Genetic Diversity of Whiteflies Colonizing Crops and Their Associated Endosymbionts in Three Agroecological Zones of Cameroon
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Whitefly Sampling and Population Study
2.1.1. Field Survey
2.1.2. Data Recording and Storage
2.2. Genetic Diversity of Sampled Whiteflies
2.2.1. DNA Extraction
2.2.2. MtCOI Amplification and Sequencing
2.2.3. Kompetitive Allele-Specific PCR (KASP)
2.3. Infectivity of Endosymbionts in Whiteflies
Screening for the Presence of Endosymbionts by PCR
3. Results
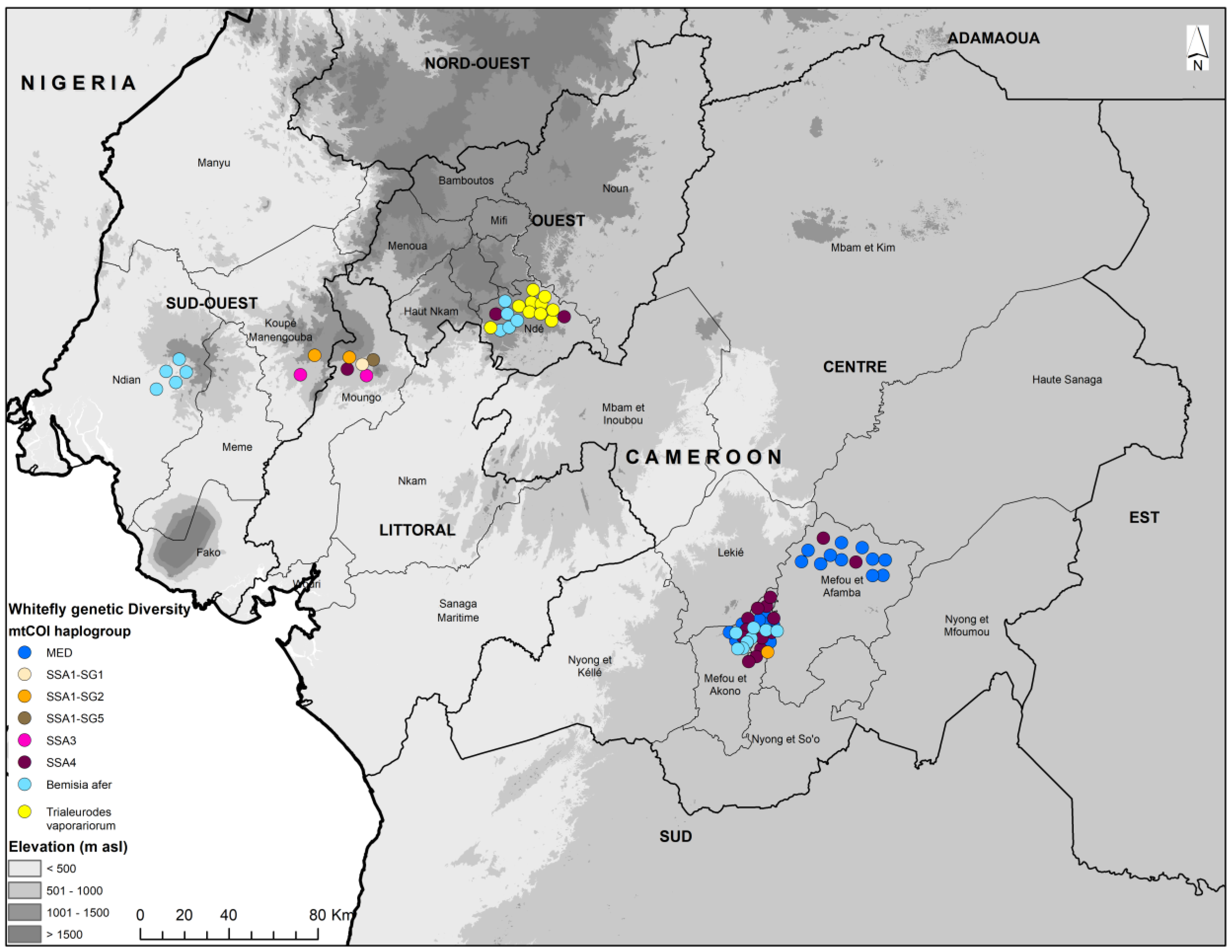
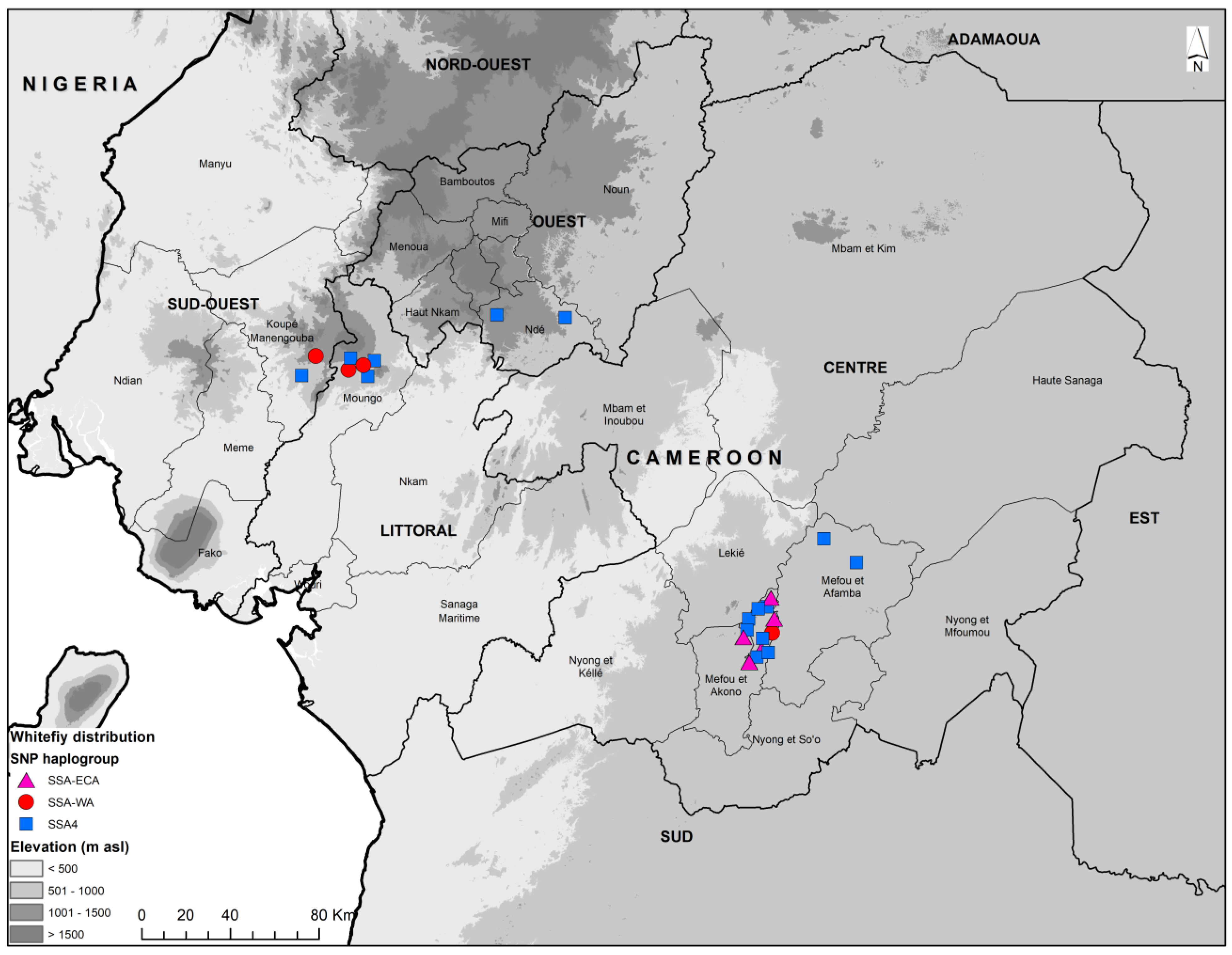
3.1. Whitefly Abundance
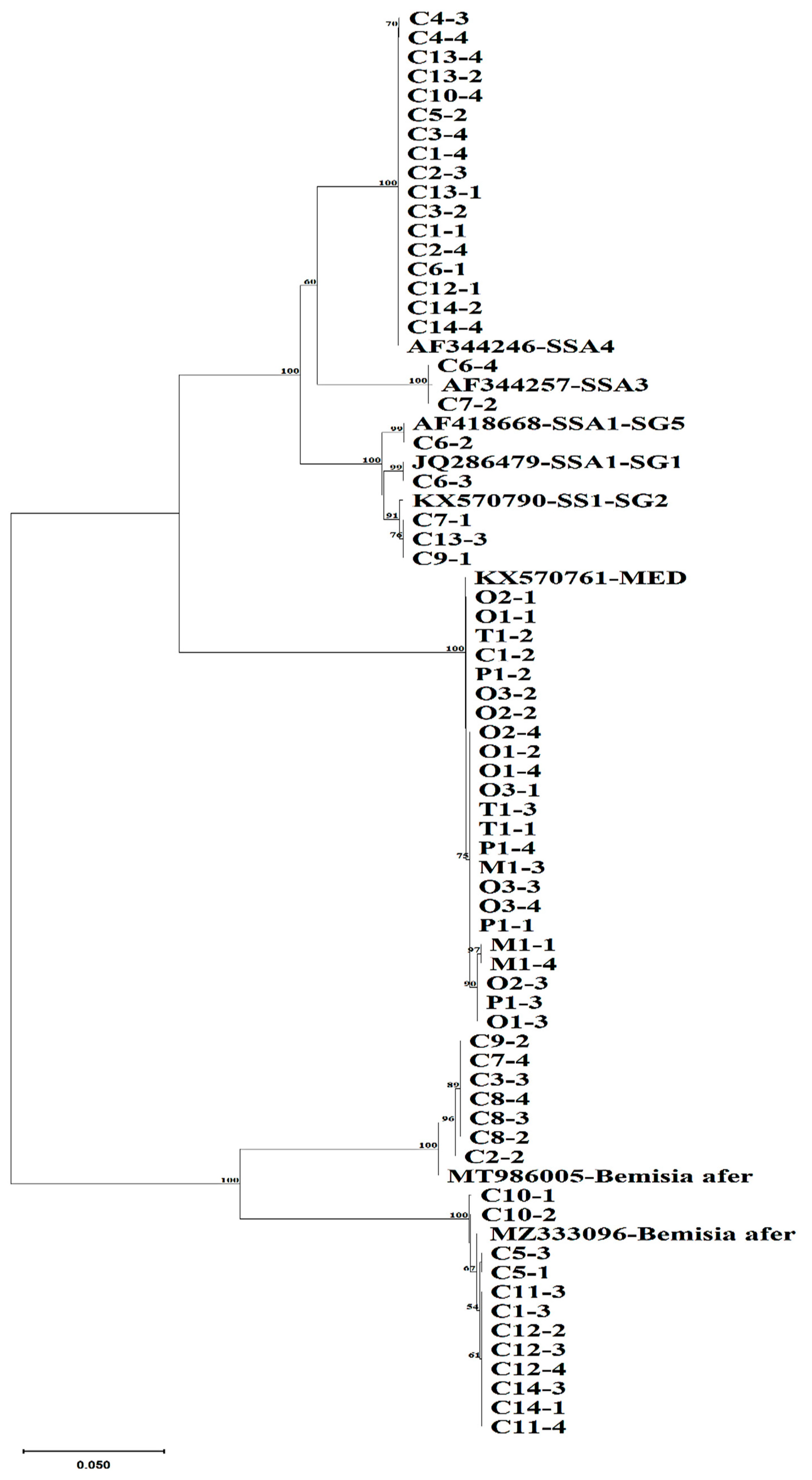
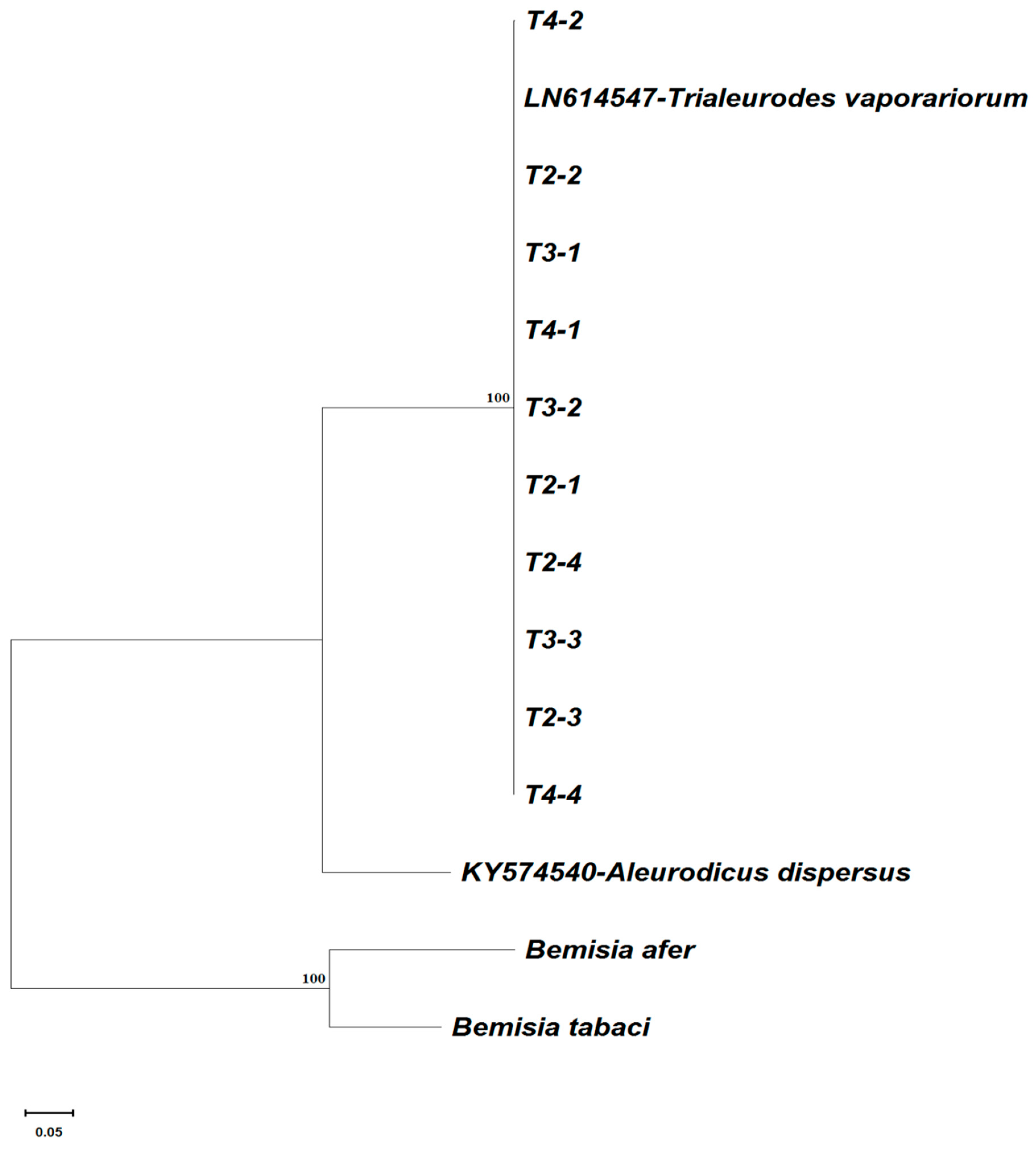
3.2. mtCOI Mitotypes of Whiteflies Colonizing Five Crop Plants in Cameroon
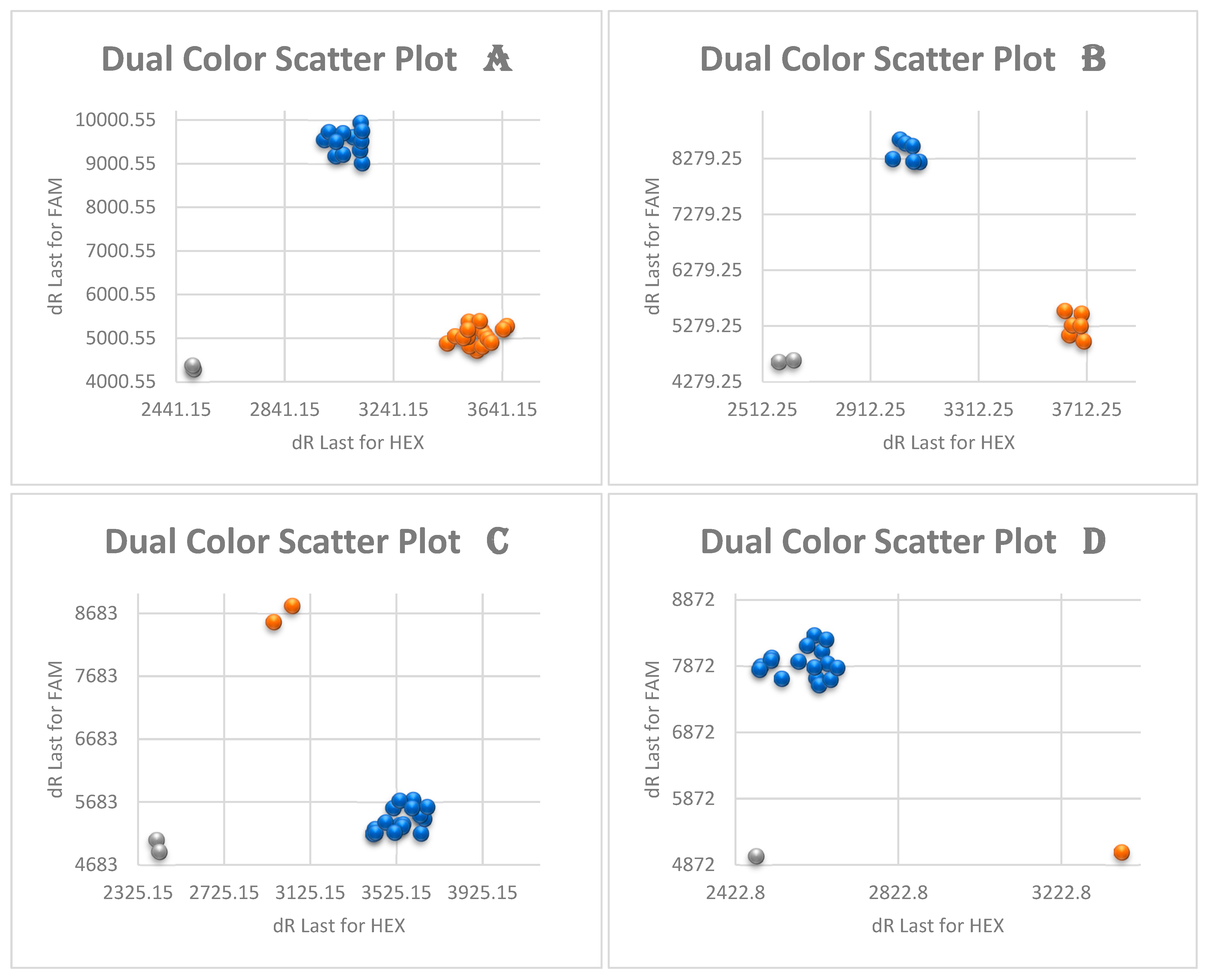
3.3. Kompetitive Allele Specific PCR Analysis (KASP)
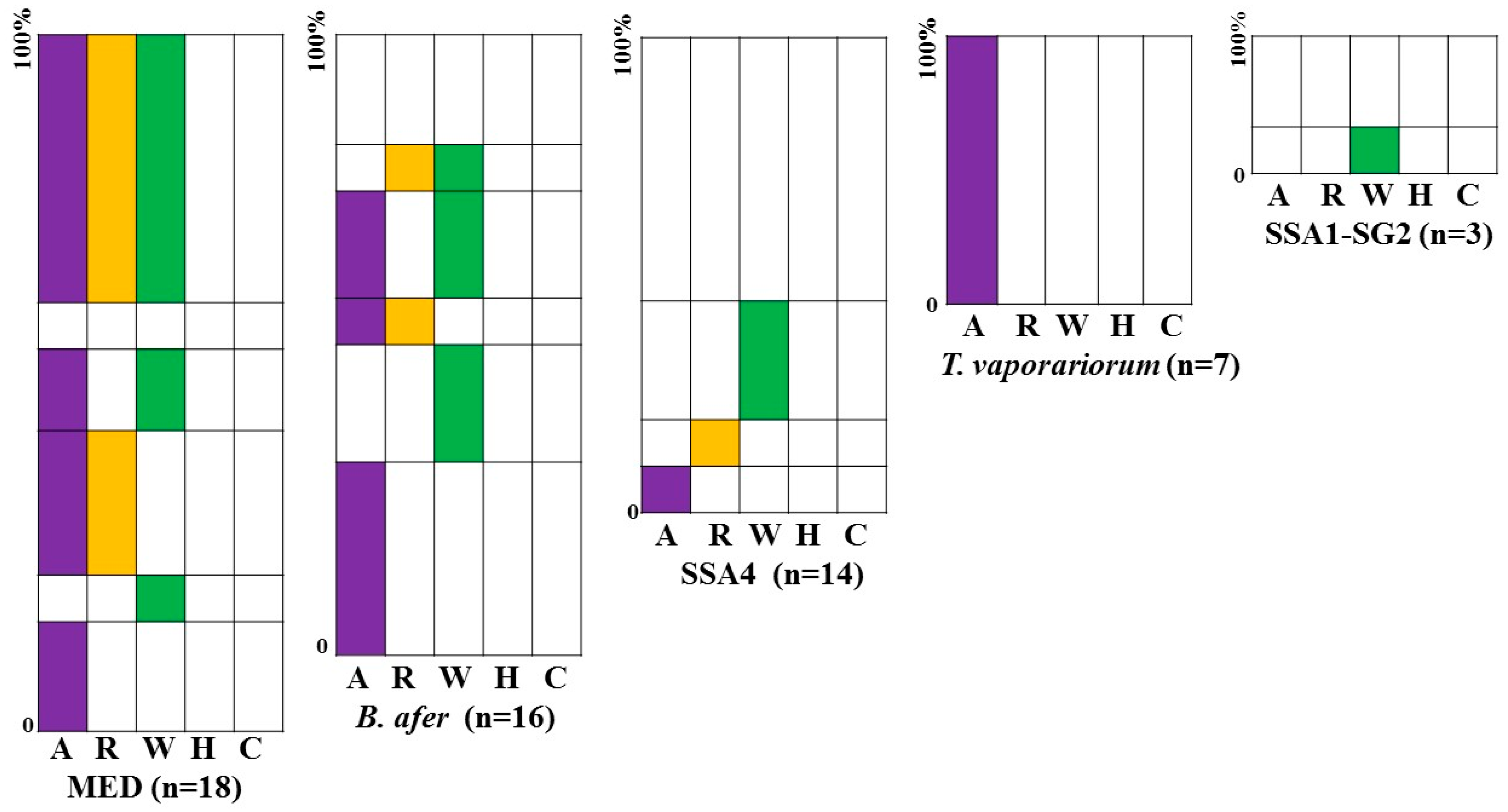
3.4. Frequency of Infection of Whiteflies by Three Endosymbionts
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gilbertson, R.L.; Batuman, O.; Webster, C.G.; Adkins, S. Role of the Insect Supervectors Bemisia tabaci and Frankliniella occidentalis in the Emergence and Global Spread of Plant Viruses. Annu. Rev. Virol. 2015, 2, 67–93. [Google Scholar] [CrossRef]
- Dalmon, A.; Halkett, F.; Granier, M.; Delatte, H.; Peterschmitt, M. Genetic Structure of the Invasive Pest Bemisia tabaci: Evidence of Limited but Persistent Genetic Differentiation in Glasshouse Populations. Heredity 2008, 100, 316–325. [Google Scholar] [CrossRef] [PubMed]
- De Barro, P.J.; Liu, S.-S.; Boykin, L.M.; Dinsdale, A.B. Bemisia tabaci: A Statement of Species Status. Annu. Rev. Entomol. 2011, 56, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Gnankiné, O.; Bassolé, I.H.N.; Chandre, F.; Glitho, I.; Akogbeto, M.; Dabiré, R.K.; Martin, T. Insecticide Resistance in Bemisia tabaci Gennadius (Homoptera: Aleyrodidae) and Anopheles Gambiae Giles (Diptera: Culicidae) Could Compromise the Sustainability of Malaria Vector Control Strategies in West Africa. Acta Tropica 2013, 128, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Lapidot, M.; Legg, J.P.; Wintermantel, W.M.; Polston, J.E. Management of Whitefly-Transmitted Viruses in Open-Field Production Systems. Adv. Virus Res. 2014, 90, 147–206. [Google Scholar] [CrossRef] [PubMed]
- Henneberry, T.J.; Faust, R.M. Introduction. In Classical Biological Control of Bemisia tabaci in the United States—A Review of Interagency Research and Implementation; Gould, J., Hoelmer, K., Goolsby, J., Eds.; Progress in Biological Control; Springer: Dordrecht, The Netherlands, 2008; pp. 1–15. ISBN 978-1-4020-6740-2. [Google Scholar]
- Legg, J.P.; Shirima, R.; Tajebe, L.S.; Guastella, D.; Boniface, S.; Jeremiah, S.; Nsami, E.; Chikoti, P.; Rapisarda, C. Biology and Management of Bemisia Whitefly Vectors of Cassava Virus Pandemics in Africa. Pest Manag. Sci. 2014, 70, 1446–1453. [Google Scholar] [CrossRef]
- Alemandri, V.; De Barro, P.; Bejerman, N.; Caro, E.B.A.; Dumón, A.D.; Mattio, M.F.; Rodriguez, S.M.; Truol, G. Species Within the Bemisia tabaci (Hemiptera: Aleyrodidae) Complex in Soybean and Bean Crops in Argentina. J. Econ. Entomol. 2012, 105, 48–53. [Google Scholar] [CrossRef]
- Parrella, G.; Scassillo, L.; Giorgini, M. Evidence for a New Genetic Variant in the Bemisia tabaci Species Complex and the Prevalence of the Biotype Q in Southern Italy. J. Pest Sci. 2012, 85, 227–238. [Google Scholar] [CrossRef]
- Boykin, L.M.; Shatters, R.G.; Rosell, R.C.; McKenzie, C.L.; Bagnall, R.A.; De Barro, P.; Frohlich, D.R. Global Relationships of Bemisia tabaci (Hemiptera: Aleyrodidae) Revealed Using Bayesian Analysis of Mitochondrial COI DNA Sequences. Mol. Phylogenetics Evol. 2007, 44, 1306–1319. [Google Scholar] [CrossRef]
- Delatte, H.; Rémy, B.; Nathalie, B.; Anne-Laure, G.; Traoré, R.S.; Jean-Michel, L.; Bernard, R. Species and Endosymbiont Diversity of Bemisia tabaci (Homoptera: Aleyrodidae) on Vegetable Crops in Senegal. Insect Sci. 2015, 22, 386–398. [Google Scholar] [CrossRef]
- Legg, J.P.; French, R.; Rogan, D.; Okao-Okuja, G.; Brown, J.K. A Distinct Bemisia tabaci (Gennadius) (Hemiptera: Sternorrhyncha: Aleyrodidae) Genotype Cluster Is Associated with the Epidemic of Severe Cassava Mosaic Virus Disease in Uganda. Mol. Ecol. 2002, 11, 1219–1229. [Google Scholar] [CrossRef]
- Taquet, A.; Delatte, H.; Barrès, B.; Simiand, C.; Grondin, M.; Jourdan-Pineau, H. Insecticide Resistance and Fitness Cost in Bemisia Tabaci (Hemiptera: Aleyrodidae) Invasive and Resident Species in La Réunion Island. Pest Manag. Sci. 2020, 76, 1235–1244. [Google Scholar] [CrossRef] [PubMed]
- Kontsedalov, S.; Zchori-Fein, E.; Chiel, E.; Gottlieb, Y.; Inbar, M.; Ghanim, M. The Presence of Rickettsia Is Associated with Increased Susceptibility of Bemisia Tabaci (Homoptera: Aleyrodidae) to Insecticides. Pest Manag. Sci. 2008, 64, 789–792. [Google Scholar] [CrossRef]
- Morin, S.; Ghanim, M.; Sobol, I.; Czosnek, H. The GroEL Protein of the Whitefly Bemisia tabaci Interacts with the Coat Protein of Transmissible and Nontransmissible Begomoviruses in the Yeast Two-Hybrid System. Virology 2000, 276, 404–416. [Google Scholar] [CrossRef] [PubMed]
- Gottlieb, Y.; Zchori-Fein, E.; Mozes-Daube, N.; Kontsedalov, S.; Skaljac, M.; Brumin, M.; Sobol, I.; Czosnek, H.; Vavre, F.; Fleury, F.; et al. The Transmission Efficiency of Tomato Yellow Leaf Curl Virus by the Whitefly Bemisia tabaci Is Correlated with the Presence of a Specific Symbiotic Bacterium Species. J. Virol. 2010, 84, 9310–9317. [Google Scholar] [CrossRef]
- Brumin, M.; Kontsedalov, S.; Ghanim, M. Rickettsia Influences Thermotolerance in the Whitefly Bemisia tabaci B Biotype. Insect Sci. 2011, 18, 57–66. [Google Scholar] [CrossRef]
- Gueguen, G.; Vavre, F.; Gnankine, O.; Peterschmitt, M.; Charif, D.; Chiel, E.; Gottlieb, Y.; Ghanim, M.; Zchori-Fein, E.; Fleury, F. Endosymbiont Metacommunities, mtDNA Diversity and the Evolution of the Bemisia tabaci (Hemiptera: Aleyrodidae) Species Complex. Mol. Ecol. 2010, 19, 4365–4376. [Google Scholar] [CrossRef]
- Raina, H.S.; Singh, A.; Popli, S.; Pandey, N.; Rajagopal, R. Infection of Bacterial Endosymbionts in Insects: A Comparative Study of Two Techniques Viz PCR and FISH for Detection and Localization of Symbionts in Whitefly, Bemisia tabaci. PLoS ONE 2015, 10, e0136159. [Google Scholar] [CrossRef] [PubMed]
- Thao, M.L.; Baumann, P. Evolutionary Relationships of Primary Prokaryotic Endosymbionts of Whiteflies and Their Hosts. Appl. Environ. Microbiol. 2004, 70, 3401–3406. [Google Scholar] [CrossRef]
- Sloan, D.B.; Moran, N.A. Endosymbiotic Bacteria as a Source of Carotenoids in Whiteflies. Biol. Lett. 2012, 8, 986–989. [Google Scholar] [CrossRef]
- Moran, A.N.; Telang, A. Bacteriocyte-Associated Symbionts of Insects. Biosciences 1998, 48, 295–304. [Google Scholar] [CrossRef]
- Baumann, P. Biology of Bacteriocyte-Associated Endosymbionts of Plant Sap-Sucking Insects. Annu. Rev. Microbiol. 2005, 59, 155–189. [Google Scholar] [CrossRef]
- Gottlieb, Y.; Ghanim, M.; Chiel, E.; Gerling, D.; Portnoy, V.; Steinberg, S.; Tzuri, G.; Horowitz, A.R.; Belausov, E.; Mozes-Daube, N.; et al. Identification and Localization of a Rickettsia Sp. in Bemisia tabaci (Homoptera: Aleyrodidae). Appl. Environ. Microbiol. 2006, 72, 3646–3652. [Google Scholar] [CrossRef] [PubMed]
- Zchori-Fein, E.; Brown, J.K. Diversity of Prokaryotes Associated with Bemisia tabaci (Gennadius) (Hemiptera: Aleyrodidae). Ann. Entomol. Soc. Am. 2002, 95, 711–718. [Google Scholar] [CrossRef]
- Nirgianaki, A.; Banks, G.K.; Frohlich, D.R.; Veneti, Z.; Braig, H.R.; Miller, T.A.; Bedford, I.D.; Markham, P.G.; Savakis, C.; Bourtzis, K. Wolbachia Infections of the Whitefly Bemisia tabaci. Curr. Microbiol. 2003, 47, 0093–0101. [Google Scholar]
- Weeks, A.R.; Velten, R.; Stouthamer, R. Incidence of a New Sex–Ratio Distorting Endosymbiotic Bacterium among Arthropods. Proc. Biol. Sci. 2003, 270, 1857–1865. [Google Scholar] [CrossRef] [PubMed]
- Everett, K.D.; Thao, M.; Horn, M.; Dyszynski, G.E.; Baumann, P. Novel Chlamydiae in Whiteflies and Scale Insects: Endosymbionts ‘Candidatus Fritschea Bemisiae’ Strain Falk and ‘Candidatus Fritschea Eriococci’ Strain Elm. Int. J. Syst. Evol. Microbiol. 2005, 55, 1581–1587. [Google Scholar] [CrossRef] [PubMed]
- Zchori-Fein, E.; Perlman, S.J. Distribution of the Bacterial Symbiont Cardinium in Arthropods. Mol. Ecol. 2004, 13, 2009–2016. [Google Scholar] [CrossRef]
- Chiel, E.; Gottlieb, Y.; Zchori-Fein, E.; Mozes-Daube, N.; Katzir, N.; Inbar, M.; Ghanim, M. Biotype-Dependent Secondary Symbiont Communities in Sympatric Populations of Bemisia tabaci. Bull. Entomol. Res. 2007, 97, 407–413. [Google Scholar] [CrossRef]
- Su, Q.; Xie, W.; Wang, S.; Wu, Q.; Liu, B.; Fang, Y.; Xu, B.; Zhang, Y. The Endosymbiont Hamiltonella Increases the Growth Rate of Its Host Bemisia tabaci during Periods of Nutritional Stress. PLoS ONE 2014, 9, e89002. [Google Scholar] [CrossRef]
- Himler, A.G.; Adachi-Hagimori, T.; Bergen, J.E.; Kozuch, A.; Kelly, S.E.; Tabashnik, B.E.; Chiel, E.; Duckworth, V.E.; Dennehy, T.J.; Zchori-Fein, E.; et al. Rapid Spread of a Bacterial Symbiont in an Invasive Whitefly Is Driven by Fitness Benefits and Female Bias. Science 2011, 332, 254–256. [Google Scholar] [CrossRef]
- Shan, H.W.; Zhang, C.R.; Yan, T.T.; Tang, H.Q.; Wang, X.W.; Liu, S.S.; Liu, Y.Q. Temporal Changes of Symbiont Density and Host Fitness after Rifampicin Treatment in a Whitefly of the Bemisia tabaci Species Complex. Insect Sci. 2016, 23, 200–214. [Google Scholar] [CrossRef] [PubMed]
- Hendry, T.A.; Hunter, M.S.; Baltrus, D.A. The Facultative Symbiont Rickettsia Protects an Invasive Whitefly against Entomopathogenic Pseudomonas Syringae Strains. Appl. Environ. Microbiol. 2014, 80, 7161–7168. [Google Scholar] [CrossRef]
- Gottlieb, Y.; Ghanim, M.; Gueguen, G.; Kontsedalov, S.; Vavre, F.; Fleury, F.; Zchori-Fein, E. Inherited Intracellular Ecosystem: Symbiotic Bacteria Share Bacteriocytes in Whiteflies. FASEB J. 2008, 22, 2591–2599. [Google Scholar] [CrossRef] [PubMed]
- Morin, S.; Ghanim, M.; Zeidan, M.; Czosnek, H.; Verbeek, M.; Van den Heuvel, J.F. A GroEL Homologue from Endosymbiotic Bacteria of the Whitefly Bemisia tabaci Is Implicated in the Circulative Transmission of Tomato Yellow Leaf Curl Virus. Virology 1999, 256, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Rana, V.S.; Singh, S.T.; Priya, N.G.; Kumar, J.; Rajagopal, R. Arsenophonus GroEL Interacts with CLCuV and Is Localized in Midgut and Salivary Gland of Whitefly B. tabaci. PLoS ONE 2012, 7, e42168. [Google Scholar] [CrossRef]
- Bing, X.L.; Yang, J.; Zchori-Fein, E.; Wang, X.W.; Liu, S.S. Characterization of a Newly Discovered Symbiont of the Whitefly Bemisia tabaci (Hemiptera: Aleyrodidae). Appl. Environ. Microbiol. 2013, 79, 569–575. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Bouvaine, S.; Maruthi, M. Prevalence and Genetic Diversity of Endosymbiotic Bacteria Infecting Cassava Whiteflies in Africa. BMC Microbiol. 2015, 15, 93. [Google Scholar] [CrossRef]
- Skaljac, M.; Kanakala, S.; Zanic, K.; Puizina, J.; Pleic, I.L.; Ghanim, M. Diversity and Phylogenetic Analyses of Bacterial Symbionts in Three Whitefly Species from Southeast Europe. Insects 2017, 8, 113. [Google Scholar] [CrossRef]
- Chu, D.; Gao, C.S.; De Barro, P.; Zhang, Y.J.; Wan, F.H.; Khan, I.A. Further Insights into the Strange Role of Bacterial Endosymbionts in Whitefly, Bemisia tabaci: Comparison of Secondary Symbionts from Biotypes B and Q in China. Bull. Entomol. Res. 2011, 101, 477–486. [Google Scholar] [CrossRef]
- Singh, S.T.; Priya, N.G.; Kumar, J.; Rana, V.S.; Ellango, R.; Joshi, A.; Priyadarshini, G.; Asokan, R.; Rajagopal, R. Diversity and Phylogenetic Analysis of Endosymbiotic Bacteria from Field Caught Bemisia tabaci from Different Locations of North India Based on 16S rDNA Library Screening. Infect. Genet. Evol. 2012, 12, 411–419. [Google Scholar] [CrossRef] [PubMed]
- Gorsane, F.; Ben Halima, A.; Ben Khalifa, M.; Bel-Kadhi, M.S.; Fakhfakh, H. Molecular Characterization of Bemisia tabaci Populations in Tunisia: Genetic Structure and Evidence for Multiple Acquisition of Secondary Symbionts. Environ. Entomol. 2011, 40, 809–817. [Google Scholar] [CrossRef] [PubMed]
- Su, Q.; Pan, H.; Liu, B.; Chu, D.; Xie, W.; Wu, Q.; Wang, S.; Xu, B.; Zhang, Y. Insect Symbiont Facilitates Vector Acquisition, Retention, and Transmission of Plant Virus. Sci. Rep. 2013, 3, 1367. [Google Scholar] [CrossRef] [PubMed]
- Kliot, A.; Cilia, M.; Czosnek, H.; Ghanim, M. Infection of the Whitefly Bemisia tabaci with Rickettsia Spp. Alters Its Interactions with Tomato Yellow Leaf Curl Virus. J. Virol. 2014, 88, 5652–5660. [Google Scholar] [CrossRef]
- Ghosh, S.; Bouvaine, S.; Richardson, S.C.W.; Ghanim, M.; Maruthi, M.N. Fitness Costs Associated with Infections of Secondary Endosymbionts in the Cassava Whitefly Species Bemisia tabaci. J. Pest Sci. 2018, 91, 17–28. [Google Scholar] [CrossRef]
- Skaljac, M.; Zanic, K.; Ban, S.G.; Kontsedalov, S.; Ghanim, M. Co-Infection and Localization of Secondary Symbionts in Two Whitefly Species. BMC Microbiol. 2010, 10, 142. [Google Scholar] [CrossRef]
- Sseruwagi, P.; Sserubombwe, W.S.; Legg, J.P.; Ndunguru, J.; Thresh, J.M. Methods of Surveying the Incidence and Severity of Cassava Mosaic Disease and Whitefly Vector Populations on Cassava in Africa: A Review. Virus Res. 2004, 100, 129–142. [Google Scholar] [CrossRef]
- Eni, A.O.; Efekemo, O.P.; Onile-ere, O.A.; Pita, J.S. South West and North Central Nigeria: Assessment of Cassava Mosaic Disease and Field Status of African Cassava Mosaic Virus and East African Cassava Mosaic Virus. Ann. Appl. Biol. 2021, 178, 466–479. [Google Scholar] [CrossRef]
- Soro, M.; Tiendrebeogo, F.; Pita, J.S.; Traore, E.T.; Some, K.; Tibiri, E.B.; Neya, J.B.; Mutuku, J.M.; Simpore, J.; Kone, D. Epidemiological Assessment of Cassava Mosaic Disease in Burkina Faso. Plant Pathol. 2021, 70, 2207–2216. [Google Scholar] [CrossRef]
- Mugerwa, H.; Seal, S.; Wang, H.-L.; Patel, M.V.; Kabaalu, R.; Omongo, C.A.; Alicai, T.; Tairo, F.; Ndunguru, J.; Sseruwagi, P.; et al. African Ancestry of New World, Bemisia tabaci-Whitefly Species. Sci. Rep. 2018, 8, 2734. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Wosula, E.N.; Chen, W.; Amour, M.; Fei, Z.; Legg, J.P. KASP Genotyping as a Molecular Tool for Diagnosis of Cassava-Colonizing Bemisia tabaci. Insects 2020, 11, 305. [Google Scholar] [CrossRef] [PubMed]
- Casinga, C.M.; Wosula, E.N.; Sikirou, M.; Shirima, R.R.; Munyerenkana, C.M.; Nabahungu, L.N.; Bashizi, B.K.; Ugentho, H.; Monde, G.; Legg, J.P. Diversity and Distribution of Whiteflies Colonizing Cassava in Eastern Democratic Republic of Congo. Insects 2022, 13, 849. [Google Scholar] [CrossRef] [PubMed]
- Misaka, B.C.; Wosula, E.N.; Marchelo-d’Ragga, P.W.; Hvoslef-Eide, T.; Legg, J.P. Genetic Diversity of Bemisia tabaci (Gennadius) (Hemiptera: Aleyrodidae) Colonizing Sweet Potato and Cassava in South Sudan. Insects 2020, 11, 58. [Google Scholar] [CrossRef]
- Chen, W.; Wosula, E.N.; Hasegawa, D.K.; Casinga, C.; Shirima, R.R.; Fiaboe, K.K.M.; Hanna, R.; Fosto, A.; Goergen, G.; Tamò, M.; et al. Genome of the African Cassava Whitefly Bemisia tabaci and Distribution and Genetic Diversity of Cassava-Colonizing Whiteflies in Africa. Insect Biochem. Mol. Biol. 2019, 110, 112–120. [Google Scholar] [CrossRef]
- Wosula, E.N.; Chen, W.; Fei, Z.; Legg, J.P. Unravelling the Genetic Diversity among Cassava Bemisia tabaci Whiteflies Using NextRAD Sequencing. Genome Biol. Evol. 2017, 9, 2958–2973. [Google Scholar] [CrossRef]
- Shirima, R.R.; Wosula, E.N.; Hamza, A.A.; Mohammed, N.A.; Mouigni, H.; Nouhou, S.; Mchinda, N.M.; Ceasar, G.; Amour, M.; Njukwe, E.; et al. Epidemiological Analysis of Cassava Mosaic and Brown Streak Diseases, and Bemisia tabaci in the Comoros Islands. Viruses 2022, 14, 2165. [Google Scholar] [CrossRef]
- Lobin, K.K.; Jaunky, V.C.; Taleb-Hossenkhan, N. A Meta-Analysis of Climatic Conditions and Whitely Bemisia tabaci Population: Implications for Tomato Yellow Leaf Curl Disease. J. Basic Appl. Zool. 2022, 83, 57. [Google Scholar] [CrossRef]
- Ahmad, S.; Saljoqi, A.-U.-R. Population Assessment of Bemisia tabaci (Gennadius.) and Disease Occurrence of Begomovirus in Okra, Abelmoschus esculentus L. J. Saudi Soc. Agriculral Sci. 2022, 21, 77–86. [Google Scholar] [CrossRef]
- Manzano, M.R.; Van Lenteren, J.C. Life History Parameters of Trialeurodes Vaporariorum (Westwood) (Hemiptera: Aleyrodidae) at Different Environmental Conditions on Two1 Bean Cultivars. Neotrop. Entomol. 2009, 38, 452–458. [Google Scholar] [CrossRef]
- Gamarra, H.A.; Fuentes, S.; Morales, F.J.; Glover, R.; Malumphy, C.; Barker, I. Bemisia afer Sensu Lato, a Vector of Sweet Potato Chlorotic Stunt Virus. Plant Dis. 2010, 94, 510–514. [Google Scholar] [CrossRef] [PubMed]
- Sseruwagi, P.; Legg, J.P.; Maruthi, M.N.; Colvin, J.; Rey, M.E.C.; Brown, J.K. Genetic Diversity of Bemisia tabaci (Gennadius) (Hemiptera: Aleyrodidae) Populations and Presence of the B Biotype and a Non-B Biotype That Can Induce Silverleaf Symptoms in Squash, in Uganda. Ann. Appl. Biol. 2005, 147, 253–265. [Google Scholar] [CrossRef]
- Hadjistylli, M.; Roderick, G.K.; Brown, J.K. Global Population Structure of a Worldwide Pest and Virus Vector: Genetic Diversity and Population History of the Bemisia tabaci Sibling Species Group. PLoS ONE 2016, 11, e0165105. [Google Scholar] [CrossRef]
- Guo, X.; Rao, Q.; Zhang, F.; Luo, C.; Zhang, H.; Gao, X. Diversity and Genetic Differentiation of the Whitefly Bemisia tabaci Species Complex in China Based on mtCOI and cDNA-AFLP Analysis. J. Integr. Agric. 2012, 11, 206–214. [Google Scholar] [CrossRef]
- Laarif, A.; Saleh, D.; Clouet, C.; Gauthier, N. Regional Co-Occurrence between Distinct Bemisia tabaci Species in Tunisia with New Insights into the Role of Host Plants. Phytoparasitica 2015, 43, 135–150. [Google Scholar] [CrossRef]
- Shadmany, M.; Boykin, L.M.; Muhamad, R.; Omar, D. Genetic Diversity of Bemisia tabaci (Hemiptera: Aleyrodidae) Species Complex Across Malaysia. J. Econ. Entomol. 2019, 112, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Horowitz, A.R.; Denholm, I.; Gorman, K.; Cenis, J.L.; Kontsedalov, S.; Ishaaya, I. Biotype Q of Bemisia tabaci Identified in Israel. Phytoparasitica 2003, 31, 94–98. [Google Scholar] [CrossRef]
- Horowitz, A.R.; Ishaaya, I. Dynamics of Biotypes B and Q of the Whitefly Bemisia tabaci and Its Impact on Insecticide Resistance. Pest Manag. Sci. 2014, 70, 1568–1572. [Google Scholar] [CrossRef]
- Roditakis, E.; Grispou, M.; Morou, E.; Kristoffersen, J.B.; Roditakis, N.; Nauen, R.; Vontas, J.; Tsagkarakou, A. Current Status of Insecticide Resistance in Q Biotype Bemisia tabaci Populations from Crete. Pest Manag. Sci. 2009, 65, 313–322. [Google Scholar] [CrossRef]
- Wang, Z.; Yan, H.; Yang, Y.; Wu, Y. Biotype and Insecticide Resistance Status of the Whitefly Bemisia tabaci from China. Pest Manag. Sci. 2010, 66, 1360–1366. [Google Scholar] [CrossRef]
- Huang, L.; Shi, X.; Shi, J.; Zhang, Z.; Fang, Y.; Zhang, Z.; Pan, Q.; Zheng, L.; Gao, Y.; Zhang, D.; et al. Tomato Chlorosis Virus Infection Facilitates Bemisia tabaci MED Reproduction by Elevating Vitellogenin Expression. Insects 2021, 12, 101. [Google Scholar] [CrossRef] [PubMed]
- Berry, S.D.; Fondong, V.N.; Rey, C.; Rogan, D.; Fauquet, C.M.; Brown, J.K. Molecular Evidence for Five Distinct Bemisia Tabaci (Homoptera: Aleyrodidae) Geographic Haplotypes Associated with Cassava Plants in Sub-Saharan Africa. Ann. Entomol. Soc. Am. 2004, 97, 852–859. [Google Scholar] [CrossRef]
- Tocko-Marabena, B.K.; Silla, S.; Simiand, C.; Zinga, I.; Legg, J.; Reynaud, B.; Delatte, H. Genetic Diversity of Bemisia tabaci Species Colonizing Cassava in Central African Republic Characterized by Analysis of Cytochrome c Oxidase Subunit I. PLoS ONE 2017, 12, e0182749. [Google Scholar] [CrossRef] [PubMed]
- Sseruwagi, P.; Maruthi, M.N.; Colvin, J.; Rey, M.E.C.; Brown, J.K.; Legg, J.P. Colonization of Non-Cassava Plant Species by Cassava Whiteflies (Bemisia tabaci) in Uganda. Entomol. Exp. Appl. 2006, 119, 145–153. [Google Scholar] [CrossRef]
- Mugerwa, H.; Rey, M.E.C.; Alicai, T.; Ateka, E.; Atuncha, H.; Ndunguru, J.; Sseruwagi, P. Genetic Diversity and Geographic Distribution of Bemisia tabaci (Gennadius) (Hemiptera: Aleyrodidae) Genotypes Associated with Cassava in East Africa. Ecol. Evol. 2012, 2, 2749–2762. [Google Scholar] [CrossRef]
- Manani, D.M.; Ateka, E.M.; Nyanjom, S.R.G.; Boykin, L.M. Phylogenetic Relationships among Whiteflies in the Bemisia tabaci (Gennadius) Species Complex from Major Cassava Growing Areas in Kenya. Insects 2017, 8, 25. [Google Scholar] [CrossRef]
- Casinga, C.M.; Shirima, R.R.; Mahungu, N.M.; Tata-Hangy, W.; Bashizi, K.B.; Munyerenkana, C.M.; Ughento, H.; Enene, J.; Sikirou, M.; Dhed’a, B.; et al. Expansion of the Cassava Brown Streak Disease Epidemic in Eastern Democratic Republic of Congo. Plant Dis. 2021, 105, 2177–2188. [Google Scholar] [CrossRef]
- Tajebe, L.S.; Boni, S.B.; Guastella, D.; Cavalieri, V.; Lund, O.S.; Rugumamu, C.P.; Rapisarda, C.; Legg, J.P. Abundance, Diversity and Geographic Distribution of Cassava Mosaic Disease Pandemic-Associated Bemisia tabaci in Tanzania. J. Appl. Entomol. 2015, 139, 627–637. [Google Scholar] [CrossRef]
- de Moraes, L.A.; Muller, C.; Bueno, R.C.O.; Santos, A.d.F.; Bello, V.H.; De Marchi, B.R.; Watanabe, L.F.M.; Marubayashi, J.M.; Santos, B.R.; Yuki, V.A.; et al. Distribution and Phylogenetics of Whiteflies and Their Endosymbiont Relationships after the Mediterranean Species Invasion in Brazil. Sci. Rep. 2018, 8, 14589. [Google Scholar] [CrossRef]
- Akintola, A.A.; Hwang, H.-S.; Khatun, M.F.; Ande, A.T.; Lee, K.-Y. Genetic Diversity of Bemisia tabaci Cryptic Species in Nigeria and Their Relationships with Endosymbionts and Acquired Begomoviruses. J. Asia-Pac. Entomol. 2020, 23, 1003–1009. [Google Scholar] [CrossRef]
- Karut, K.; Castle, S.J.; Karut, Ş.T.; Karaca, M.M. Secondary Endosymbiont Diversity of Bemisia tabaci and Its Parasitoids. Infect. Genet. Evol. 2020, 78, 104104. [Google Scholar] [CrossRef] [PubMed]
- El Hamss, H.; Maruthi, M.N.; Ally, H.M.; Omongo, C.A.; Wang, H.-L.; Van Brunschot, S.; Colvin, J.; Delatte, H. Spatio-Temporal Changes in Endosymbiont Diversity and Composition in the African Cassava Whitefly, Bemisia tabaci SSA1. Front. Microbiol. 2022, 13, 986226. [Google Scholar] [CrossRef] [PubMed]
- Santos-Garcia, D.; Juravel, K.; Freilich, S.; Zchori-Fein, E.; Latorre, A.; Moya, A.; Morin, S.; Silva, F.J. To B or Not to B: Comparative Genomics Suggests Arsenophonus as a Source of B Vitamins in Whiteflies. Front. Microbiol. 2018, 9, 2254. [Google Scholar] [CrossRef] [PubMed]
- Milenovic, M.; Gouttepifre, A.; Eickermann, M.; Junk, J.; Rapisarda, C. Plant-Mediated Rifampicin Treatment of Bemisia tabaci Disrupts but Does Not Eliminate Endosymbionts. Sci. Rep. 2022, 12, 20766. [Google Scholar] [CrossRef] [PubMed]
| Region | Average Mean Temperature (°C) | Annual Relative Humidity (%) | Average Rainfall (mm) | Agroecological Zone |
|---|---|---|---|---|
| Southwest | 22.6–33.6 | 66.1–86.2 | 2000–3700 | Monomodal rainforest |
| Littoral | 22.6–33.6 | 66.1–86.2 | 2000–3700 | Monomodal rainforest (humid forest) |
| West | 22.6–33.1 | 76.4–84.5 | 1800–2100 | Western highland |
| Center | 17.3–31.6 | 60–90 | 1500–2000 | Bimodal rainforest (humid forest) |
| Target Gene | Primer Name | Sequence (5′→3′) | Reference | Amplicon Length | Annealing Temperature |
|---|---|---|---|---|---|
| Portiera 16S rDNA | 28F 1098R | TGCAAGTCGAGCGGCATCAT AAAGTTCCCGCCTTATGCGT | [25] | 1050 bp | 58 °C |
| Arsenophonus 23S rDNA | Ars23S-1 Ars23S-2 | CGTTTGATGAATTCATAGTCAAA GGTCCTCCAGTTAGTGTTACCCAAC | [30] | 750 bp | 58 °C |
| Rickettsia 16S rDNA | Rb-F Rb-R | GCTCAGAACGAACGCTATC GAAGGAAAGCATCTCTGC | [24] | 960 bp | 58 °C |
| Wolbachia wsp gene | 81F 471R | TGGTCCAATAAGTGATGAAGAAACAAAAATTAAACGCTACTCCA | [39] | 600 bp | 53 °C |
| Cardinium 16S rDNA | Card-F Card-R | TAGACACACACGAAAGTTCATGT GCATGCAATCTACTTTACACTGG | [39] | 650 bp | 57 °C |
| Hamiltonella 16S rDNA | Hb-F Hb-R | TGAGTAAAGTCTGGGAATCTGG AGTTCAAGACCGCAACCTC | [18] | 730 bp | 58 °C |
| Region | Division | Agroecological Zone | Number of Fields | Crop (Number of Fields) | Whitefly Abundance (Mean) |
|---|---|---|---|---|---|
| Southwest | Dian Koupemanengouba | Monomodal rainforest | 2 | Cassava | 12.5 |
| Littoral | Moungo | Monomodal rainforest (humid forest) | 3 | Cassava | 2.9 |
| West | Nde | Western highland | 5 | Cassava (2) Tomato (3) | 5.9 37.3 |
| Center | Mefou et Afamba Mefou et Akono Mfoundi | Bimodal rainforest (humid forest) | 14 | Cassava (8) Tomato (1) Okra (3) Melon (1) Pepper (1) | 10.7 22.0 31.6 60.0 18.0 |
| KASP Haplogroup | Mitotypes (mtCOI) | References |
|---|---|---|
| SSA-ECA | SSA1-SG1 SSA1-SG2, SSA1-SG1/SG2, SSA4 | [54,55,56,57]; in this study |
| SSA-ESA | SSA1-SG3, SSA1-SG2 | [54,56,57,58]; |
| SSA-WA | SSAA1-SG1, SSA1-SG5, SSA2, SSA4 | [56,57]; in this study |
| SSA-CA | SSA1-SG1, SSA1-SG2 | [56] |
| SSA2 | SSA2, SSA3, SSA4 | [54,56,57] |
| SSA4 | SSA4, SSA1-SG2, SSA1-SG5 | [56,57]; in this study |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kepngop, L.R.K.; Wosula, E.N.; Amour, M.; Ghomsi, P.G.T.; Wakam, L.N.; Kansci, G.; Legg, J.P. Genetic Diversity of Whiteflies Colonizing Crops and Their Associated Endosymbionts in Three Agroecological Zones of Cameroon. Insects 2024, 15, 657. https://doi.org/10.3390/insects15090657
Kepngop LRK, Wosula EN, Amour M, Ghomsi PGT, Wakam LN, Kansci G, Legg JP. Genetic Diversity of Whiteflies Colonizing Crops and Their Associated Endosymbionts in Three Agroecological Zones of Cameroon. Insects. 2024; 15(9):657. https://doi.org/10.3390/insects15090657
Chicago/Turabian StyleKepngop, Lanvin R. K., Everlyne N. Wosula, Massoud Amour, Pierre G. T. Ghomsi, Louise N. Wakam, Germain Kansci, and James P. Legg. 2024. "Genetic Diversity of Whiteflies Colonizing Crops and Their Associated Endosymbionts in Three Agroecological Zones of Cameroon" Insects 15, no. 9: 657. https://doi.org/10.3390/insects15090657






