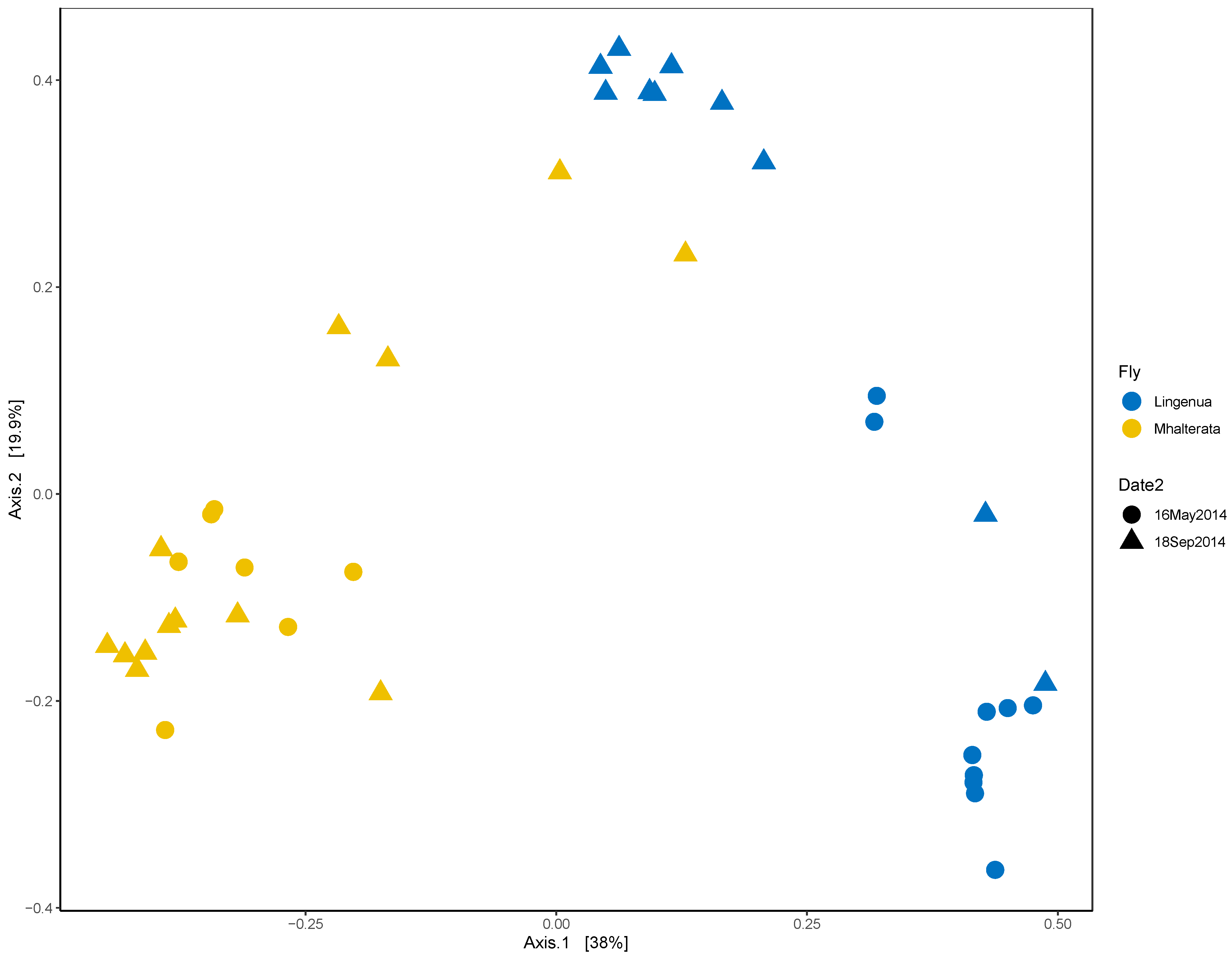
3. Results
3.1. Bacterial Community Composition
3.2. Bacterial Diversity
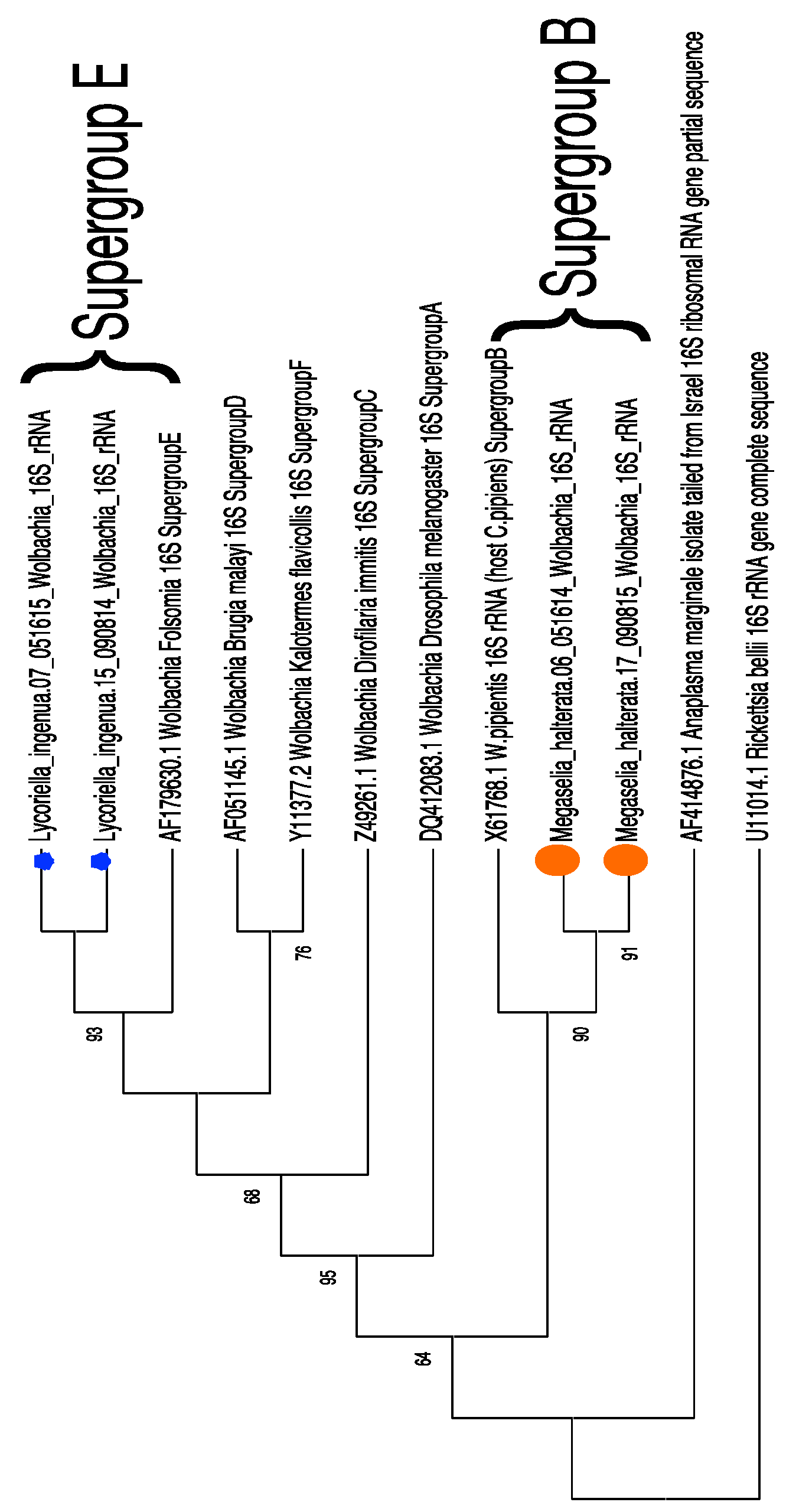
3.3. Sequence Confirmation and Phylogenetic Placement of Wolbachia Sequences
4. Discussion
4.1. Limitations of the Study
4.2. Future Studies
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Grimaldi, D.; Jaenike, J. Competition in Natural Populations of Mycophagous Drosophila. Ecology 1984, 65, 1113–1120. [Google Scholar] [CrossRef]
- Jakovlev, J. Fungal Hosts of Mycetophilids (Diptera: Sciaroidea Excluding Sciaridae): A Review. Mycology 2012, 3, 11–23. [Google Scholar] [CrossRef]
- Bunyard, B.A. Biodiversity and Ecology of Mycophagous Diptera in Northeastern Ohio. Proc. Entomol. Soc. Wash. 2003, 105, 847–858. [Google Scholar]
- Stamets, P.; Chilton, J.S. The Mushroom Cultivator: A Practical Guide to Growing Mushrooms at Home; Agarikon Press: Houston, TX, USA; Western Distribution by Homestead Book Co.: Olympia, WA, USA; Seattle, WA, USA, 1983; ISBN 978-0-9610798-0-2. [Google Scholar]
- Willette, A.; Van Slambrook, L.; Hollingsworth, C. Mushroom Pests: Mushroom-Mushroom Fly. In Pacific Northwest Insect Management Handbook [Online]; Oregon State University: Corvallis, OR, USA, 2023. [Google Scholar]
- Wuest, P.J.; Bengtson, G.D.; Schisler, L.C. Pennsylvania State University College of Agricultural Sciences Penn State Handbook for Commercial Mushroom Growers: A Compendium of Scientific and Technical Information Useful to Mushroom Farmers; Penn State, College of Agricultural Sciences University Park: University Park, PA, USA, 2003. [Google Scholar]
- Jess, S.; Bingham, J.F.W. Biological Control of Sciarid and Phorid Pests of Mushroom with Predatory Mites from the Genus Hypoaspis (Acari: Hypoaspidae) and the Entomopathogenic Nematode Steinernema feltiae. Bull. Entomol. Res. 2004, 94, 159–167. [Google Scholar] [CrossRef]
- Scheepmaker, J.W.A.; Geels, F.P.; Van Griensven, L.J.L.D.; Smits, P.H. Substrate Dependent Larval Development and Emergence of the Mushroom Pests Lycoriella auripila and Megaselia halterata. Entomol. Exp. Appl. 1996, 79, 329–334. [Google Scholar] [CrossRef]
- Mazin, M.; Andreadis, S.S.; Jenkins, N.E.; Cloonan, K.R.; Baker, T.C.; Rajotte, E.G. Activity and Distribution of the Mushroom Phorid Fly, Megaselia halterata, in and around Commercial Mushroom Farms. Entomol. Exp. Appl. 2019, 167, 389–395. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global Patterns of 16S rRNA Diversity at a Depth of Millions of Sequences per Sample. Proc. Natl. Acad. Sci. USA 2011, 108, 4516–4522. [Google Scholar] [CrossRef] [PubMed]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Callahan, B.J.; Sankaran, K.; Fukuyama, J.A.; McMurdie, P.J.; Holmes, S.P. Bioconductor Workflow for Microbiome Data Analysis: From Raw Reads to Community Analyses. F1000Research 2016, 5, 1492. [Google Scholar] [CrossRef]
- Lahti, L.; Shetty, S. Microbiome R Package; 2012–2019. Available online: http://microbiome.github.io (accessed on 21 May 2024).
- Ernst, F.G.M.; Shetty, S.; Borman, T.; Lahti, L. Mia: Microbiome Analysis. 2023. Available online: https://github.com/microbiome/mia (accessed on 21 May 2024).
- Barnett, D.; Arts, I.; Penders, J. microViz: An R Package for Microbiome Data Visualization and Statistics. J. Open Source Softw. 2021, 6, 3201. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Werren, J.H.; Windsor, D.M. Wolbachia Infection Frequencies in Insects: Evidence of a Global Equilibrium? Proc. R. Soc. Lond. B Biol. Sci. 2000, 267, 1277–1285. [Google Scholar] [CrossRef]
- Sakamoto, J.M.; Silva Diaz, G.E.; Wagner, E.A. Bacterial Communities of Ixodes Scapularis from Central Pennsylvania, USA. Insects 2020, 11, 718. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More Models, New Heuristics and Parallel Computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Abou-Zeid, M.A. Pathogenic Variation in Isolates of Pseudomonas Causing the Brown Blotch of Cultivated Mushroom, Agaricus bisporus. Braz. J. Microbiol. Publ. Braz. Soc. Microbiol. 2012, 43, 1137–1146. [Google Scholar] [CrossRef]
- Eastwood, D.; Green, J.; Grogan, H.; Burton, K. Viral Agents Causing Brown Cap Mushroom Disease of Agaricus bisporus. Appl. Environ. Microbiol. 2015, 81, 7125–7134. [Google Scholar] [CrossRef] [PubMed]
- Largeteau, M.L.; Savoie, J.-M. Microbially Induced Diseases of Agaricus bisporus: Biochemical Mechanisms and Impact on Commercial Mushroom Production. Appl. Microbiol. Biotechnol. 2010, 86, 63–73. [Google Scholar] [CrossRef]
- McGee, C.F. Microbial Ecology of the Agaricus bisporus Mushroom Cropping Process. Appl. Microbiol. Biotechnol. 2018, 102, 1075–1083. [Google Scholar] [CrossRef]
- Rinker, D.L.; Finley, R.J.; Wuest, P.J.; Snetsinger, R.; Tetrault, R.; Royse, D.J. The influence of phorid and sciarid populations on mushroom yield at commercial mushroom farms in Pennsylvania. Proc. Pa. Acad. Sci. 1984, 58, 70–72. [Google Scholar]
- Konecka, E.; Olszanowski, Z. Wolbachia Supergroup E Found in Hypochthonius rufulus (Acari: Oribatida) in Poland. Infect. Genet. Evol. 2021, 91, 104829. [Google Scholar] [CrossRef] [PubMed]
- Konecka, E.; Olszanowski, Z.; Koczura, R. Wolbachia of Phylogenetic Supergroup E Identified in Oribatid Mite Gustavia microcephala (Acari: Oribatida). Mol. Phylogenet Evol. 2019, 135, 230–235. [Google Scholar] [CrossRef] [PubMed]
- Schütte, C.; Dicke, M. Verified and Potential Pathogens of Predatory Mites (Acari: Phytoseiidae). Exp. Appl. Acarol. 2008, 46, 307–328. [Google Scholar] [CrossRef] [PubMed]
- Majumder, R.; Sutcliffe, B.; Taylor, P.W.; Chapman, T.A. Microbiome of the Queensland Fruit Fly through Metamorphosis. Microorganisms 2020, 8, 795. [Google Scholar] [CrossRef] [PubMed]
- Delmas, J. Cultivation in Western Countries: Growing in Caves. In The Biology and Cultivation of Edible Mushrooms; Elsevier: New York, NY, USA; Academic Press, Inc.: New York, NY, USA, 1978; pp. 251–298. ISBN 978-0-12-168050-3. [Google Scholar]
- Insect Pest Control for the Amateur Mushroom Grower; E-347; Bureau of Entomology and Plant Quarantine: USA, 1935.
- Shamshad, A. The Development of Integrated Pest Management for the Control of Mushroom Sciarid Flies, Lycoriella ingenua (Dufour) and Bradysia ocellaris (Comstock), in Cultivated Mushrooms: IPM for Control of Sciarid Flies in Mushrooms. Pest Manag. Sci. 2010, 66, 1063–1074. [Google Scholar] [CrossRef] [PubMed]
- Snetsinger, R.; Wuest, P. A Historical Perspective on Mushroom Arthropod Pest Control. In Developments in Crop Science; Elsevier: Amsterdam, The Netherlands, 1987; Volume 10, pp. 641–648. ISBN 978-0-444-42747-2. [Google Scholar]
- Beyer, D.M.; Rinker, D. Nematode Problems in Mushroom Cultivation and Their Sustainable Management. In Nematode Diseases of Crops and Their Sustainable Management; Elsevier: Amsterdam, The Netherlands, 2023; pp. 337–347. ISBN 978-0-323-91226-6. [Google Scholar]
- Andreadis, S.S.; Cloonan, K.R.; Bellicanta, G.S.; Paley, K.; Pecchia, J.; Jenkins, N.E. Efficacy of Beauveria bassiana Formulations against the Fungus Gnat Lycoriella ingenua. Biol. Control 2016, 103, 165–171. [Google Scholar] [CrossRef]
- Cloonan, K.R.; Andreadis, S.S.; Chen, H.; Jenkins, N.E.; Baker, T.C. Attraction, Oviposition and Larval Survival of the Fungus Gnat, Lycoriella ingenua, on Fungal Species Isolated from Adults, Larvae, and Mushroom Compost. PLoS ONE 2016, 11, e0167074. [Google Scholar] [CrossRef]
- Liu, S.; Xie, J.; Cheng, J.; Li, B.; Chen, T.; Fu, Y.; Li, G.; Wang, M.; Jin, H.; Wan, H.; et al. Fungal DNA Virus Infects a Mycophagous Insect and Utilizes It as a Transmission Vector. Proc. Natl. Acad. Sci. USA 2016, 113, 12803–12808. [Google Scholar] [CrossRef]
| SampleID | Fly 1 | Date | SampleNo |
|---|---|---|---|
| C09Ph051614Mh01 | M. halterata | 16 May 2014 | MhSp01 |
| C10Ph051614Mh02 | M. halterata | 16 May 2014 | MhSp02 |
| C11Ph051614Mh03 | M. halterata | 16 May 2014 | MhSp03 |
| C12Ph051614Mh04 | M. halterata | 16 May 2014 | MhSp04 |
| D01Ph051614Mh05 | M. halterata | 16 May 2014 | MhSp05 |
| D02Ph051614Mh06 | M. halterata | 16 May 2014 | MhSp06 |
| D03Ph051614Mh07 | M. halterata | 16 May 2014 | MhSp07 |
| D04Ph091814Mh04 | M. halterata | 18 September 2014 | MhFa01 |
| D05Ph091814Mh05 | M. halterata | 18 September 2014 | MhFa02 |
| D06Ph091814Mh06 | M. halterata | 18 September 2014 | MhFa3 |
| D07Ph091814Mh11 | M. halterata | 18 September 2014 | MhFa04 |
| D08Ph091814Mh12 | M. halterata | 18 September 2014 | MhFa05 |
| D09Ph091814Mh13 | M. halterata | 18 September 2014 | MhFa06 |
| D10Ph091814Mh14 | M. halterata | 18 September 2014 | MhFa07 |
| D11Ph091814Mh15 | M. halterata | 18 September 2014 | MhFa08 |
| D12Ph091814Mh16 | M. halterata | 18 September 2014 | MhFa09 |
| E01Ph091814Mh17 | M. halterata | 18 September 2014 | MhFa10 |
| E02Ph091814Mh18 | M. halterata | 18 September 2014 | MhFa11 |
| E03Ph091814Mh19 | M. halterata | 18 September 2014 | MhFa12 |
| E04Ph091814Mh20 | M. halterata | 18 September 2014 | MhFa13 |
| E05Sc051614Li01 | L. ingenua | 16 May 2014 | LiSp01 |
| E06Sc051614Li02 | L. ingenua | 16 May 2014 | LiSp02 |
| E07Sc051614Li03 | L. ingenua | 16 May 2014 | LiSp03 |
| E08Sc051614Li04 | L. ingenua | 16 May 2014 | LiSp04 |
| E09Sc051614Li05 | L. ingenua | 16 May 2014 | LiSp05 |
| E10Sc051614Li06 | L. ingenua | 16 May 2014 | LiSp06 |
| E11Sc051614Li07 | L. ingenua | 16 May 2014 | LiSp07 |
| E12Sc051614Li08 | L. ingenua | 16 May 2014 | LiSp08 |
| F01Sc051614Li09 | L. ingenua | 16 May 2014 | LiSp09 |
| F02Sc051614Li10 | L. ingenua | 16 May 2014 | LiSp10 |
| F03Sc091814Li01 | L. ingenua | 18 September 2014 | LiFa01 |
| F04Sc091814Li02 | L. ingenua | 18 September 2014 | LiFa02 |
| F05Sc091814Li03 | L. ingenua | 18 September 2014 | LiFa03 |
| F06Sc091814Li04 | L. ingenua | 18 September 2014 | LiFa04 |
| F07Sc091814Li05 | L. ingenua | 18 September 2014 | LiFa05 |
| F08Sc091814Li06 | L. ingenua | 18 September 2014 | LiFa06 |
| F09Sc091814Li07 | L. ingenua | 18 September 2014 | LiFa07 |
| F10Sc091814Li08 | L. ingenua | 18 September 2014 | LiFa08 |
| F11Sc091814Li09 | L. ingenua | 18 September 2014 | LiFa09 |
| F12Sc091814Li10 | L. ingenua | 18 September 2014 | LiFa10 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sakamoto, J.M.; Shikano, I.; Rasgon, J.L. Microbiomes of Two Pest Fly Species of Pennsylvania Mushroom Houses. Insects 2024, 15, 525. https://doi.org/10.3390/insects15070525
Sakamoto JM, Shikano I, Rasgon JL. Microbiomes of Two Pest Fly Species of Pennsylvania Mushroom Houses. Insects. 2024; 15(7):525. https://doi.org/10.3390/insects15070525
Chicago/Turabian StyleSakamoto, Joyce M., Ikkei Shikano, and Jason L. Rasgon. 2024. "Microbiomes of Two Pest Fly Species of Pennsylvania Mushroom Houses" Insects 15, no. 7: 525. https://doi.org/10.3390/insects15070525
APA StyleSakamoto, J. M., Shikano, I., & Rasgon, J. L. (2024). Microbiomes of Two Pest Fly Species of Pennsylvania Mushroom Houses. Insects, 15(7), 525. https://doi.org/10.3390/insects15070525





