Effects of Bacillus thuringiensis Treatment on Expression of Detoxification Genes in Chlorantraniliprole-Resistant Plutella xylostella
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Insects
2.2. Insecticides
2.3. Leaf-Dip Bioassays
2.4. Enzyme Assays
2.5. Quantitative Reverse-Transcription Polymerase Chain Reaction (qRT-PCR)
2.6. Functional Verification of CYP6B7 by RNAi
2.7. Effects of Resistant-Strain Exposure to a Sublethal Dose of Bt-G033
3. Results
3.1. Resistance to Chlorantraniliprole (CAP) and Bt-G033A of DBM Strains
3.2. Specific Activity of Detoxification Enzymes in CAP-Resistant and -Susceptible DBM Strains
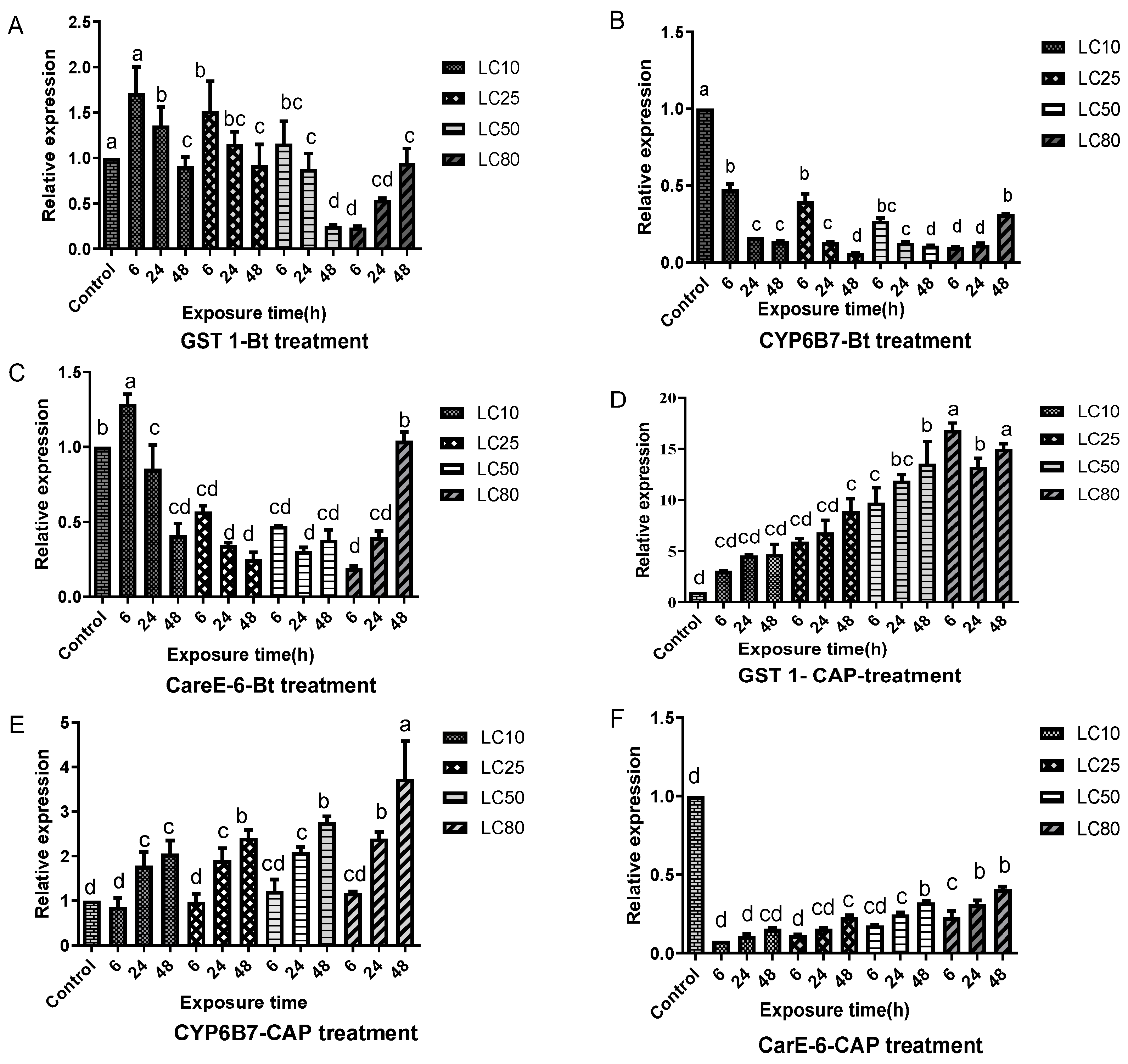
3.3. Differences in Detoxification Enzyme Gene Expression between Strains
3.4. Functional Test for Participation of CYP6B7 in Resistance to CAP
3.5. Sublethal Bt-G033 Pretreatment Does Not Significantly Increase Susceptibility to CAP in Resistant DBM
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zalucki, M.P.; Shabbir, A.; Silva, R.; Adamson, D.; Shu-Sheng, L.; Furlong, M.J. Estimating the economic cost of one of the world’s major insect pests, Plutella xylostella (Lepidoptera: Plutellidae): Just how long is a piece of string? J. Econ. Entomol. 2012, 105, 1115–1129. [Google Scholar] [CrossRef] [PubMed]
- Lahm, G.P.; Cordova, D.; Barry, J.D. New and selective ryanodine receptor activators for insect control. Bioorganic Med. Chem. 2009, 17, 4127–4133. [Google Scholar] [CrossRef]
- Cordova, D.; Benner, E.; Sacher, M.; Rauh, J.; Sopa, J.; Lahm, G.; Selby, T.; Stevenson, T.; Flexner, L.; Gutteridge, S. Anthranilic diamides: A new class of insecticides with a novel mode of action, ryanodine receptor activation. Pestic. Biochem. Physiol. 2006, 84, 196–214. [Google Scholar] [CrossRef]
- Lahm, G.P.; Selby, T.P.; Freudenberger, J.H.; Stevenson, T.M.; Myers, B.J.; Seburyamo, G.; Smith, B.K.; Flexner, L.; Clark, C.E.; Cordova, D. Insecticidal anthranilic diamides: A new class of potent ryanodine receptor activators. Bioorganic Med. Chem. Lett. 2005, 15, 4898–4906. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, R.; Zhu, B.; Gao, X.; Liang, P. Overexpression of cytochrome P450 CYP6BG1 may contribute to chlorantraniliprole resistance in Plutella xylostella (L.). Pest Manag. Sci. 2018, 74, 1386–1393. [Google Scholar] [CrossRef]
- Ribeiro, L.; Wanderley-Teixeira, V.; Ferreira, H.; Teixeira, Á.A.; Siqueira, H. Fitness costs associated with field-evolved resistance to chlorantraniliprole in Plutella xylostella (Lepidoptera: Plutellidae). Bull. Entomol. Res. 2014, 104, 88–96. [Google Scholar] [CrossRef]
- Troczka, B.; Zimmer, C.T.; Elias, J.; Schorn, C.; Bass, C.; Davies, T.E.; Field, L.M.; Williamson, M.S.; Slater, R.; Nauen, R. Resistance to diamide insecticides in diamondback moth, Plutella xylostella (Lepidoptera: Plutellidae) is associated with a mutation in the membrane-spanning domain of the ryanodine receptor. Insect Biochem. Mol. Biol. 2012, 42, 873–880. [Google Scholar] [CrossRef]
- Wang, X.; Wu, Y. High levels of resistance to chlorantraniliprole evolved in field populations of Plutella xylostella. J. Econ. Entomol. 2012, 105, 1019–1023. [Google Scholar] [CrossRef]
- Wang, Q.; Rui, C.; Wang, L.; Nahiyoon, S.A.; Huang, W.; Zhu, J.; Ji, X.; Yang, Q.; Yuan, H.; Cui, L. Field-evolved resistance to 11 insecticides and the mechanisms involved in Helicoverpa armigera (Lepidoptera: Noctuidae). Pest Manag. Sci. 2021, 77, 5086–5095. [Google Scholar] [CrossRef]
- Teng, H.; Zuo, Y.; Yuan, J.; Fabrick, J.A.; Wu, Y.; Yang, Y. High frequency of ryanodine receptor and cytochrome P450 CYP9A186 mutations in insecticide-resistant field populations of Spodoptera exigua from China. Pestic. Biochem. Physiol. 2022, 186, 105153. [Google Scholar] [CrossRef]
- Zhu, W.; Wang, J.; Zhang, Y. The Mechanism of Chlorantraniliprole Resistance and Detoxification in Trichogramma chilonis (Hymenoptera: Trichogrammatidae). J. Insect Sci. 2022, 22, 7. [Google Scholar] [CrossRef] [PubMed]
- Shan, J.; Sun, X.; Li, R.; Zhu, B.; Liang, P.; Gao, X. Identification of ABCG transporter genes associated with chlorantraniliprole resistance in Plutella xylostella (L.). Pest Manag. Sci. 2021, 77, 3491–3499. [Google Scholar] [CrossRef] [PubMed]
- Shabbir, M.Z.; Yang, X.; Batool, R.; Yin, F.; Kendra, P.E.; Li, Z.-Y. Bacillus thuringiensis and chlorantraniliprole trigger the expression of detoxification-related genes in the larval midgut of Plutella xylostella. Front. Physiol. 2021, 12, 780255. [Google Scholar] [CrossRef] [PubMed]
- Yin, F.; Lin, Q.; Wang, X.; Li, Z.; Feng, X.; Shabbir, M.Z. The glutathione S-transferase (PxGST2L) may contribute to the detoxification metabolism of chlorantraniliprole in Plutella xylostella (L.). Ecotoxicology 2021, 30, 1007–1016. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.-D.; Xia, F.; Lin, Q.-S.; Chen, H.-Y.; Li, Z.-Y.; Yin, F.; Liang, P.; Gao, X.-W. Biochemical mechanism of chlorantraniliprole resistance in the diamondback moth, Plutella xylostella Linnaeus. J. Integr. Agric. 2014, 13, 2452–2459. [Google Scholar] [CrossRef]
- Wang, X.; Khakame, S.K.; Ye, C.; Yang, Y.; Wu, Y. Characterisation of field-evolved resistance to chlorantraniliprole in the diamondback moth, Plutella xylostella, from China. Pest Manag. Sci. 2013, 69, 661–665. [Google Scholar] [CrossRef] [PubMed]
- Younas, A.; Wakil, W.; Khan, Z.; Shaaban, M.; Prager, S.M. The efficacy of Beauveria bassiana, jasmonic acid and chlorantraniliprole on larval populations of Helicoverpa armigera in chickpea crop ecosystems. Pest Manag. Sci. 2017, 73, 418–424. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Chen, Q.; Wang, J.; Hu, C.; Lu, J.; Luo, X.; Sun, D. Novel chlorantraniliprole derivatives as potential insecticides and probe to chlorantraniliprole binding site on ryanodine receptor. Bioorganic Med. Chem. Lett. 2014, 24, 1987–1992. [Google Scholar] [CrossRef]
- Xiao, Y.; Liu, K.; Zhang, D.; Gong, L.; He, F.; Soberón, M.; Bravo, A.; Tabashnik, B.E.; Wu, K. Resistance to Bacillus thuringiensis mediated by an ABC transporter mutation increases susceptibility to toxins from other bacteria in an invasive insect. PLoS Pathog. 2016, 12, e1005450. [Google Scholar] [CrossRef]
- Sarfraz, M.; Keddie, A.B.; Dosdall, L.M. Biological control of the diamondback moth, Plutella xylostella: A review. Biocontrol Sci. Technol. 2005, 15, 763–789. [Google Scholar] [CrossRef]
- Wang, G.; Zhang, J.; Song, F.; Wu, J.; Feng, S.; Huang, D. Engineered Bacillus thuringiensis GO33A with broad insecticidal activity against lepidopteran and coleopteran pests. Appl. Microbiol. Biotechnol. 2006, 72, 924–930. [Google Scholar] [CrossRef] [PubMed]
- Tabashnik, T.; Cushing, N. Leaf residue vs. topical bioassays for assessing insecticide resistance in the diamond-back moth, Plutella xylostella L. FAO Plant Prot. Bull. (FAO) 1987, 35, 11–14. [Google Scholar]
- Van Asperen, K. A study of housefly esterases by means of a sensitive colorimetric method. J. Insect Physiol. 1962, 8, 401–416. [Google Scholar] [CrossRef]
- Habig, W.H.; Pabst, M.J.; Jakoby, W.B. Glutathione S-transferases: The first enzymatic step in mercapturic acid formation. J. Biol. Chem. 1974, 249, 7130–7139. [Google Scholar] [CrossRef] [PubMed]
- Qian, L.; Cao, G.; Song, J.; Yin, Q.; Han, Z. Biochemical mechanisms conferring cross-resistance between tebufenozide and abamectin in Plutella xylostella. Pestic. Biochem. Physiol. 2008, 91, 175–179. [Google Scholar] [CrossRef]
- Rodenhouse, N.; Best, L.; O’Connor, R.; Bollinger, E.; O’Connor, R.; Bollinger, E.; O’Connor, R.; Bollinger, E. SAS 9.1.3; SAS Institute: Cary, NC, USA, 2004. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhang, Y. Identification and characterization of multiple glutathione S-transferase genes from the diamondback moth, Plutella xylostella. Pest Manag. Sci. 2015, 71, 592–600. [Google Scholar] [CrossRef] [PubMed]
- Crickmore, N. Bacillus thuringiensis resistance in Plutella—Too many trees? Curr. Opin. Insect Sci. 2016, 15, 84–88. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Liang, P.; Zhou, X.; Gao, X. Novel mutations and mutation combinations of ryanodine receptor in a chlorantraniliprole resistant population of Plutella xylostella (L.). Sci. Rep. 2014, 4, 6924. [Google Scholar] [CrossRef]
- Sparks, T.C.; Nauen, R. IRAC: Mode of action classification and insecticide resistance management. Pestic. Biochem. Physiol. 2015, 121, 122–128. [Google Scholar] [CrossRef]
- Sparks, T.C. Insecticide mixtures-uses, benefits and considerations. Pest Manag. Sci. 2024. ahead of print. [Google Scholar] [CrossRef]
- Madgwick, P.G.; Kanitz, R. Beyond redundant kill: A fundamental explanation of how insecticide mixtures work for resistance management. Pest Manag. Sci. 2023, 79, 495–506. [Google Scholar] [CrossRef] [PubMed]
- Khaliq, A.; Attique, M.N.; Sayyed, A.H. Evidence for resistance to pyrethroids and organophosphates in Plutella xylostella (Lepidoptera: Plutellidae) from Pakistan. Bull. Entomol. Res. 2007, 97, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Jiang, D.; Yu, Z.; He, Y.; Wang, F.; Gu, Y.; Davies, T.G.E.; Fan, Z.; Wang, X.; Wu, Y. Key role of the ryanodine receptor I4790K mutation in mediating diamide resistance in Plutella Xylostella. Insect Biochem. Mol. Biol. 2024, 168, 104–107. [Google Scholar] [CrossRef] [PubMed]
- Jouraku, A.; Kuwazaki, S.; Miyamoto, K.; Uchiyama, M.; Kurokawa, T.; Mori, E.; Mori, M.X.; Mori, Y.; Sonoda, S. Ryanodine receptor mutations (G4946E and I4790K) differentially responsible for diamide insecticide resistance in diamondback moth, Plutella xylostella L. Insect Biochem. Mol. Biol. 2020, 118, 103308. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Tian, Z.; Li, R.; Ni, S.; Sun, H.; Yin, F.; Li, Z.; Zhang, Y.; Li, Y. Key Contributions of the Overexpressed Plutella xylostella Sigma Glutathione S-Transferase 1 Gene (PxGSTs1) in the Resistance Evolution to Multiple Insecticides. J. Agric. Food Chem. 2024, 72, 2560–2572. [Google Scholar] [CrossRef] [PubMed]
- Mallott, M.D.; Hamm, S.; Troczka, B.J.; Randall, E.; Pym, A.; Grant, C.E.; Baxter, S.; Vogol, H.; Shelton, A.M.; Field, L.M.; et al. A flavin-dependent monooxgenase confers resistance to chlorantraniliprole in the diamondback moth, plutella xylostella. Insect Biochem. Mol. Biol. 2019, 115, 103247. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Shan, C.; Li, F.; Liang, P.; Smagghe, G.; Gao, X. Transcription factor FTZ-F1 and cis-acting elements mediate expression of CYP6BG1 conferring resistance to chlorantraniliprole in Plutella Xylostella. Pest Manag. Sci. 2019, 75, 1172–1180. [Google Scholar] [CrossRef] [PubMed]
- Dai, W.; Zhu, B.; Tuinen, M.V.; Zhu, T.; Shang, D.; Almeida, P.; Liang, P.; Ullah, H.; Ban, L. Genome-wide scans and transcriptomic analyses characterize selective changes as a result of chlorantraniliprole resistance in Plutella xylostella. Int. J. Mol. Sci. 2022, 23, 12245. [Google Scholar] [CrossRef]
- Nauen, R.; Bass, C.; Feyereisen, R.; Vontas, J. The Role of Cytochrome P450s in Insect Toxicology and Resistance. Annu. Rev. Entomol. 2022, 67, 105–124. [Google Scholar] [CrossRef]
- Anita, M.; Bautista, M.; Miyata, T.; Miura, K.; Tanaka, T. RNA interference-mediated knockdown of a cytochrome P450, CYP6BG1, from the diamondback moth, Plutella xylostella, reduces larval resistance to permethrin. Insect Biochem. Mol. Biol. 2009, 39, 38–46. [Google Scholar]
- Do Nascimento, A.R.B.; Rodrigues, J.G.; Kanno, R.H.; de Amaral, F.S.A.E.; Malaquias, J.B.; Silva-Brandão, K.L.; Cônsoli, F.L.; Omoto, C. Susceptibility monitoring and comparative gene expression of susceptible and resistant strains of Spodoptera frugiperda to lambda-cyhalothrin and chlorpyrifos. Pest Manag. Sci. 2023, 79, 2206–2219. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Li, X.; Cang, X.; Guo, J.; Shen, X.; Wu, K. Influence of seasonal migration on the development of the insecticide resistance of oriental armyworm (Mythimna separata) to λ-cyhalothrin. Pest Manag. Sci. 2022, 78, 1194–1205. [Google Scholar] [CrossRef] [PubMed]
- Perini, C.R.; Tabuloc, C.A.; Chiu, J.C.; Zalom, F.G.; Stacke, R.F.; Bernardi, O.; Nelson, D.R.; Guedes, J.C. Transcriptome Analysis of Pyrethroid-Resistant Chrysodeixis includens (Lepidoptera: Noctuidae) Reveals Overexpression of Metabolic Detoxification Genes. J. Econ. Entomol. 2021, 114, 274–283. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Mei, Y.; Liu, R.; Chen, X.; Li, D.; Wang, C. Transcriptome analysis of Spodoptera litura reveals the molecular mechanism to pyrethroids resistance. Pestic. Biochem. Physiol. 2020, 169, 104649. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.G.; Jiang, S.S.; Mota-Sanchez, D.; Wang, W.; Li, X.R.; Gao, Y.L.; Lu, X.P.; Yang, X.Q. Cytochrome P450-Mediated λ-Cyhalothrin-Resistance in a Field Strain of Helicoverpa armigera from Northeast China. J. Agric. Food Chem. 2019, 67, 3546–3553. [Google Scholar] [CrossRef] [PubMed]
- Hou, W.T.; Staehelin, C.; Elzaki, M.E.A.; Hafeez, M.; Luo, Y.S.; Wang, R.L. Functional analysis of CYP6AE68, a cytochrome P450 gene associated with indoxacarb resistance in Spodoptera litura (Lepidoptera: Noctuidae). Pestic. Biochem. Physiol. 2021, 178, 104946. [Google Scholar] [CrossRef] [PubMed]
- Riveron, J.M.; Irving, H.; Ndula, M.; Barnes, K.G.; Ibrahim, S.S.; Paine, M.J.; Wondji, C.S. Directionally selected cytochrome P450 alleles are driving the spread of pyrethroid resistance in the major malaria vector Anopheles funestus. Proc. Natl. Acad. Sci. USA 2013, 110, 252–257. [Google Scholar] [CrossRef] [PubMed]
- Pu, J.; Sun, H.; Wang, J.; Wu, M.; Wang, K.; Denholm, I.; Han, Z. Multiple cis-acting elements involved in up-regulation of a cytochrome P450 gene conferring resistance to deltamethrin in smal brown planthopper, Laodelphax striatellus (Fallén). Insect Biochem. Mol. Biol. 2016, 78, 20–28. [Google Scholar] [CrossRef]
- Tang, T.; Zhao, C.; Feng, X.; Liu, X.; Qiu, L. Knockdown of several components of cytochrome P450 enzyme systems by RNA interference enhances the susceptibility of Helicoverpa armigera to fenvalerate. Pest Manag. Sci. 2012, 68, 1501–1511. [Google Scholar] [CrossRef]


| Insecticide | Population | N a | LC50 (μg/mL) | 95% CI b | χ2 (df) | RR c |
|---|---|---|---|---|---|---|
| CAP | DBM-S | 150 | 0.05 | 0.044–0.056 | 0.9(3) | 1 |
| GZ-R | 150 | 27.4 | 21.3–34.8 | 0.8(3) | 548 | |
| HZ-R | 150 | 52.84 | 45.67–62.37 | 0.7(3) | 1057 | |
| Bt-G033A | DBM-S | 150 | 0.012 | 0.01–0.0142 | 0.7(3) | 1 |
| GZ-R | 150 | 0.08 | 0.07–0.088 | 0.8(3) | 7 | |
| HZ-R | 150 | 0.15 | 0.1–0.25 | 0.7(3) | 12 |
| Strain | Esterase Mmol/min/mg Protein ± SE | a AR | GST Mmol/min/mg Protein ± SE | AR | MFO nmol/min/mg Protein ± SE | AR |
|---|---|---|---|---|---|---|
| DBM-S | 0.014 ± 0.005 a | 1 | 0.12 ± 0.01 a | 1 | 3.9 ± 0.02 a | 1 |
| GZ-R | 0.027 ± 0.001 b | 1.9 | 0.43 ± 0.03 b | 3.5 | 5.6 ± 0.04 b | 1.4 |
| HZ-R | 0.040 ± 0.003 c | 2.8 | 0.20 ± 0.03 c | 1.6 | 4.8 ± 0.01 c | 1.2 |
| Strain | Treatment | Larval Mortality (%) |
|---|---|---|
| GZ-R | Bt + CAP | 45.0 ± 1.6 ab |
| Water + CAP | 54.0 ± 1.9 b | |
| Bt | 16.7 ± 6.7 c | |
| Water | 6.7 ± 3.3 de | |
| HZ-R | Bt + CAP | 45.0 ± 1.6 ab |
| Water + CAP | 40.0 ± 2.2 b | |
| Bt | 10.0 ± 5.8 cd | |
| Water | 0.0 ± 0.0 e |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zolfaghari, M.; Yin, F.; Jurat-Fuentes, J.L.; Xiao, Y.; Peng, Z.; Wang, J.; Yang, X.; Li, Z.-Y. Effects of Bacillus thuringiensis Treatment on Expression of Detoxification Genes in Chlorantraniliprole-Resistant Plutella xylostella. Insects 2024, 15, 595. https://doi.org/10.3390/insects15080595
Zolfaghari M, Yin F, Jurat-Fuentes JL, Xiao Y, Peng Z, Wang J, Yang X, Li Z-Y. Effects of Bacillus thuringiensis Treatment on Expression of Detoxification Genes in Chlorantraniliprole-Resistant Plutella xylostella. Insects. 2024; 15(8):595. https://doi.org/10.3390/insects15080595
Chicago/Turabian StyleZolfaghari, Maryam, Fei Yin, Juan Luis Jurat-Fuentes, Yong Xiao, Zhengke Peng, Jiale Wang, Xiangbing Yang, and Zhen-Yu Li. 2024. "Effects of Bacillus thuringiensis Treatment on Expression of Detoxification Genes in Chlorantraniliprole-Resistant Plutella xylostella" Insects 15, no. 8: 595. https://doi.org/10.3390/insects15080595








