Identification of Rickettsia spp. in Ticks Removed from Tick-Bitten Humans in Northwestern Spain
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Tick Collection and Identification
2.2. DNA Extraction, Amplification, Purification and Sequencing
2.3. Sequence Edition, Alignment and Phylogenetic Analyses
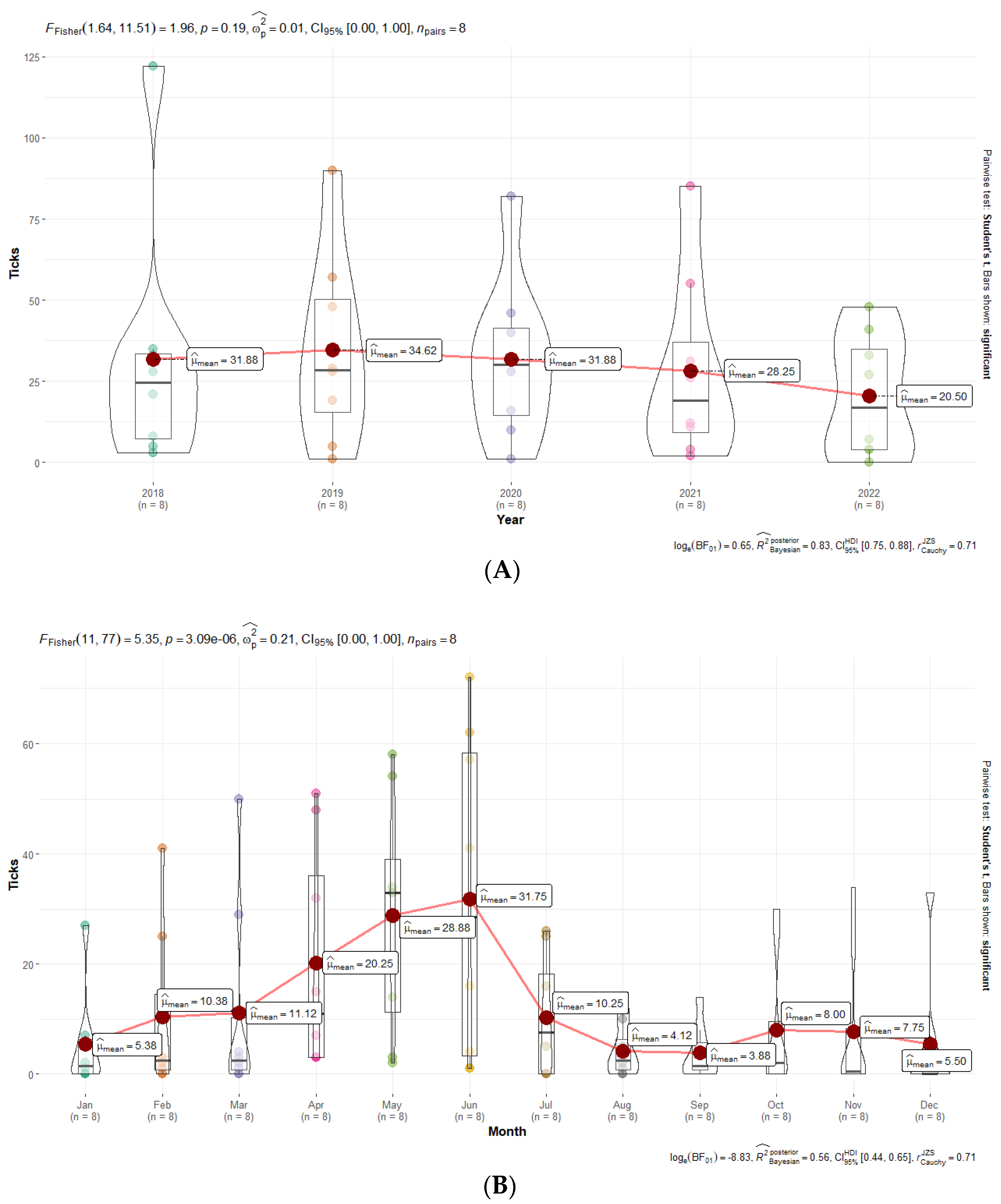
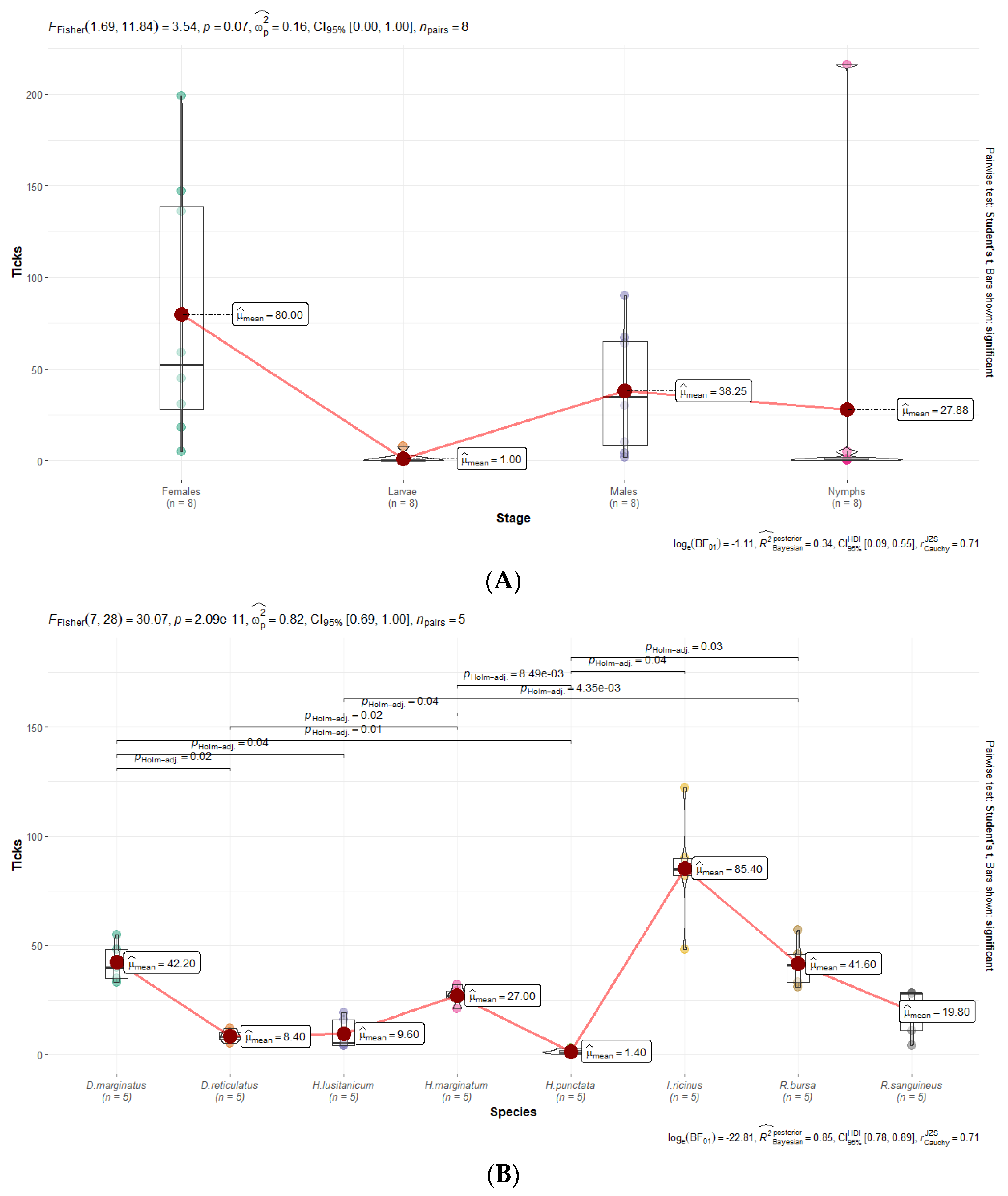
2.4. Statistical and Graphical Analyses
3. Results
3.1. Tick Identification
3.2. Molecular Detection of Rickettsia
3.3. Sequences and BLAST (Identification of Rickettsia Species)
3.4. Phylogenetic Analyses
3.5. Distribution of Rickettsia Species within the Sampling Area
3.6. Statistical Analyses
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Oteo, J.A.; Portillo, A. Tick-borne rickettsioses in Europe. Ticks Tick Borne Dis. 2012, 3, 271–278. [Google Scholar] [CrossRef] [PubMed]
- Remesar, S.; Fernández, P.D.; Venzal, J.M.; Pérez-Creo, A.; Prieto, A.; Estrada-Peña, A.; López, C.M.; Panadero, R.; Fernández, G.; Díez-Baños, P.; et al. Tick species diversity and population dynamics of Ixodes ricinus in Galicia (north-western Spain). Ticks Tick Borne Dis. 2019, 10, 132–137. [Google Scholar] [CrossRef] [PubMed]
- Portillo, A.; Santibáñez, S.; García-Álvarez, L.; Palomar, A.M.; Oteo, J.A. Rickettsioses in Europe. Microbes Infect. 2015, 17, 834–838. [Google Scholar] [CrossRef] [PubMed]
- Guccione, C.; Colomba, C.; Tolomeo, M.; Trizzino, M.; Iaria, C.; Cascio, A. Rickettsiales in Italy. Pathogens 2021, 10, 181. [Google Scholar] [CrossRef] [PubMed]
- Remesar, S.; Cano-Terriza, D.; Morrondo, P.; Jiménez-Ruiz, S.; López, C.M.; Jiménez-Martín, D.; Díaz, P.; Paniagua, J.; García-Bocanegra, I. Molecular detection of Rickettsia spp. in wild ungulates and their ticks in Mediterranean areas of southwestern Spain. Zoonoses Public Health 2023, 70, 485–497. [Google Scholar] [CrossRef] [PubMed]
- Maitre, A.; Wu-Chuang, A.; Mateos-Hernández, L.; Foucault-Simonin, A.; Moutailler, S.; Paoli, J.C.; Falchi, A.; Díaz-Sánchez, A.A.; Banović, P.; Obregón, D.; et al. Rickettsia helvetica infection is associated with microbiome modulation in Ixodes ricinus collected from humans in Serbia. Sci. Rep. 2022, 12, 11464. [Google Scholar] [CrossRef] [PubMed]
- El Karkouri, K.; Ghigo, E.; Raoult, D.; Fournier, P.-E. Genomic evolution and adaptation of arthropod-associated Rickettsia. Sci. Rep. 2022, 12, 3807. [Google Scholar] [CrossRef] [PubMed]
- Fournier, P.-E.; Raoult, D. Current knowledge on phylogeny and taxonomy of Rickettsia spp. Ann. N. Y. Acad. Sci. 2009, 1166, 1–11. [Google Scholar] [CrossRef]
- Merhej, V.; Raoult, D. Rickettsial evolution in the light of comparative genomics. Biol. Rev. Camb. Philos. Soc. 2011, 86, 379–405. [Google Scholar] [CrossRef]
- Igolkina, Y.; Nikitin, A.; Verzhutskaya, Y.; Gordeyko, N.; Tikunov, A.; Epikhina, T.; Tikunova, N.; Rar, V. Multilocus genetic analysis indicates taxonomic status of “Candidatus Rickettsia mendelii” as a separate basal group. Ticks Tick Borne Dis. 2023, 14, 102104. [Google Scholar] [CrossRef]
- Moerbeck, L.; Domingos, A.; Antunes, S. Tick-borne rickettsioses in the Iberian Peninsula. Pathogens 2022, 11, 1377. [Google Scholar] [CrossRef] [PubMed]
- Romaní Vidal, A.; Fernández-Martínez, B.; Herrador, Z.; León Gómez, I.; Gómez Barroso, D. Spatial and temporal trends of Mediterranean spotted fever in Spain, 2005–2015. Ticks Tick Borne Dis 2020, 11, 101353. [Google Scholar] [CrossRef] [PubMed]
- Vieira Lista, M.C.; Belhassen-García, M.; Vicente Santiago, M.B.; Sánchez-Montejo, J.; Pedroza Pérez, C.; Monsalve Arteaga, L.C.; Herrador, Z.; del Álamo-Sanz, R.; Benito, A.; Soto López, J.D. Identification and distribution of human-biting ticks in Northwestern Spain. Insects 2022, 13, 469. [Google Scholar] [CrossRef]
- Fernández-Soto, P. Garrapatas Que Parasitan a Las Personas en Castilla y León, Determinación Por Serología de su Parasitismo y Detección Molecular de Los Patógenos Que Albergan; Universidad de Salamanca: Salamanca, Spain, 2003; p. 296. [Google Scholar]
- Gil-Collado, J.; Guillen, J.; Zapatero, L. Claves para la identificacion de los Ixodoidea espanoles (adultos). Rev. Ibérica Parasitol. 1979, 39, 107–118. [Google Scholar]
- Guglielmone, A.A.; Robbins, R.G.; Apanaskevich, D.A.; Petney, T.N.; Estrada-Peña, A.; Horak, I.G. The Hard Ticks of the World (Acari: Ixodida: Ixodidae); Springer: Dordrecht, The Netherlands, 2014; Volume 10, pp. 978–994. [Google Scholar]
- Estrada-Peña, A.; Martinez, J.M.; Sanchez Acedo, C.; Quilez, J.; Del Cacho, E. Phenology of the tick, Ixodes ricinus, in its southern distribution range (central Spain). Med. Vet. Entomol. 2004, 18, 387–397. [Google Scholar] [CrossRef]
- Dantas-Torres, F.; Latrofa, M.S.; Annoscia, G.; Giannelli, A.; Parisi, A.; Otranto, D. Morphological and genetic diversity of Rhipicephalus sanguineus sensu lato from the New and Old Worlds. Parasit. Vectors 2013, 6, 213. [Google Scholar] [CrossRef]
- Fournier, P.-E.; Roux, V.; Raoult, D. Phylogenetic analysis of spotted fever group rickettsiae by study of the outer surface protein rOmpA. Int. J. Syst. Evol. Microbiol. 1998, 48, 839–849. [Google Scholar] [CrossRef] [PubMed]
- Roux, V.; Rydkγna, E.; Eremeeva, M.; Raoult, D. Citrate synthase gene comparison, a new tool for phylogenetic analysis, and its application for the Rickettsiae. Int. J. Syst. Evol. Microbiol. 1997, 47, 252–261. [Google Scholar] [CrossRef]
- Regnery, R.L.; Spruill, C.L.; Plikaytis, B.D. Genotypic identification of rickettsiae and estimation of intraspecies sequence divergence for portions of two rickettsial genes. J. Bacteriol. 1991, 173, 1576–1589. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. MBE 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Huelsenbeck, J.P.; Larget, B.; Alfaro, M.E. Bayesian phylogenetic model selection using reversible jump Markov chain Monte Carlo. MBE 2004, 21, 1123–1133. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef]
- Rambaut, A. FigTree v 1.4.4. 2018. Available online: https://github.com/rambaut/figtree/releases/tag/v1.4.4 (accessed on 7 December 2023).
- Jeffreys, H. The theory of probability. Oxford Classic Texts. In The Physical Sciences, 3rd ed.; Oxford University Press: Oxford, UK, 1961. [Google Scholar]
- Patil, I. Visualizations with statistical details: The’ggstatsplot’approach. J. Open Source Softw. 2021, 6, 3167. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2024. [Google Scholar]
- Kolde, R.; Kolde, M.R. Package ‘pheatmap’. R Package 2015, 1, 790. [Google Scholar]
- Field, A. Discovering Statistics Using IBM SPSS Statistics, 4th ed.; Sage: London, UK, 2013. [Google Scholar]
- de Sousa, R.; Barata, C.; Vitorino, L.; Santos-Silva, M.; Carrapato, C.; Torgal, J.; Walker, D.; Bacellar, F. Rickettsia sibirica isolation from a patient and detection in ticks, Portugal. Emerg. Infect. Dis. 2006, 12, 1103–1108. [Google Scholar] [CrossRef] [PubMed]
- Aubry, C.; Socolovschi, C.; Raoult, D.; Parola, P. Bacterial agents in 248 ticks removed from people from 2002 to 2013. Ticks Tick Borne Dis. 2016, 7, 475–481. [Google Scholar] [CrossRef] [PubMed]
- de la Fuente, J.; Estrada-Peña, A.; Rafael, M.; Almazán, C.; Bermúdez, S.; Abdelbaset, A.E.; Kasaija, P.D.; Kabi, F.; Akande, F.A.; Ajagbe, D.O.; et al. Perception of ticks and tick-borne diseases worldwide. Pathogens 2023, 12, 1258. [Google Scholar] [CrossRef]
- Piotrowski, M.; Rymaszewska, A. Expansion of tick-borne rickettsioses in the world. Microorganisms 2020, 8, 1906. [Google Scholar] [CrossRef]
- Verhoeve, V.I.; Fauntleroy, T.D.; Risteen, R.G.; Driscoll, T.P.; Gillespie, J.J. Cryptic genes for interbacterial antagonism distinguish Rickettsia species infecting blacklegged ticks from other Rickettsia pathogens. Front. Cell Infect. Microbiol. 2022, 12, 515. [Google Scholar] [CrossRef] [PubMed]
- Otranto, D.; Dantas-Torres, F.; Giannelli, A.; Latrofa, M.S.; Cascio, A.; Cazzin, S.; Ravagnan, S.; Montarsi, F.; Zanzani, S.A.; Manfredi, M.T.; et al. Ticks infesting humans in Italy and associated pathogens. Parasit. Vectors 2014, 7, 328. [Google Scholar] [CrossRef] [PubMed]
- Audino, T.; Pautasso, A.; Bellavia, V.; Carta, V.; Ferrari, A.; Verna, F.; Grattarola, C.; Iulini, B.; Pintore, M.D.; Bardelli, M.; et al. Ticks infesting humans and associated pathogens: A cross-sectional study in a 3-year period (2017–2019) in northwest Italy. Parasit. Vectors 2021, 14, 136. [Google Scholar] [CrossRef] [PubMed]
- Kubiak, K.; Dmitryjuk, M.; Dziekońska-Rynko, J.; Siejwa, P.; Dzika, E. The risk of exposure to ticks and tick-borne pathogens in a spa town in northern Poland. Pathogens 2022, 11, 542. [Google Scholar] [CrossRef] [PubMed]
- Jahfari, S.; Hofhuis, A.; Fonville, M.; van der Giessen, J.; van Pelt, W.; Sprong, H. Molecular detection of tick-borne pathogens in humans with tick bites and erythema migrans, in the Netherlands. PLoS Negl. Trop. Dis. 2016, 10, e0005042. [Google Scholar] [CrossRef] [PubMed]
- Banović, P.; Díaz-Sánchez, A.A.; Simin, V.; Foucault-Simonin, A.; Galon, C.; Wu-Chuang, A.; Mijatović, D.; Obregón, D.; Moutailler, S.; Cabezas-Cruz, A. Clinical aspects and detection of emerging rickettsial pathogens: A “one health” approach study in Serbia, 2020. Front. Microbiol. 2022, 12, 797399. [Google Scholar] [CrossRef] [PubMed]
- Gargili, A.; Palomar, A.M.; Midilli, K.; Portillo, A.; Kar, S.; Oteo, J.A. Rickettsia species in ticks removed from humans in Istanbul, Turkey. Vector Borne Zoonotic Dis. 2012, 12, 938–941. [Google Scholar] [CrossRef] [PubMed]
- Lindblom, A.; Wallménius, K.; Sjöwall, J.; Fryland, L.; Wilhelmsson, P.; Lindgren, P.E.; Forsberg, P.; Nilsson, K. Prevalence of Rickettsia spp. in ticks and serological and clinical outcomes in tick-bitten individuals in Sweden and on the Åland Islands. PLoS ONE 2016, 11, e0166653. [Google Scholar] [CrossRef] [PubMed]
- Andersson, M.O.; Marga, G.; Banu, T.; Dobler, G.; Chitimia-Dobler, L. Tick-borne pathogens in tick species infesting humans in Sibiu County, central Romania. Parasitol. Res. 2018, 117, 1591–1597. [Google Scholar] [CrossRef]
- Merino, F.J.; Nebreda, T.; Serrano, J.L.; Fernández-Soto, P.; Encinas, A.; Pérez-Sánchez, R. Tick species and tick-borne infections identified in population from a rural area of Spain. Epidemiol. Infect. 2005, 133, 943–949. [Google Scholar] [CrossRef]
- Oteo, J.; Portillo, A.; Santibáñez, S.; Pérez-Martínez, L.; Blanco, J.; Jiménez, S.; Ibarra, V.; Pérez-Palacios, A.; Sanz, M. Prevalence of spotted fever group Rickettsia species detected in ticks in La Rioja, Spain. Ann. N. Y. Acad. Sci. 2006, 1078, 320–323. [Google Scholar] [CrossRef] [PubMed]
- Ibarra, V.; Oteo, J.; Portillo, A.; Santibanez, S.; Blanco, J.; Metola, L.; Eiros, J.; Pérez-Martínez, L.; Sanz, M. Rickettsia slovaca infection: DEBONEL/TIBOLA. Ann. N. Y. Acad. Sci. 2006, 1078, 206–214. [Google Scholar] [CrossRef] [PubMed]
- Toledo, A.; Olmeda, A.S.; Escudero, R.; Jado, I.; Valcárcel, F.; Casado-Nistal, M.A.; Rodríguez-Vargas, M.; Gil, H.; Anda, P. Tick-borne zoonotic bacteria in ticks collected from central Spain. Am. J. Trop. Med. Hyg. 2009, 81, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Aguirrebengoa, K.; Portillo, A.; Santibáñez, S.; Marín, J.J.; Montejo, M.; Oteo, J.A. Human Rickettsia sibirica mongolitimonae infection, Spain. Emerg. Infect. Dis. 2008, 14, 528–529. [Google Scholar] [CrossRef]
- Mura, A.; Masala, G.; Tola, S.; Satta, G.; Fois, F.; Piras, P.; Rolain, J.-M.; Raoult, D.; Parola, P. First direct detection of rickettsial pathogens and a new rickettsia, ‘Candidatus Rickettsia barbariae’, in ticks from Sardinia, Italy. Clin. Microbiol. Infect. 2008, 14, 1028–1033. [Google Scholar]

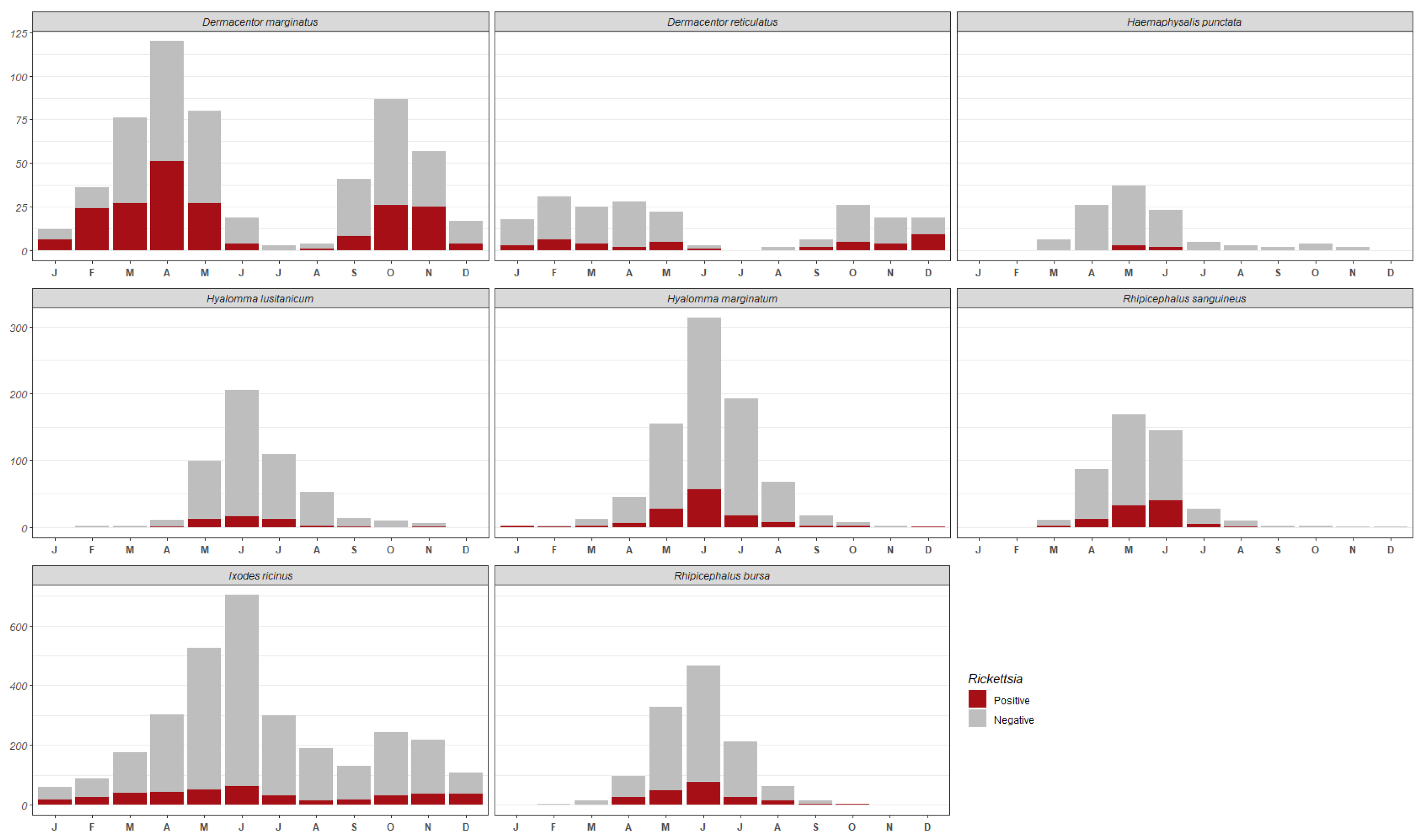
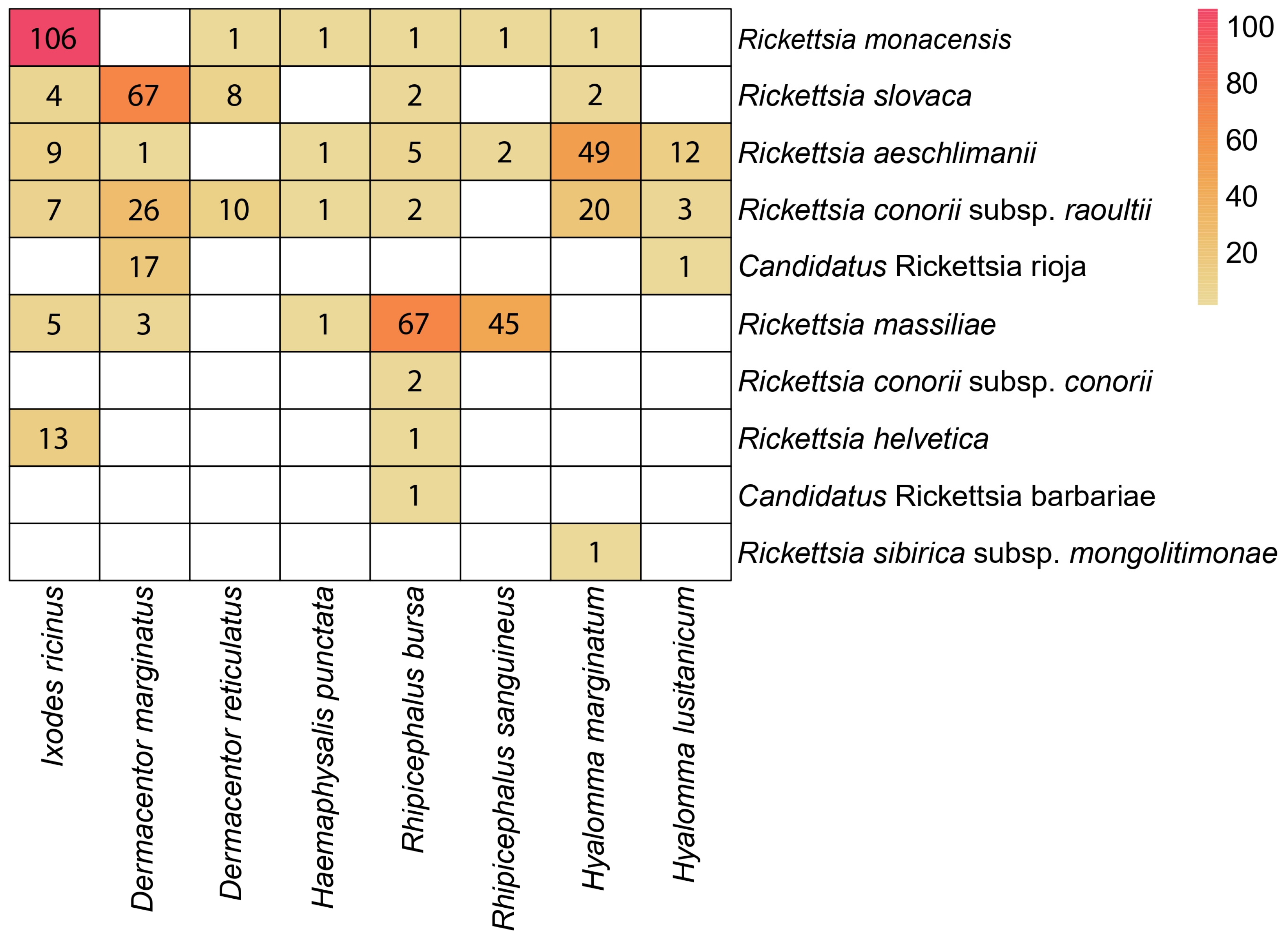
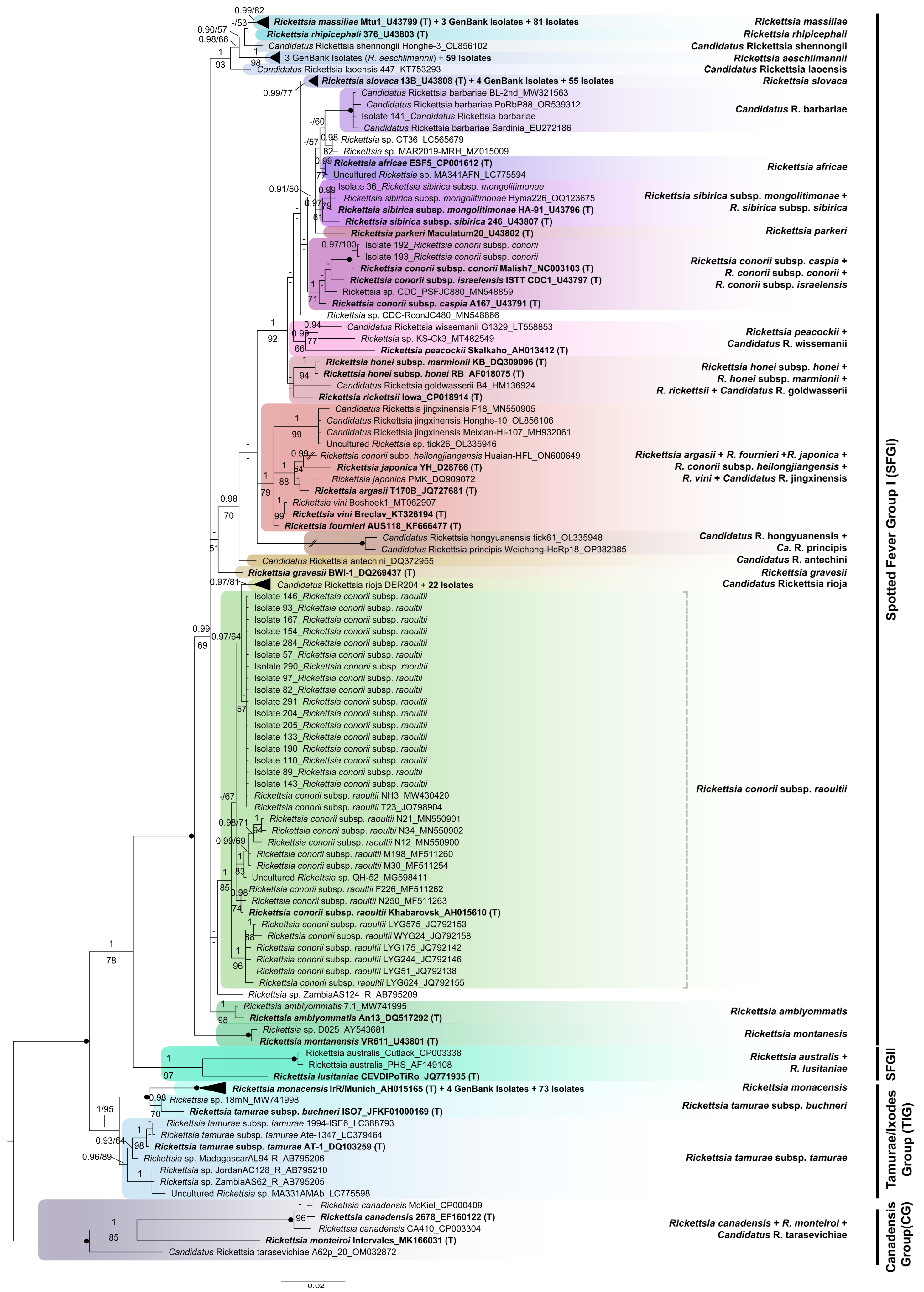

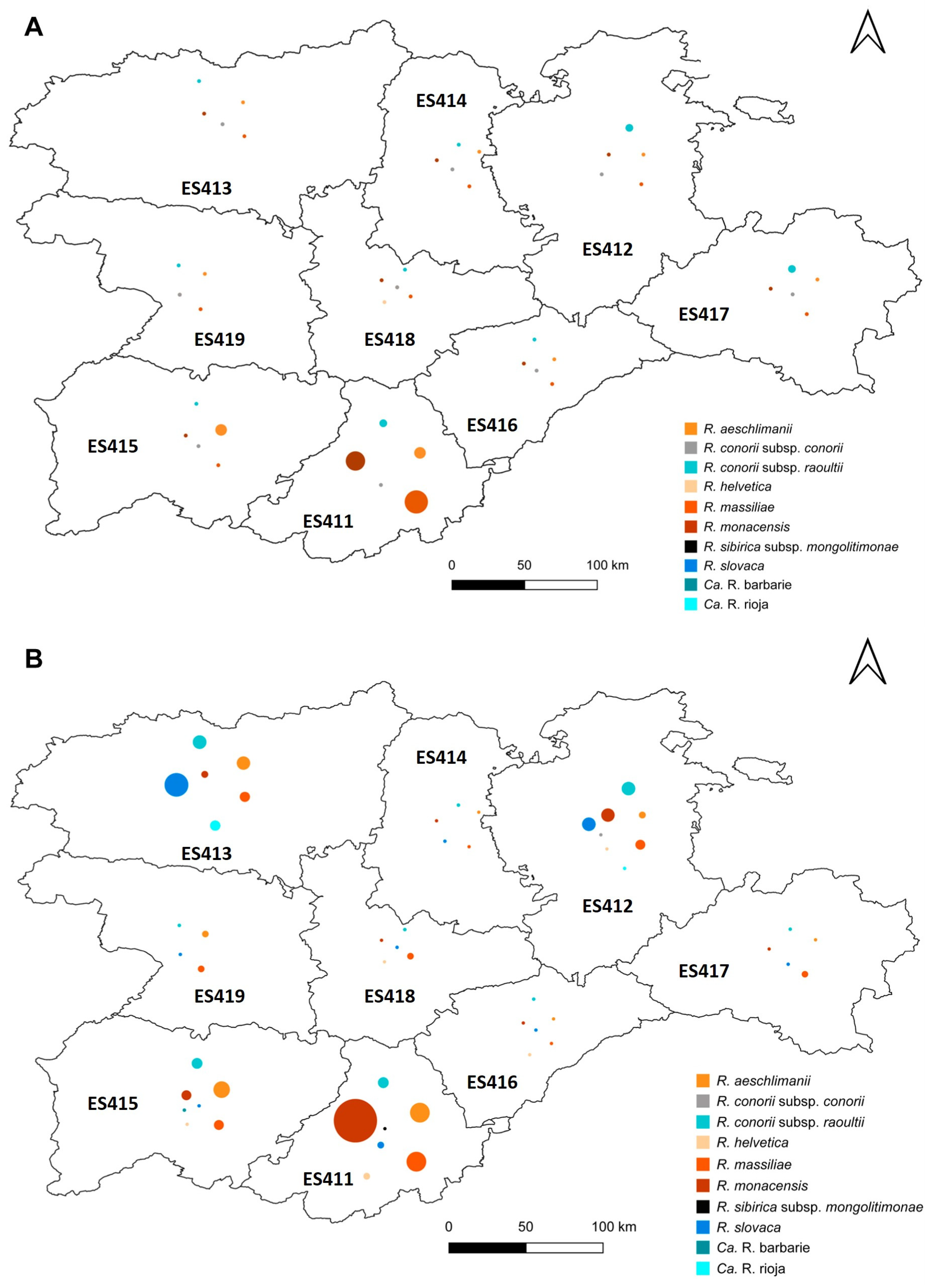
| Tick Species | No. Specimens Analysed (%) | No. Positive for Rickettsia | Percentage of Infected Specimens per Species |
|---|---|---|---|
| Dermacentor marginatus (Sulzer, 1776) | 588 (7.95%) | 211 | 35.88% |
| Dermacentor reticulatus (Fabricius, 1794) | 208 (2.81%) | 42 | 20.19% |
| Haemaphysalis punctata (Canestrini and Fanzago, 1877) | 121 (1.64%) | 7 | 5.79% |
| Hyalomma lusitanicum (Koch, 1844) | 531 (7.18%) | 48 | 9.04% |
| Hyalomma marginatum (Koch, 1844) | 874 (11.82%) | 135 | 15.45% |
| Ixodes ricinus (Linnaeus, 1758) | 3269 (44.19%) | 427 | 13.06% |
| Rhipicephalus bursa (Canestrini and Fanzago, 1877) | 1299 (17.56%) | 208 | 16.01% |
| Rhipicephalus sanguineus (Latreille, 1806) | 507 (6.85%) | 99 | 19.53% |
| Total | 7397 (100%) | 1177 | 15.91% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vieira Lista, M.C.; Vicente Santiago, M.B.; Soto-López, J.D.; García-Martín, J.M.; Álamo-Sanz, R.; Belhassen-García, M.; Muro, A. Identification of Rickettsia spp. in Ticks Removed from Tick-Bitten Humans in Northwestern Spain. Insects 2024, 15, 571. https://doi.org/10.3390/insects15080571
Vieira Lista MC, Vicente Santiago MB, Soto-López JD, García-Martín JM, Álamo-Sanz R, Belhassen-García M, Muro A. Identification of Rickettsia spp. in Ticks Removed from Tick-Bitten Humans in Northwestern Spain. Insects. 2024; 15(8):571. https://doi.org/10.3390/insects15080571
Chicago/Turabian StyleVieira Lista, María Carmen, María Belén Vicente Santiago, Julio David Soto-López, Joaquina María García-Martín, Rufino Álamo-Sanz, Moncef Belhassen-García, and Antonio Muro. 2024. "Identification of Rickettsia spp. in Ticks Removed from Tick-Bitten Humans in Northwestern Spain" Insects 15, no. 8: 571. https://doi.org/10.3390/insects15080571
APA StyleVieira Lista, M. C., Vicente Santiago, M. B., Soto-López, J. D., García-Martín, J. M., Álamo-Sanz, R., Belhassen-García, M., & Muro, A. (2024). Identification of Rickettsia spp. in Ticks Removed from Tick-Bitten Humans in Northwestern Spain. Insects, 15(8), 571. https://doi.org/10.3390/insects15080571







