Simple Summary
This study surveyed the diversity of yeast species in the guts of various insects from three Bulgarian National Parks, Vitosha, Rila, and Pirin. Insects from a wide range of taxonomic groups, including Coleoptera, Orthoptera, Lepidoptera, Hymenoptera, Dermaptera, and Collembola, were collected. Yeast strains were identified using DNA barcoding of ribosomal markers. This study found 89 ascomycetous and 18 basidiomycetous yeast isolates. Forty-two percent of the yeast isolates represented novel yeast species. Our study confirms that insects remain a rich source of unknown yeast species.
Abstract
In this study, we conducted a comprehensive survey aimed at assessing the diversity of yeast species inhabiting the guts of various insect species collected mainly from two Bulgarian National Parks, namely, Rila, and Pirin. The insect specimens encompass a broad taxonomic spectrum, including representatives from Coleoptera, Orthoptera, Lepidoptera, Hymenoptera, Dermaptera, Isopoda, and Collembola. Yeast strains were identified with DNA barcoding using the ribosomal markers, specifically, the D1/D2 domains of the ribosomal large subunit (LSU) and the internal transcribed spacers regions ITS 1 + 2 (ITS). The analysis unveiled the presence of 89 ascomycetous and 18 basidiomycetous yeast isolates associated with the insect specimens. Furthermore, our study identified 18 hitherto unknown yeast species.
1. Introduction
In various regions around the world, including Bulgaria, environmental and climate changes have led to a rapid decline in biodiversity [1]. This critical situation emphasizes the necessity for conducting comprehensive biodiversity research and conservation efforts. Yeasts have ecological, medical, and biotechnological significance, inhabiting various natural ecosystems, such as soil, water, plants, and animals [2,3]. Insects, constituting the largest phylum by species diversity with an estimated 5.5 million species, stand out among animal taxa as a well-known but not well explored habitat for yeasts [4]. In forest ecosystems, insects play a pivotal role in nutrient cycling, pollination, and plant seed dispersal. Yeasts and insects engage in various symbiotic interactions. This includes mutualism, wherein yeasts contribute to nutrient breakdown through enzymatic processes [5], furnish insects with nutrients and essential vitamins, and provide antimicrobial defense [6,7,8]. In exchange, insects offer a secure niche for yeast reproduction, act as transportation agents, and facilitate access to food sources in natural environments [6,9]. Despite the well-documented importance of these associations, our knowledge of yeast biodiversity associated with insects in various regions of our planet remains limited. Most studies on yeast biodiversity have been concentrated in regions such as Western Europe, Japan, and North America [10,11]. Moreover, most yeast strains maintained in world culture collections were isolated from food products, plants, and human sources [3].
Studies on yeasts associated with insects have revealed the presence of many unknown species in insect guts [12,13,14,15,16,17,18,19,20,21,22]. It has been suggested that further exploration of insects in less-studied regions may lead to the discovery of additional novelties [11]. For instance, recent publications [23,24] have unveiled a significant diversity of non-Saccharomyces yeast species associated with dung beetles collected from Botswana, a scarcely studied region in Africa.
Bulgaria, one of Europe’s most biologically diverse countries, owes its rich biodiversity to a varied climate and geography. It is located in the eastern part of the Balkan Peninsula and belongs to the Holarctic floristic region. The country’s complex geological history, mountains with highly dissected topography, river valleys, and basin fields, as well as the influence of marine basins in the east and south, contribute to a diverse climate that creates conditions for varied vegetation and rich flora. Approximately 35% of Bulgaria’s land area is covered by forests, housing around 4030 vascular plant species [25], including 172 endemics [26]. The country also hosts over 4870 known fungal species [27], along with 20,900 species of insects [28]. This wealth of flora and fauna provides potential habitats for numerous yet undiscovered yeast species with possible important biotechnological properties.
The systematics of yeasts has evolved alongside major advancements in biological science. Initially, microscopy facilitated yeast species identification through the examination of phenotypic markers, including cell and colony morphology. However, these markers often lack resolution due to the extensive biodiversity of yeast. Advances in biochemistry enabled the utilization of biochemical characteristics such as the assimilation and fermentation of various carbohydrates, as well as the assimilation of nitrogen compounds. Progress in molecular biology has enabled the use of proteins, DNA, and RNA as markers. In 1951, the complete amino acid sequence of the insulin molecule was determined [29]. For the first time, it was shown that every protein has a unique sequence with 20 different amino acids at each position. The number of protein characteristics, for comparison, equals to 20N (20 amino acids, N length of the protein). In 1962, based on measurements of the rate of mutations in hemoglobin molecules of closely related species, the concept of a molecular clock was introduced [30]. This concept is based on the idea that mutations accumulate at a fairly consistent rate, serving as a “clock” that can be used to estimate the time since two species shared a common ancestor. From then on, it became possible to calculate the evolutionary distance between organisms and construct phylogenetic trees based on molecular sequences. In 1977, biology was revolutionized by the proposition of ribosomal RNA (rRNA) as a universal barcoding marker for the identification of organisms and the determination of genetic relationships among them [31]. The rRNA marker has superior discriminative power compared to proteins; for example, in the case of 18S rRNA, the number of characteristics is equal to 4 1789 (4 different nucleotides, 1789 the length of Saccharomyces cerevisiae 18S rRNA).
Pioneering studies by Cletus Kurtzman on the sequence analysis of the D1/D2 region of the LSU (~600 nucleotides) of 500 known yeast species demonstrated that conspecific yeast strains may exhibit up to three nucleotide differences, and a separate species may differ by six or more nucleotides [32,33]. This approach was also applied to the delimitation of basidiomycetous yeast species by Jack Fell [34]. Along with the LSU marker, the ITS region is widely used for yeast identification [35,36,37,38,39,40]. The LSU and ITS ribosomal markers are effective for the identification of most yeasts, but there are cases when they were found to be unable to discriminate closely related yeast species. To overcome this, multigene analysis using additional protein-coding gene sequences has been suggested [41,42]. Sequence identity cut-off values for the LSU and ITS markers were determined based on the analysis of a large dataset, including 9000 CBS yeast strains belonging to 2000 known yeast species [43].
The present study aims to contribute to our understanding of yeast biodiversity by focusing on the yeasts associated with insects collected in Bulgaria. We used combined sequence analyses of the ITS and LSU ribosomal DNA markers for the identification of the isolated yeast strains [37,38,39,40].
2. Materials and Methods
2.1. Insect Collection and Isolation of Yeasts
The collection of insects was conducted utilizing entomological methodologies involving searches conducted beneath stones, logs, and tree bark, as well as on mushrooms and within decaying wood. Furthermore, a thorough entomological examination of plant leaves and flowers was undertaken. The insect sampling encompassed a wide spectrum of plant communities, including xerophilic, deciduous, mixed, and coniferous forests, as well as wetlands, arid grasslands characterized by herbaceous vegetation, and open riparian shrub and forest ecosystems. The altitudinal gradient spanned from 70 to 1600 m above sea level.
To prevent any potential cross-contamination, each individual insect specimen was transported within separate sterile containers from the respective collection sites to the laboratory. Following the initial taxonomic identification, which took place at the National Museum of Natural History in Sofia, those insects designated for yeast isolation purposes were subsequently transported to the Institute of Microbiology, Bulgarian Academy of Sciences, located in Sofia. The insects were stored at −80 °C. Prior to the dissection process, the insects underwent a thorough cleansing procedure involving a wash with 70% ethanol, followed by rinsing with 0.7% sterile saline solution. As a quality control measure, the saline wash was cultured on an acidified yeast extract–malt extract agar (YM agar, pH 4.0–4.2) plate, serving as a negative control. To aseptically extract the gut contents from the insect bodies, the dissection procedure was carried out under a microscope, utilizing a sterile Petri dish. Subsequently, the gut contents were gently crushed within a sterile Eppendorf tube containing 0.7% sterile saline. The crushed gut contents were then plated on acidified YM agar plates (pH-3.4) and incubated at ambient room temperature. Following an incubation period of three days, colonies displaying a yeast-like morphology were selectively isolated and sub-cultured as necessary until pure yeast cultures were obtained, following established protocols [44].
2.2. Yeast Identification
Morphological, biochemical, and physiological characterization of the isolates was performed according to established methodsls [44,45,46,47].
2.3. Yeast Strain Deposit in Culture Collections
New yeast strains obtained in this study were deposited in the CBS culture collection of the Westerdijk Fungal Biodiversity Institute (CBS), Utrecht, The Netherlands, and the yeast culture collection of the National Bank for Industrial Microorganisms and Cell Cultures (NBIMCC), Sofia, Bulgaria.
2.4. DNA Barcoding Analysis
The LSU and ITS regions were amplified directly from individual yeast colonies without extracting DNA from cells [14]. The primer sets V9G and LS266 were used for the amplification of the LSU and ITS sequences [48]. A cell suspension of one loopful of cells in 50 μL of autoclaved water was heated at 95 °C for 5 min, and 2 μL of the supernatant was used as a template in 25 μL PCR mixture containing 7.5 μL of PCR master mix (Fermentas, Darmstadt, Germany), 0.5 μM of each primer. The amplified products were purified using a QIAquick purification kit (QiaGen, Germantown, MD, USA) according to the manufacturer’s instructions. Direct sequencing of the LSU and ITS regions was performed using primers NL1/NL4 [49] and ITS1/ITS4 [50], respectively, with a Taq DyeDeoxy terminator cycle sequencing kit (Applied Biosystems, Foster City, CA, USA) by Macrogen Inc. (Seoul, Republic of Korea), according to the manufacturer’s protocol. Purified sequencing reaction mixtures were separated with a 3730XL automated DNA analyzer (Applied Biosystems, Foster City, Foster City, CA, USA). The program BLAST from NCBI (https://www.ncbi.nlm.nih.gov, accessed on 1 July 2024) was used to compare the sequences to known LSU and ITS sequences for the assignment of the closest related taxa [51]. All the LSU and ITS sequences from this study were submitted to the GenBank database under the accession numbers listed in Tables S1 and S2 (Supplementary Materials).
3. Results and Discussion
3.1. Insect Collection and Distribution of Yeasts across Insects Orders
Insects were collected from diverse regions across Bulgaria during the following three periods: May 2008, April to September 2009, and July to September 2012 (Figure 1). Additional pertinent details concerning the precise locations, altitudes, and seasons of collection are outlined in Table 1.
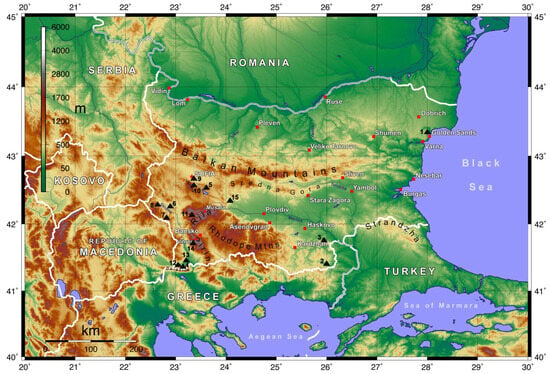
Figure 1.
Insect collection sites. The numbers 1–15 correspond to the entries in column 1 of Table 1. Red squares denote major settlements, the red pentagram indicates the capital city, and red triangles highlight the country’s highest peaks.

Table 1.
Insect collection sites in Bulgaria: site number on the map above, region, the year of collection and altitude.
More than 170 insect specimens were collected. They belonged to various insect orders, including Coleoptera, Orthoptera, Lepidoptera, Hymenoptera, Dermaptera, and Collembola. The isolation of yeast strains from the insect guts yielded a total of 107 strains, distributed across 28 insect families. The breakdown is as follows: 14% from longhorn beetles (Cerambycidae), 13% from unidentified Coleoptera spp., 10% from darkling beetles (Tenebrionidae), 8% from ground beetles Carabidae, 7% from scarab beetles (Scarabaeidae), 6% from Cetoniidae, 5% from Orthoptera. 3.7% from Hymenoptera, 3.7% from leaf beetles (Chrysomelidae), 2.8% from Ciidae, and 2% each from Staphylinidae, Ruthelidae, Geotrupidae, Buprestidae, Melandryidae, Dermaptera, Lepidoptera, and Hexapoda. The twelve remaining insects collectively contributed 10.8%.
In most instances, each individual insect specimen was found to harbor a single yeast species. However, it is worth noting that there were exceptions, with certain insects containing as many as three distinct yeast species.
Bolitophagus reticulatus (Tenebrionoidea) found in National park Rila contained two species, while Bolitophagus interruptus found in the Podgorie area contained three species; two species were isolated from Cetonia aurata (Cetoniidae) found in East Rhodopes, two species were recovered from Sminthuridae sp. (Collembola) found on a fungus in Nature park Vitosha, as well as from an unknown Orthoptera spp. collected from a plant on the bank of the Struma River (Supplementary Materials, Table S1).
3.2. Ascomycetous Yeast Associated with Insects
DNA barcoding analysis revealed that 89 isolates of yeasts were affiliated with ascomycetous yeast genera—Candida, Debaryomyces, Diutina, Exophiala, Hanseniaspora, Kazachstania, Kluyveromyces, Lachanceae, Metschnikowia, Meyerozyma, Nakaseomyces, Ogataea, Pichia, Priceomyces, Saccharomyces, Scheffersomyces, Starmerella, Wickerhamiella and Wickerhamomyces.
Genus Scheffersomyces Kurtzman and M. Suzuki (2010). Seventeen yeast isolates of the genus Scheffersomyces were obtained from various insects. Ten isolates were obtained from Cerambycidae beetles, while two isolates came from beetles of the Lucanidae and Tenebronidae families and an unidentified Zygaenidae moth (Supplementary Materials, Table S1). Four isolates originated from an unidentified Coleoptera spp. and the larva of Cetoniidae sp. Five yeast strains were identified as Scheffersomyces stipitis. Twelve strains belonged to Scheffersomyces insectosa. Scheffersomyces yeasts have been isolated from wood-infesting insect larvae and their frass, from the guts of lignicolous beetles, and from rotten wood in Europe and North and Central America [52]. Members of the genus Scheffersomyces, Sch. stipitis, and Sch. shehatae are of particular interest due to their ability for the fermentative production of ethanol from D-xylose, a major pentose found in plant biomass [53]. A biodiversity survey conducted in the USA and Panama showed the consistent presence of Scheffersomyces yeasts in the gut of Odontotaenius disjunctus (Passalidae) [54,55]. A symbiotic relationship was proposed, wherein the yeast Sch. stipitis metabolizes D-xylose to the advantage of the associated insects [54]. Another survey of yeast associated with passalids in Guatemala and Thailand showed that Scheffersomyces yeast was the most abundant species in the gut of both wood roaches and Guatemalan passalids, while in Thai passalids, the most abundant yeasts were closely related to Candida insectamans [56]. Scheffersomyces yeasts were also shown to occur in the gut of the North American wood roach Cryptocercus spp. (Blattodea: Cryptocercidae) [57].
Genus Suhomyces M. Blackwell and Kurtzman (2016). Five strains of Suhomyces atakaporum were found in the gut of beetles belonging to various families, including Tenebrionidae, Cerambycidae, Erotylidae, and Melandryidae (Supplementary Materials, Table S1). Suhomyces tanzawaensis was isolated from mosses in Japan [58], and it had no known close relatives for a long time. An additional five species were isolated from soil, rotten wood, mushroom fruiting bodies, frass of moss larva, and a Bostrichidae beetle collected in the USA, Australia, and South Africa [59]. Sixteen new species, including Su. atakaporum, were discovered from fungus-feeding beetles collected in Panama and the USA [14]. The species form a well-supported monophyletic clade and were isolated from 11 Coleoptera families, with the majority coming from Tenebrionidae and Erotylidae beetles. A yeast biodiversity survey conducted in Thailand showed that almost half of the 128 collected Thai strains were members of the Su. tanzawaensis clade. Three species, Suhomyces panamericana, Suhomyces anneliseae, and Suhomyces chickasaworum, common in the Western Hemisphere, also occurred in Thailand [56].
Genus Debaryomyces Lodder and Kreger-van Rij (1952): Two yeast isolates obtained from the beetle species Lachnaia sexpunctata (Chrysomelidae) and an unidentified grasshopper (Orthoptera) (Table S1) were identified as Debaryomyces fabryi. It has been demonstrated that ribosomal markers are insufficient for distinguishing species of Debaryomyces hansenii, D. fabryi, and Debaryomyces subglobosus [42]. Therefore, for accurate identification of these isolates, analysis of an additional marker, the actin gene (ACT1), is necessary. [42]. Previous studies reported on the isolation of Debaryomyces yeasts from Central European bumblebees [60] and the beetle species Melanotus villosus (Elateridae) collected in the UK [61]. These data highlight the coexistence of Debaryomyces species within different host organisms.
Genus Schwanniomyces Klöcker (1909): Two strains of Schwanniomyces vanrijiae were isolated, one from the larva of Cetonia aurata (Cetoniidae) and the other from the gut of Harpalus anxius (Carabidae). Schwanniomyces vanrijjiae and Schwanniomyces polymorphus were shown to be associated with wood ants, Formica aquilonia [62], and carpenter ants, Camponotus vicinus [Formicidae] [63]. Experiments conducted on Camponotus vicinus larvae, which were fed a nutrient-deficient diet and exposed to live yeast Schw. polymorphus, demonstrated that the yeasts increased brood weight and facilitated the complete development of larvae into adult ants. This led to the conclusion that yeasts provide ants with essential substances for their nutrition and development [64].
Genus Metschnikowia Kamienski (1899). Two strains of Metschnikowia pulcherrima were isolated from an unidentified butterfly species (Lepidoptera) and a bumblebee (Bombus sp.: Apidae). A strain of Metschnikowia reukaufii was isolated from a Buprestidae spp. Metschnikowia yeasts were consistently isolated from the gut of diverse insects. Woolfolk and Inglis isolated Metschnikowia pulcherrima from Chrysoperla rufilabris (Chrysopidae) populations in the USA [65]. Three novel Metschnikowia species were isolated from Chrysoperla species collected in Arizona [66]. Metschnikowia chrysoperlae was found in the gut or frass of corn-feeding caterpillars Ostrinia nubilalis (Crambidae) in Austria, as well as in corn earworm Helicoverpa armigera (Noctuidae) and corn rootworm Diabrotica virgifera (Chrysomelidae) [67]. Metschnikowia yeasts were also found in the gut of lacewings, soldier beetles, and leaf beetles in the United States and Panama [20]. A survey of yeasts associated with social wasps in the USA has shown that about 25% of identified yeast belonged to the genus Metschnikowia [68]. Recently, a new species Metschnikowia baotianmanensis was shown to be associated with a rhinoceros beetle, i.e., Allomyrina dichotoma (Scarabeidae) collected in China [69].
Genus Hypopichia von Arx and van der Walt (1976): In the genus Hypopichia, two strains of Hypopichia rhagii were isolated from Dinoptera colaris (Cerambycidae) and a larva of Buprestidae sp. In Europe, H. rhagii was solated from both the adult and larval stages of Rhagium inquisitor (Cerambycidae) and the species was considered a symbiont of this insect [70]. Subsequent studies on gut symbionts of Cerambycidae beetles in Germany [22] further confirmed the association of H. rhagii with R. inquisitor. Our findings expand this association to include additional beetle species.
Genus Saccharomyces Meyen ex Reess (1870). In the present study, one strain was identified as Saccharomyces cerevisiae and was isolated from the beetle Mimela aurata (Rutelidae) (Supplementary Materials, Table S1). The correct identification of this isolate requires the analysis of several additional markers, namely, translation elongation factor 1-α (EF1-α), mitochondrial small-subunit rDNA, and cytochrome oxidase II (COX II), because the LSU and ITS markers alone could not distinguish closely related species, namely S. cerevisiae, S. paradoxus, and S. cariocanus [41].
Saccharomyces yeasts were found in the gut of Corydalidae (Megaloptera) [21] and are well-known to be associated with Drosophila flies [71,72]. A yeast survey conducted in Europe demonstrated that social wasps (Apocrita: Hymenoptera) serve as a reservoir and as a dispersion vector of S. cerevisiae in nature [73]. The yeast was also present in the gut of rhinoceros beetles (Scarabaeidae) [69].
Genus Lachancea Kurtzman (2003). In this study, a strain of Lachancea kluyveri was isolated from the beetle Purpuricenus budensis (Cerambycidae). Lachancea thermotolerans, and L. fermentati were frequently isolated from the gut of dobsonfly (Megaloptera: Corydalidae) [21]. A study of yeasts associated with social wasps conducted in the USA revealed that approximately 30% of yeast isolates belonged to L. kluyveri, L. thermotolerans, and L. fermentati [68]. This finding emphasizes the importance of Lachancea species in diverse insects, encompassing beetles, dobsonflies, and social wasps.
Genus Hanseniaspora Zikes (1912). Two strains of Hanseniaspora uvarum were isolated, one from an unidentified Coleoptera spp. and the other from Forficula auricularia (Dermaptera: Forficulidae) found on a vine tree. In prior research, Hanseniaspora viniae yeasts were isolated from the surface and the gut of Corydalidae (Megaloptera) and Ascalaphidae (Neroptera) [21]. In another survey, Hanseniaspora was the most dominant yeast in Drosophila species in decaying fruits [72]. Interestingly, the yeast was not found in Drosophila flies that feed on flowers, suggesting that the diet of Drosophila shapes its yeast symbiont community. In another study, Hanseniaspora species were the most frequently isolated yeast from the fruit fly Bactrocera tryoni (Diptera: Tephritidae) [74]. Experimental data demonstrated that H. uvarum plays a beneficial role in the survival and fitness of the fly larvae.
Genus Starmerella Rosa and Lachance (1998). Two species, Starmerella bombicola and Starmerella bombi, were isolated from the beetle Valgus hemipterus (Scarabaeidae) and from Bombus sp., correspondingly (Table S1). Starmerella yeasts have predominantly been obtained from habitats in tropical and temperate regions of the Western Hemisphere, as well as the Australia-Pacific biogeographical zone. They exhibit an ecological association with flowers, along with their attendant bees and wasps, as well as beetle species belonging to the families Nitidulidae and Chrysomelidae [75].
Species belonging to an unaffiliated clade (Table S1). Three strains of Candida savonica were consistently isolated from Ciidae spp. found on the fungus Trametes versicolor (Polyporaceae) in two different locations (Table S1). Based on our observations, we suggest the possibility of an association between C. savonica and Ciidae spp. Single isolates of C. schatavii and C. boleticola were obtained from an unidentified grasshopper (Orthoptera) and the bracket fungus Ganoderma spp. (Ganodermataceae), respectively.
Opportunistic Yeast Pathogens: We obtained an isolate of the opportunistic pathogen Nakaseomyces glabratus (formerly Candida glabrata) from Oxythyrea funesta (Cetoniidae). Nakaseomyces glabratus is ranked as the second most prevalent causative agent of candidiasis globally [76]. Isolates of Candida maltosa and Candida zeylanoides originated from the gut of the beetles Rhaesus serricollis (Cerambycidae) and Cybister lateralimarginalis (Dytiscidae) (Table S1, Supplementary Materials).
Additionally, an isolate of the opportunistic human pathogen Meyerozyma guilliermondii was found in the gut of the beetle Bitoma crenata (Colydiidae). This yeast has been isolated from various beetles, including Cerambycidae [77], Scarabaeidae [78], Curculionidae [79], and Chrysomelidae. It has also been found in grass moss (Lepidoptera: Crambidae) [80], ants (Formicidae) [81], mining bees (Hymenoptera: Andrenidae), a mushroom-feeding fly [82], dobsonflies (Corydalidae: Megaloptera), and owlflies (Ascalaphidae: Neroptera) [19]. Recent studies have demonstrated that Me. guilliermondii was the third most frequently isolated species from the gut of dung beetles (Scarabaeidae) collected in Botswana [23]. Notably, Me. guilliermondii is the sixth most frequently isolated yeast in clinical environments, as reported by Pfaller et al. [83].
Opportunistic pathogens, including Candida orthopsilosis, C. maltosa, Candida parapsilosis, Candida tropicalis, Candida neerlandica, and Lodderomyces elongisporus, were found in the guts of different insects in the USA and Panama [19]. A study conducted in Brazil demonstrated that leaf-cutting ants (Formicidae: Attini) harbored the yeast C. parapsilosis [84]. These findings highlight the significant role of insects as major vectors for the dispersal of potential pathogenic yeasts in natural ecosystems.
Other yeast species isolated in this study: Five yeast strains belonged to the species Exophiala cideris (black yeast), Kluyveromyces dobzhanskii, Pichia membranifaciens, Teunomyces kruisii, and Yarrowia lipolitica. They were isolated from various insects as presented in Table S1.
3.3. Novel Ascomycetous Yeast Species
A DNA barcoding analysis revealed several novel species, some of which have already been validly described in our previous articles, namely, Ogatae saltuana (Curculionidae: Scolitinae sp.) [85], Priceomyces vitoshaensis (Carabidae: Pterostichus melas) [86], Nematodospora valgi (Scarabaeidae: Valgus hemipterus), and Candida cetonii (Cetoniidae: Oxythyrea funesta) [37], Yarrowia parophonii (Carabidae: Paraphonus hirsutulus and Coleoptera spp.) [87], Kazachstania molopis (Carabidae: Molops piceus) [88], Kazachstania chrysolinae (Chrysomelidae: Chrysolina polita) [38,39], Suhomyces rilaensis (Tenebrionidae: Bolitophagus interruptus; B. reticulatus) [89], and Starmerella xylocopis (Hymenoptera: Aphidae) [90].
Two isolates, 91W and 91WR (100% sequence similarity in both the LSU and ITS), originated from the beetle Carabus violaceus azurescens (Carabidae; Supplementary Materials, Table S1). A blast analysis showed that the most similar sequences in the GenBank database belong to Starmerella sorbosivorans CBS 8768T with 93% identity in the LSU (28 nucl. subst., 1 gap) and Candida spenceri CBS 11673T, with 87% identity in the ITS (27 nucl. subst., 20 gaps).
Two isolates, DZ1_1 and DZ2_1, came from the beetle Bolitophagus interruptus (Tenebrionidae) found on Ganoderma spp. (Ganodermataceae). An additional isolate, D300, was obtained from the specimen of Bolitophagus reticulatus (Tenebrionidae) found on the fungus Fomes spp. (Polyporaceae). A pairwise sequence analysis showed that the strains have 86% sequence identity (51 nucleotide substitutions and 24 gaps) in the LSU with Wickerhamiella musiphila CBS 10697T and 84% identity (62 nucleotide substitutions, 20 gaps) in ITS sequences with W. infanticola CBS 11940T.
Additionally, two isolates, DZ1_2 and DZ2_2 (100% sequence similarity in both the LSU and ITS), were isolated from the beetle species Bolitophagus reticulatus (Tenebrionidae; Table S1). The analysis showed that the strains show 99% identity (three nucl. subst. in the LSU) and 93% identity (23 nucl. subst., 12 gaps in the ITS) with Blastobotrys peoriensis CBS 10340T.
The analysis showed that nine isolates, IMB-YP, IMB-P1, IMB-PP2, IMB-P21, IMB-P5, IMB-P72, IMB-YP8, IMB-PX1, and IMB-PG2, showed 100% sequence similarity in the LSU and 99–100% sequence similarity in the ITS. The strain showed 100% sequence similarity in the LSU and 86% sequence similarity in the ITS (41 nucl. subst., 14 gaps) of the new strain IMBP43, 90% sequence similarity in the LSU of Suhomyces panamericana ATCC MYA-4318T (45 nucl. subst., 11 gaps), and 84% identity in the ITS of S. tanzawaensis CBS 7422T (46 nucl. subst., 22 gaps).
The isolate IMB-P43 showed 100% sequence similarity in the LSU and 86% sequence similarity in the ITS (41 nucl. subst., 14 gaps) of the nine strains, IMB-YP, IMB-P1, IMB-PP2, IMB-P21, IMB-P5, IMB-P72, IMB-YP8, IMB-PX1, and IMB-PG2, 94% similarity (19 nucl. subst., five gaps in the LSU) with Suhomyces ambrosiae CBS 8844T, and 91% similarity (21 nucl. subst., seven gaps in the ITS) with Suhomyces chikasaworum CBS 9830T.
A blast analysis showed that two isolates, D15 and D24 (100% sequence identity in both the LSU and ITS), have 88% sequence identity (53 nucl. subst., 11 gaps in the LSU and 52 nucl. subst., 23 gaps in the ITS) with Diutina ranongensis CBS 10861T.
3.4. Basidiomycetous Yeast Associated with Insects
The LSU and ITS sequences of 18 yeast isolates showed similarities with that of members of basidiomycetous yeast genera as retrieved from the GenBank database.
Genus Filobasidium L.S. Olive (1968). The most frequently isolated yeast species belonged to the genus Filobasidium, with the four species Filobasidium stepposus, Filobasidium oeirensis, Filobasidium chernovii, and Filobasidium wieringae. These yeasts were isolated from beetles belonging to the families Chrysomelidae, Silvanidae, Buprestidae, and Cerambycidae (Supplementary Materials, Table S2).
Genus Apiotrichum Stautz (1931). Three strains belonged to two species of the genus Apiotrichum, namely, Apiotrichum lignicola and Apiotrichum humicola, and originated from beetles of the families Geotrupidae and Staphylinidae.
Additional yeast species isolated in this study: Naganishia friedmanii, Rhodosporidium kratochvilovae, Vanrija albida, Rhodotorula mucilaginosa, and Trichosporon coremiiforme. They originated from the gut of insects belonging to Cerambycidae, an unidentified grasshopper (Orthoptera sp.), and Alexiidae sp. (Table S2).
Prior to our research, species from the genus Vanrija, i.e., V. humicola and V. musci, and from the genus Trichosporon, i.e., T. dermatis, T. moniliiforme, and T. porosum, were found in Guatemalan and Thai passalids [56]. Trichosporon species were identified in insect frass from Thailand [91] and in the guts of beetles (Scarabaeidae, Tenebrionidae) collected across diverse locations, including Panama, South Africa, and the USA [92,93,94]. The species from the genera Apiotrichum, Naganishia, Rhodotorula, and Trichosporon were also found in the guts of dung beetles collected in Botswana [23].
Cystobasidium psychroaquaticum [95], was isolated from an unidentified springtail (Collembola: Sminthuridae).
Four yeast isolates, D54, D59-1, D55, and D60-1, exhibited 99% sequence identity in the LSU (nine nucl. substit.) of Trichosporon asteroides CBS 6183 and 99% identity in ITS (four nucl. subst., two gaps) of T. faecalis CBS 4228T.
A single isolate, D162_2, originating from a Melandrydae beetle spp. showed 92% sequence similarity with the LSU (47 nucl. subst., 5 gaps) of Saitozyma flava CBS 331T and 89% similarity with ITS (15 nucleotide substitutions, 12 gaps) of Teunia cuniculi CBS 10309T and represents a novel species.
According to the currently accepted cut-off values of sequence similarity of the ITS and LSU markers for the delimitation of yeast species, the isolates presented above constitute eight new yeast species.
4. Conclusions
Our survey uncovered a remarkable diversity of yeast species associated with insects. Notably, ~42% of the yeast isolates proved to be previously unknown species, underscoring the vast potential for new discoveries in unexplored fungal populations interacting with insects globally.
Our results affirm the role of insects as vectors for the dispersal of opportunistic yeast in natural environments.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/insects15080566/s1, Table S1: List of Ascomycetous yeasts isolated in this study, locality and habitat, isolation hosts, and accession numbers of LSU and ITS sequences in GenBank database; Table S2: List of Basidiomycetous yeasts isolated in this study, locality and habitat, isolation hosts, and accession numbers of LSU and ITS sequences in GenBank database.
Author Contributions
Conceptualization, D.G. and R.D.; methodology, D.G., R.D., B.G., T.B., M.S. and M.G.; software, R.D.; validation, D.G., T.B., M.S. and M.G.; formal analysis, R.D.; investigation, D.G., R.D. and B.G.; resources, D.G. and R.D.; data curation, R.D. and D.G.; writing—original draft preparation, R.D. and D.G.; writing—review and editing, R.D., D.G., T.B. and M.G.; supervision, D.G. and R.D.; project administration, D.G. and R.D.; funding acquisition, D.G. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the grant KP-06-H31/19 from the Bulgarian National Science Fund.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The new sequences generated in this study have been deposited in GenBank under the accession numbers listed in Tables S1 and S2 of the Supplementary Materials. Additionally, raw data and other relevant materials can be requested from the corresponding author upon reasonable request.
Acknowledgments
T.B. wants to acknowledge the Distinguished Scientist Fellow Program (DSFP) of King Saud University, Riyadh, Saudi Arabia.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Dirzo, R.; Young, H.S.; Galetti, M.; Ceballos, G.; Isaac, N.J.; Collen, B. Defaunation in the Anthropocene. Science 2014, 345, 401–406. [Google Scholar] [CrossRef] [PubMed]
- Starmer, W.T.; Lachance, M.A. Yeast ecology. In The Yeasts, a Taxonomic Study, 5th ed.; Kurtzman, C.P., Fell, J.W., Boekhout, T., Eds.; Elsevier: New York, NY, USA, 2011; Volume I, pp. 65–86. [Google Scholar]
- Groenewald, M.; Boundy-Mills, K.; Čadež, N.; Endoh, R.; Jindamorakot, S.; Pohl-Albertyn, C.; Rosa, C.A.; Turchetti, B.; Yurkov, A. Census of yeasts isolated from natural ecosystem and conserved in worldwide collections. In Yeasts in Natural Ecosystems: Diversity; Buzzini, P., Lachance, M.A., Yurkov, A., Eds.; Springer: Berlin/Heidelberg, Germany, 2017; pp. 455–476. [Google Scholar]
- Stork, N.E. How many species of insects and other terrestrial arthropods are there on Earth? Annu. Rev. Entomol. 2018, 63, 31–45. [Google Scholar] [CrossRef] [PubMed]
- Vega, F.E.; Dowd, P.F. The role of yeasts as insect endosymbionts. In Insect-Fungal Associations: Ecology and Evolution; Vega, F.E., Blackwell, M., Eds.; Oxford University Press: New York, NY, USA, 2005; pp. 211–243. [Google Scholar]
- Blackwell, M. Made for Each Other: Ascomycete Yeasts and Insects. Microbiol. Spectr. 2017, 5, 945–962. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yue, L.; Zhenming, C.; Wang, X. Marine killer yeasts active against a yeast strain pathogenic to crab Portunus trituberculatus. Dis. Aquat Org. 2008, 80, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Cappelli, A.; Ulissi, U.; Valzano, M.; Damiani, C.; Epis, S.; Gabrielli, M.G.; Conti, S.; Polonelli, L.; Bandi, C.; Favia, G.; et al. A Wickerhamomyces anomalus killer strain in the malaria vector Anopheles stephensi. PLoS ONE 2014, 9, e95988. [Google Scholar] [CrossRef] [PubMed]
- Gibson, C.M.; Hunter, M.S. Extraordinarily widespread and fantastically complex: Comparative biology of endosymbiotic bacterial and fungal mutualists of insects. Ecol. Lett. 2010, 13, 223–234. [Google Scholar] [CrossRef] [PubMed]
- Boekhout, T.; Amend, A.S.; El Baidouri, F.; Gabaldón, T.; Geml, J.; Mittelbach, M.; Robert, V.; Tan, C.S.; Turchetti, B.; Vu, D.; et al. Trends in yeast diversity discovery. Fungal Divers. 2022, 114, 491–537. [Google Scholar] [CrossRef]
- Boekhout, T. Biodiversity: Gut feeling for yeasts. Nature 2004, 434, 449–451. [Google Scholar] [CrossRef] [PubMed]
- Kurtzman, C.P. Three new ascomycetous yeasts from insect-associated arboreal habitat. Can. J. Microbiol. 2000, 46, 50–58. [Google Scholar] [CrossRef]
- Suh, S.O.; Blackwell, M. Three new beetle-associated yeast species in the Pichia guilliermondii clade. FEMS Yeast Res. 2004, 4, 87–95. [Google Scholar] [CrossRef]
- Suh, S.O.; McHugh, J.V.; Blackwell, M. Expansion of the Candida tanzawaensis yeast clade: 16 novel Candida species from basidiocarp-feeding beetles. Int. J. Syst. Evol. Microbiol. 2004, 54, 2409–2429. [Google Scholar] [CrossRef] [PubMed]
- Suh, S.O.; McHugh, J.V.; Pollock, D.D.; Blackwell, M. The beetle gut: A hyperdiverse source of novel yeasts. Mycol. Res. 2005, 109, 261–265. [Google Scholar] [CrossRef] [PubMed]
- Suh, S.O.; Nguyen, N.H.; Blackwell, M. Nine new Candida species near C. membranifaciens isolated from insects. Mycol. Res. 2005, 109, 1045–1056. [Google Scholar] [CrossRef] [PubMed]
- Suh, S.O.; Blackwell, M. Four new yeasts in the Candida mesenterica clade associated with basidiocarp-feeding beetles. Mycologia 2005, 97, 167–177. [Google Scholar] [CrossRef] [PubMed]
- Suh, S.O.; Nguyen, N.H.; Blackwell, M. A yeast clade near Candida kruisii uncovered: Nine novel Candida species associated with basidioma-feeding beetles. Mycol. Res. 2006, 110, 1379–1394. [Google Scholar] [CrossRef] [PubMed]
- Suh, S.O.; Nguyen, N.H.; Blackwell, M. Yeasts isolated from plant-associated beetles and other insects: Seven novel Candida species near Candida albicans. FEMS Yeast Res. 2008, 8, 88–102. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, N.H.; Suh, S.O.; Erbil, C.K.; Blackwell, M. Metschnikowia noctiluminum sp. nov., Metschnikowia corniflorae sp. nov., and Candida chrysomelidarum sp. nov., isolated from green lacewings and beetles. Mycol. Res. 2006, 110, 346–356. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, N.H.; Suh, S.O.; Blackwell, M. Five novel yeast species associated with Neuroptera and other insects. Mycologia 2007, 99, 842–858. [Google Scholar] [CrossRef]
- Grünwald, S.; Pilhofer, M.; Höll, W. Microbial associations in gut systems of wood- and bark-inhabiting longhorned beetles [Coleoptera: Cerambycidae]. Syst. Appl. Microbiol. 2010, 33, 25–34. [Google Scholar] [CrossRef]
- Nwaefuna, A.E.; Boekhout, T.; Garcia-Aloy, M.; Vrhovsek, U.; Zhou, N. Diversity of dung beetle-associated yeasts from pristine environments of Botswana. Yeast 2023, 40, 182–196. [Google Scholar] [CrossRef]
- Makopa, T.P.; Ncube, T.; Alwasel, S.; Boekhout, T.; Zhou, N. Yeast-insect interactions in southern Africa: Tapping the diversity of yeasts for modern bioprocessing. Yeast 2024, 41, 330–348. [Google Scholar] [CrossRef] [PubMed]
- Asoyov, B.; Petrova, A. Synopsis of the higher flora of Bulgaria. In Chorological and Floristic Elements; Bulgarian Biodiversity Foundation: Sofia, Bulgaria, 2006. [Google Scholar]
- Petrova, A.; Velchev, V. List of the Bulgarian endemic plants. In Atlas of Bulgarian Endemic Plants; Petrova, A., Ed.; Gea-Libris: Sofia, Bulgaria, 2006; pp. 362–369. [Google Scholar]
- Red Data Book of the Republic of Bulgaria. Volume 1, Plants & Fungi. Available online: http://e-ecodb.bas.bg/rdb/en/vol1/ (accessed on 1 July 2024).
- Hubenov, Z. Entomofaunistic diversity of Bulgaria. Acta Entomol. Bulg. 2005, 11, 118–132. [Google Scholar]
- Sanger, F.; Tuppy, H. The amino-acid sequence in the phenylalanyl chain of insulin. I. The identification of lower peptides from partial hydrolysates. Biochem. J. 1951, 49, 463–481. [Google Scholar] [CrossRef] [PubMed]
- Zuckerkandl, E.; Pauling, L. Molecules as documents of evolutionary history. J. Theor. Biol. 1965, 8, 357–366. [Google Scholar] [CrossRef] [PubMed]
- Woese, C.R.; Fox, G.E. Phylogenetic structure of the prokaryotic domain: The primary kingdoms. Proc. Natl. Acad. Sci. USA 1977, 74, 5088–5090. [Google Scholar] [CrossRef] [PubMed]
- Kurtzman, C.P.; Robnett, C.J. Identification of clinically important ascomycetous yeasts based on nucleotide divergence in the 5’ end of the large-subunit (26s) ribosomal DNA gene. J. Clin. Microbiol. 1997, 35, 1216–1223. [Google Scholar] [CrossRef] [PubMed]
- Kurtzman, C.P.; Robnett, C.J. Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie Van Leeuwenhoek 1998, 73, 331–371. [Google Scholar] [CrossRef] [PubMed]
- Fell, J.W.; Boekhout, T.; Fonseca, A.; Scorzetti, G.; Statzell-Tallman, A. Biodiversity and systematics of basidiomycetous yeasts as determined by large-subunit rDNA D1/D2 domain sequence analysis. Int. J. Syst. Evol. Microbiol. 2000, 3, 1351–1371. [Google Scholar] [CrossRef] [PubMed]
- Gouliamova, D.E.; Hennebert, G.L.; Smith, M.T.; van der Walt, J.P. Diversity and affinities among species and strains of Lipomyces. Antonie Van Leeuwenhoek 1998, 74, 283–291. [Google Scholar] [CrossRef]
- Schoch, C.L.; Seifert, K.A.; Huhndorf, S.; Robert, V.; Spouge, J.L.; Levesque, C.A.; Chen, W.; Fungal Barcoding Consortium; Fungal Barcoding Consortium Author List; Bolchacova, E.; et al. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. USA 2012, 109, 6241–6246. [Google Scholar] [CrossRef]
- Gouliamova, D.E.; Dimitrov, R.A.; Smith, M.T.; Groenewald, M.; Stoilova-Disheva, M.M.; Guéorguiev, B.V.; Boekhout, T. DNA barcoding revealed Nematodospora valgi gen. nov., sp. nov. and Candida cetoniae sp. nov. in the Lodderomyces clade. Fungal Biol. 2016, 120, 179–190. [Google Scholar] [CrossRef] [PubMed]
- Dimitrov, R.A.; Gouliamova, D.E. Genetic and Phenotypic cut-off values for species and genera discrimination of the Kazachstania clade. CR Acad. Bulg. Sci. 2019, 72, 350–357. [Google Scholar]
- Gouliamova, D.; Dimitrov, R. Kazachstania chrysolinae and Kazachstania bozae two new yeast species of the genus Kazachstania. transfer of four Kazachstania species to Grigorovia gen. nov. as new combinations. CR Acad. Bulg. Sci. 2020, 73, 48–57. [Google Scholar]
- Dimitrov, R.A.; Gouliamova, D.E. Phylogenetic Cut-off Values for Species and Genera Discrimination of Yeast. Acta Microbiol. Bul. 2022, 38, 207–215. [Google Scholar]
- Kurtzman, C.P. Phylogenetic circumscription of Saccharomyces, Kluyveromyces and other members of the Saccharomycetaceae, and the proposal of the new genera Lachancea, Nakaseomyces, Naumovia, Vanderwaltozyma and Zygotorulaspora. FEMS Yeast Res. 2003, 4, 233–245. [Google Scholar] [CrossRef] [PubMed]
- Groenewald, M.; Daniel, H.M.; Robert, V.; Poot, G.A.; Smith, M.T. Polyphasic re-examination of Debaryomyces hansenii strains and reinstatement of D. hansenii, D. fabryi and D. subglobosus. Persoonia 2008, 21, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Vu, D.; Groenewald, M.; De Vries, M.; Gehrmann, T.; Stielow, B.; Eberhardt, U.; Al-Hatmi, A.; Groenewald, J.Z.; Cardinali, G.; Houbraken, J.; et al. Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Stud. Mycol. 2019, 92, 135–154. [Google Scholar] [CrossRef] [PubMed]
- Kurtzman, C.P.; Fell, J.W.; Boekhout, T.; Robert, V. Methods for isolation, phenotypic characterization and maintenance of yeasts. In The Yeasts, a Taxonomic Study, 5th ed.; Kurtzman, C.P., Fell, J.W., Boekhout, T., Eds.; Elsevier: New York, NY, USA, 2011; Volume I, pp. 87–110. [Google Scholar]
- Yarrow, D. Methods for the isolation, maintenance and identification of yeasts. In The Yeasts: A Taxonomic Study, 4th ed.; Kurtzman, C.P., Fell, J.W., Eds.; Elsevier: Amsterdam, The Netherlands, 1998; pp. 77–100. [Google Scholar]
- Boekhout, T.; Bai, F.-Y.; Daniel, H.-M.; Groenewald, M.; Robert, V.; Tan, S.; Yurkov, A. The Yeasts Trust Database. 2024. Available online: https://theyeasts.org/ (accessed on 1 July 2024).
- Gams, W.; Hoekstra, E.S.; Aptroot, A. CBS Course of Mycology; Centraalbureau Voor Schimmelcultures: Utrecht, The Netherlands, 1998; p. 153. [Google Scholar]
- Gerrits van den Ende, A.H.G.; De Hoog, G.S. Variability and molecular diagnostics of the neurotropic species Cladophialophora bantiana. Stud. Mycol. 1999, 43, 151–162. [Google Scholar]
- O’Donnell, K. Fusarium and its near relatives. In The Fungal Holomorph: Mitotic, Meiotic and Pleomorphic Speciation in Fungal Systematics; Reynolds, D.R., Taylor, J.W., Eds.; CAB InterNational: Wallingford, UK, 1993; pp. 225–233. [Google Scholar]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols—A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Kurtzman, C.P. Scheffersomyces Kurtzman & M. Suzuki (2010). In The Yeasts, a Taxonomic Study, 5th ed.; Kurtzman, C.P., Fell, J.W., Boekhout, T., Eds.; Elsevier: New York, NY, USA, 2011; Volume 2, pp. 773–779. [Google Scholar]
- Jeffries, T.W.; Grigoriev, I.V.; Grimwood, J.; Laplaza, J.M.; Aerts, A.; Salamov, A.; Schmutz, J.; Lindquist, E.; Dehal, P.; Shapiro, H.; et al. Genome sequence of the lignocellulose-bioconverting and xylose-fermenting yeast Pichia stipitis. Nat. Biotechnol. 2007, 25, 319–326. [Google Scholar] [CrossRef]
- Suh, S.O.; Marshall, C.J.; McHugh, J.V.; Blackwell, M. Wood ingestion by passalid beetles in the presence of xylose-fermenting gut yeasts. Mol. Ecol. 2003, 12, 3137–3145. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Suh, S.O.; Blackwell, M. Microorganisms in the gut of beetles: Evidence from molecular cloning. J. Invertebr. Pathol. 2003, 84, 226–233. [Google Scholar] [CrossRef]
- Urbina, H. Diversity of yeasts associated with wood and the gut of wood-feeding insects. In Dissertation; Louisiana State University and Agricultural and Mechanical College: Baton Rouge, LA, USA, 2012. [Google Scholar]
- Urbina, H.; Frank, R.; Blackwell, M. Scheffersomyces cryptocercus: A new xylose-fermenting yeast associated with the gut of wood roaches and new combinations in the Sugiyamaella yeast clade. Mycologia 2013, 105, 650–660. [Google Scholar] [CrossRef]
- Nakase, T.; Itoh, M.; Takematsu, A.; Komagata, K. Candida tanzawaensis, a new species of yeast isolated from moss collected in Japan. Trans. Mycol. Soc. 1988, 29, 331–338. [Google Scholar]
- Kurtzman, C.P. Six new anamorphic ascomycetous yeasts near Candida tanzawaensis. FEMS Yeast Res. 2001, 1, 177–185. [Google Scholar]
- Brysch-Herzberg, M.; Lachance, M.A. Candida bombiphila sp. nov., a new asexual yeast species in the Wickerhamiella clade. Int. J. Syst. Evol. Microbiol. 2004, 54, 1857–1859. [Google Scholar] [CrossRef]
- Ravella, S.R.; Donovan, N.; James, S.A.; Shivaji, S.; Arunasri, K.; Bond, C.J.; Roberts, I.N.; Hobbs, P.J. Candida northwykensis sp. nov., a novel yeast isolated from the gut of the click beetle Melanotus villosus. Curr. Microbiol. 2011, 63, 115–120. [Google Scholar]
- Maksimova, I.A.; Glushakova, A.M.; Kachalkin, A.V.; Chernov, I.Y.; Panteleeva, S.N.; Reznikova, Z.I. Yeast communities of Formica aquilonia colonies. Microbiology 2016, 85, 124–129. [Google Scholar] [CrossRef]
- Mankowski, M.E.; Morrell, J.J. Yeasts associated with the infrabuccal pocket and colonies of the carpenter ant Camponotus vicinus. Mycologia 2004, 96, 226–231. [Google Scholar] [CrossRef]
- Mankowski, M.E.; Morrell, J.J.; Lebow, P.K. Effects on Brood Development in the Carpenter Ant Camponotus vicinus Mayr after Exposure to the Yeast Associate Schwanniomyces polymorphus Kloecker. Insects 2021, 12, 520. [Google Scholar] [CrossRef]
- Woolfolk, S.; Inglis, G.D. Microorganisms associated with field-collected Chrysoperla rufilabris (Neuroptera: Chrysopidae) adults with emphasis on yeast symbionts. Biol. Control 2004, 29, 155–168. [Google Scholar] [CrossRef]
- Suh, S.O.; Gibson, C.M.; Blackwell, M. Metschnikowia chrysoperlae sp. nov., Candida picachoensis sp. nov. and Candida pimensis sp. nov., isolated from the green lacewings Chrysoperla comanche and Chrysoperla carnea (Neuroptera: Chrysopidae). Int. J. Syst. Evol. Microbiol. 2004, 54, 1883–1890. [Google Scholar] [CrossRef] [PubMed]
- Molnár, O.; Prillinger, H. Analysis of yeast isolates related to Metschnikowia pulcherrima using the partial sequences of the large subunit rDNA and the actin gene; description of Metschnikowia andauensis sp. nov. Syst. Appl. Microbiol. 2005, 28, 717–726. [Google Scholar] [CrossRef] [PubMed]
- Jimenez, S.I.; Carroll, C.; Babcock, T.; Derstine, N.; Hadwin, A.; Moore, M.; Gries, G. Yeasts Harbored by Vespine Wasps in the Pacific Northwest. Environ. Entomol. 2017, 46, 217–225. [Google Scholar] [CrossRef] [PubMed]
- Chai, C.Y.; Huang, L.N.; Cheng, H.; Liu, W.J.; Hui, F.L. Metschnikowia baotianmanensis f.a., sp. nov., a new yeast species isolated from the gut of the rhinoceros beetle Allomyrina dichotoma. Int. J. Syst. Evol. Microbiol. 2019, 69, 3087–3092. [Google Scholar] [CrossRef] [PubMed]
- Jurzitza, G.; Kuehlwein, H.; Kreger-van Rij, N.J. On the systematology of various symbionts of Cerambycidae. Arch. Mikrobiol. 1960, 36, 229–243. [Google Scholar] [CrossRef] [PubMed]
- Morais, P.B.; Hagler, A.N.; Rosa, C.A.; Mendonca-Hagler, L.C.; Klaczko, L.B. Yeasts associated with Drosophila in tropical forests of Rio de Janeiro, Brazil. Can. J. Microbiol. 1992, 38, 1150–1155. [Google Scholar] [CrossRef] [PubMed]
- Chandler, J.A.; Eisen, J.A.; Kopp, A. Yeast communities of diverse Drosophila species: Comparison of two symbiont groups in the same hosts. Appl. Environ. Microbiol. 2012, 78, 7327–7336. [Google Scholar] [CrossRef] [PubMed]
- Stefanini, I.; Dapporto, L.; Legras, J.L.; Calabretta, A.; Di Paola, M.; De Filippo, C.; Viola, R.; Capretti, P.; Polsinelli, M.; Turillazzi, S.; et al. Role of social wasps in Saccharomyces cerevisiae ecology and evolution. Proc. Natl. Acad. Sci. USA 2012, 109, 13398–13403. [Google Scholar] [CrossRef]
- Piper, A.M.; Farnier, K.; Linder, T.; Speight, R.; Cunningham, J.P. Two Gut-Associated Yeasts in a Tephritid Fruit Fly have Contrasting Effects on Adult Attraction and Larval Survival. J. Chem. Ecol. 2017, 43, 891–901. [Google Scholar] [CrossRef]
- Lachance, M.-A. Starmerella Rosa & Lachance (1998). In The Yeasts, a Taxonomic Study, 5th ed.; Kurtzman, C.P., Fell, J.W., Boekhout, T., Eds.; Elsevier: New York, NY, USA, 2011; Volume 2, pp. 811–816. [Google Scholar]
- Pfaller, M.A.; Diekema, D.J. Epidemiology of invasive candidiasis: A persistent public health problem. Clin. Microbiol. Rev. 2007, 20, 133–163. [Google Scholar] [CrossRef] [PubMed]
- Nardon, P.; Grenier, A.M. Endosymbiosis in Coleoptera: Biological, biochemical, and genetic aspects. In Insect Endocytobiosis: Morphology, Physiology, Genetics, Evolution; CRC Press: Boca Raton, FL, USA, 1989; pp. 175–216. [Google Scholar]
- Vishniac, H.S.; .Johnson, D.T. Development of a Yeast Flora in The Adult Green June Beetle (Cotinis Nitida, Scarabaeidae). Mycologia, 1990; 82, 471–479. [Google Scholar]
- Rivera, F.N.; Gonzalez, E.; Gomez, Z.; Lopez, N.; Hernandez-Rodriguez, C.; Berkov, A.; Zuniga, G. Gut-associated yeast in bark beetles of the genus Dendroctonus Erichson (Coleoptera: Curculionidae: Scolytinae). Biol. J. Linn. Soc. 2009, 98, 325–342. [Google Scholar] [CrossRef][Green Version]
- Molnar, O.; Wuczkowski, M.; Prillinger, H. Yeast biodiversity in the guts of several pests on maize; comparison of three methods: Classical isolation, cloning and DGGE. Mycol. Prog. 2008, 7, 111–123. [Google Scholar] [CrossRef]
- Zacchi, I.; Vaughn-Martini, A. Yeasts associated with insects in agricultural areas of Perugia, Italy. Annu. Rev. Microbiol. 2002, 52, 237–244. [Google Scholar]
- Gusmão, D.S.; Santos, A.V.; Marini, D.C.; Bacci, M.; Berbert-Molina, M.A.; Lemos, F.J. Culture-dependent and culture-independent characterization of microorganisms associated with Aedes aegypti (Diptera: Culicidae) (L.) and dynamics of bacterial colonization in the midgut. Acta Trop. 2010, 115, 275–281. [Google Scholar] [CrossRef] [PubMed]
- Pfaller, M.A.; Diekema, D.J.; Mendez, M.; Kibbler, C.; Erzsebet, P.; Chang, S.C.; Gibbs, D.L.; Newell, V.A.; the Global Antifungal Surveillance Group. Candida guilliermondii, an opportunistic fungal pathogen with decreased susceptibility to fluconazole: Geographic and temporal trends from the ARTEMIS DISK antifungal surveillance program. J. Clin. Microbiol. 2006, 44, 3551–3556. [Google Scholar]
- Pagnocca, F.C.; Rodrigues, A.; Nagamoto, N.S.; Bacci, M. Yeasts and filamentous fungi carried by the gynes of leaf-cutting ants. Antonie Van Leeuwenhoek 2008, 94, 517–526. [Google Scholar] [CrossRef] [PubMed]
- Péter, G.; Dlauchy, D.; Tornai-Lehoczki, J.; Gouliamova, D.; Kurtzman, C.P. Ogataea saltuana sp. nov., a novel methanol-assimilating yeast species. Antonie Van Leeuwenhoek 2011, 100, 375–383. [Google Scholar] [CrossRef] [PubMed]
- Gouliamova, D.E.; Dimitrov, R.A.; Smith, M.T.; Stoilova-Disheva, M.M.; Groenewald, M. Priceomyces vitoshaensis. Fungal Planet Description Sheets. Persoonia 2016, 36, 429. [Google Scholar]
- Gouliamova, D.E.; Dimitrov, R.A.; Guéorguiev, B.; Smith, M.T.; Groenewald, M. Yarrowia parophoni. Fungal Planet Description Sheets. Persoonia 2017, 39, 288–289. [Google Scholar]
- Gouliamova, D.E.; Dimitrov, R.A. Kazachstania molopis. Fungal Planet Description Sheets. Persoonia 2019, 42, 421. [Google Scholar]
- Dimitrov, R.A.; Gouliamova, D.E. Suhomyces rilaensis. Fungal Planet Description Sheets. Persoonia 2020, 45, 391. [Google Scholar]
- Gouliamova, D.E.; Dimitrov, R.A.; Groenewald, M.; Smith, M.T.; Boekhout, T. Starmerella xylocopis. Fungal Planet Description Sheets. Persoonia 2021, 46, 507. [Google Scholar]
- Nakase, T.; Jindamorakot, S.; Am-In, S.; Potacharoen, W.; Tanticharoen, M.; Sugita, T.; Kawasaki, H. Trichosporon siamense sp. nov. isolated from insect frass in Thailand. Mycoscience 2006, 47, 106–109. [Google Scholar] [CrossRef]
- Fuentefria, A.M.; Suh, S.O.; Landell, M.F.; Faganello, J.; Schrank, A.; Vainstein, M.H.; Blackwell, M.; Valente, P. Trichosporon insectorum sp. nov., a new anamorphic basidiomycetous killer yeast. Mycol. Res. 2008, 112, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Middelhoven, W.J.; Scorzetti, G.; Fell, J.W. Systematics of the anamorphic basidiomycetous yeast genus Trichosporon Behrend with the description of five novel species Trichosporon vadense, T. smithiae, T. dehoogii, T. scarabeorum and T. gamsii. Int. J. Syst. Evol. Microbiol. 2004, 54, 975–986. [Google Scholar] [CrossRef] [PubMed]
- Gujjari, P.; Suh, S.O.; Lee, C.F.; Zhou, J.J. Trichosporon xylopini sp. nov., a hemicellulose-degrading yeast isolated from the wood-inhibiting beetle Xylopinus saperdioides. Int. J. Syst. Evol. Microbiol. 2011, 61, 2538–2542. [Google Scholar] [CrossRef][Green Version]
- Yurkov, A.M.; Kachalkin, A.V.; Daniel, H.M.; Groenewald, M.; Libkind, D.; de Garcia, V.; Zalar, P.; Gouliamova, D.E.; Boekhout, T.; Begerow, D. Two yeast species Cystobasidium psychroaquaticum f.a. sp. nov. and Cystobasidium rietchieii f.a. sp. nov. isolated from natural environments, and the transfer of Rhodotorula minuta clade members to the genus Cystobasidium. Antonie Van Leeuwenhoek 2015, 107, 173–185. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).