Simple Summary
Limacodidae, known for its high species diversity in tropical and subtropical regions, includes over 300 genera and 1700 species globally, with China hosting a significant number of these. In this study, we describe a new genus and species within the family Limacodidae from Guangxi and Jiangxi, China. The new genus and species, Guium nebulum gen. et sp. nov., displays unique morphological features, including distinct wing patterns and male genital structures that differentiate it from related genera. Through an analysis of the COI mitochondrial gene, we further confirmed the genus’s monophyly.
Abstract
A new genus and species of Limacodidae, Guium nebulum gen. et sp. nov., is described based on specimens collected from Guangxi Autonomous Region and Jiangxi Province in China. The new genus shares certain morphological features, such as a well-developed labial palpus, with related genera like Tanvia Solovyev & Witt, 2009; Scopelodes Westwood, 1841; Hyphorma Walker, 1865; and Monema Walker, 1855. However, the new genus can be separated from them by the wing venation and the male genital characteristics. COI molecular marker analysis further supports the monophyly of this new genus, indicating a close relationship with Scopelodes.
1. Introduction
The family Limacodidae, within the superfamily Zygaenoidea, is one of the largest families in this group, predominantly found in tropical and subtropical regions, with a high diversity in the Afrotropical and Oriental regions. The family comprises over 300 genera and 1700 species worldwide (van Nieukerken et al. [1]). Of these, China is home to 72 genera, 264 species, and 5 subspecies, representing over 15% of the known species, as noted by Wu (2010) [2] and by Wu and Fang (2023) [3]. Phylogenetic studies on this family have been relatively limited in the past. However, Liang et al. (2024) [4] recently reconstructed a higher-level taxonomic framework for Palaearctic and Indomalayan Limacodidae, recognizing six major clades within this family.
Guangxi Autonomous Region and Jiangxi Province are both located in southern China, with favorable natural conditions and high biodiversity. Wu and Fang (2023) [3] reported 79 species of the family in Guangxi, with a tie with Sichuan Province for the second-highest species richness in China, accounting for roughly one-third of the national total. The highest species diversity was reported in Yunnan Province, with 138 species. In comparison, Jiangxi reported 62 species, slightly fewer than Guangxi and Sichuan. During the field surveys in Guangxi and Jiangxi, an unknown Limacodidae species with elongated labial palpus was identified. Based on morphological and molecular analyses, we confirmed this species is a new species belonging to an undescribed genus.
Research on Limacodidae groups with elongated labial palpus can be traced back to Hering (1931) [5], who documented that several genera, including Scopelodes Westwood, 1841; Monema Walker, 1855; Elassoptila Turner, 1902; Susica Walker, 1855 (female); Hedraea Turner, 1926; Hyphorma Walker, 1865; and Hyphormides Hering, 1931 [6,7,8,9,10], exhibit distinct elongated labial palpus. Among these, the labial palpus of Scopelodes is particularly notable for the presence of a conspicuous hair tuft at the apex. Within the Oriental region, Hyphormides and Scopelodes form a group with Hyphorma, as they share a distinctive elongation of the labial palpi (Holloway, 1986) [11]. Solovyev & Witt (2009) [12] described a new genus, Tanvia Solovyev & Witt, 2009, closely allied to Scopelodes with elongated labial palpus and an apical hair tuft. Later, Solovyev & Giusti (2017) [13] further confirmed that members of Scopelodes are related to those in the genera Tanvia and Hyphorma by their appearance.
More recently, Liang et al. (2024) [4] provided molecular evidence demonstrating that genera from the Palaearctic and Indomalayan regions, including Scopelodes, Tanvia, Monema, and Hyphorma, form a well-supported lineage within the Parasa-clade, corroborating earlier morphological studies. Building upon these findings, the present study will select representative taxa from these genera for a detailed comparative analysis with the newly described genus. Specifically, it aims to (1) formally describe the new genus and species using comprehensive morphological and molecular data, and (2) investigate its phylogenetic position within Limacodidae, with a focus on its affinities to morphologically similar genera, such as Scopelodes and Tanvia.
2. Material and Methods
2.1. Sampling
The specimens examined in this study were collected and used for DNA extraction. Initially, three legs from one side of each individual were removed, preserved in 100% alcohol, and stored at −20 °C for further analysis. For phylogenetic analysis, we selected two Zygaenidae sequences, Eterusia aedea (Linnaeus, 1763) and Histia flabellicornis (Fabricius, 1775), from BOLD/NCBI as outgroups, based on the close phylogenetic relationship between Limacodidae and Zygaenidae (Bian et al. (2020) [14]). To ascertain the phylogenetic placement of the new genus within Limacodidae, we incorporated several published sequences alongside newly sequenced data, including those of the new species. These sequences represent genera Tanvia, Scopelodes, Hyphorma, and Monema, all of which belong to the Parasa-clade as defined by Liang et al. (2024) [4]. Morphologically, the new genus shares features with these genera, such as extended labial palpi and comparable wing venations. In addition, we also included the genus Mahanta Moore, 1879 [15] as a comparative genus. Although it lacks a distinctly elongated labial palpus, it clusters with Scopelodes or Monema in the phylogenetic tree. The collecting locality, BOLD sample IDs, and GenBank accession numbers are listed in Table 1. All sequences of the new species and the newly sequenced data presented in this paper have been deposited in GenBank under Accession Numbers PQ561650–PQ561656 (Table 1).

Table 1.
Sampling information and BOLD SampleID/GenBank accession numbers of Limacodidae and outgroups used in this study.
2.2. Morphological Study
Traditional insect taxonomic methods were used for the morphological study. Specimens were collected at night using a high-pressure 220V/450W mercury lamp supplemented with an 8V/5W DC blacklight. Genitalia preparations followed the method described by Kononenko & Han (2007) [16]. Abdomens were detached with fine tweezers and soaked in a 10% KOH solution for 12–24 h. After defatting, genitalia were separated in 75% ethanol for further cleaning, dehydrated in 100% ethanol, stained, and then fixed in xylene for observation and photography. To examine the vesica, the phallus was removed, and the vesica everted using a 1 mL syringe to expose internal structures such as cornuti. Permanent slides were mounted with Canada balsam.
Adult specimens were photographed with Nikon D700 (Nikon Corporation, Tokyo, Japan) and Canon M6 Mark II (Canon Inc., Tokyo, Japan) cameras to document observable morphological traits, including body color, forewing length, wingspan, and wing patterns. Specimen measurements were taken with a ruler; forewing length was measured from apex to base, and wingspan from apex to apex. Observations and measurements of genital structures on slides were conducted under a microscope. Genitalia slide photographs were taken with an AOSVI Hk-830 microscope (Shenzhen Aoswei Optical Instruments Co., Ltd., Shenzhen, China), with images refined using Helicon Focus 7 (Helicon Soft Ltd., Kharkiv, Ukraine) and Adobe Photoshop 2020 (Adobe Inc., San Jose, CA, USA) for enhanced clarity. The collecting map was constructed using the QGIS v3.40 software.
The terminology for adult and genital structures follows Epstein (1996) [17] and Kristensen (2003) [18]. All type materials of the new species are deposited in the collection of the Northeast Forestry University (NEFU), Harbin, China.
2.3. DNA Extraction and PCR
Standard DNA extraction and amplification methods were performed. Genomic DNA was extracted from three legs of each specimen using the TaKaRa MiniBEST Universal Genomic DNA Extraction Kit Ver.5.0 (Takara Bio Inc., Shiga, Japan), following overnight incubation at 56 °C and according to the kit’s protocol. Polymerase chain reactions (PCRs) were conducted on a PCR Thermal Cycler (Hangzhou LongGene Scientific Instruments Co., Ltd., Hangzhou, China) using the primers LCO1490 and HCO2198 (Folmer et al. (1994) [19]) to amplify a 658 bp fragment of the mitochondrial cytochrome c oxidase I (COI) gene, as listed in Table 2.

Table 2.
PCR primers used to sequence COI genes of limacodid species in this study.
The total reaction volume was 25 μL, consisting of 0.5 μL template DNA, 10 μL ddH2O, 12.5 μL 2× Rapid Taq Master Mix (Vazyme Biotech Co., Ltd., Nanjing, China), and 1 μL each of forward and reverse primers (synthesized by Sangon Biotech from Changchun, China). The PCR program included an initial denaturation at 95 °C for 3 min; 35 cycles of 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 1 min; with a final extension at 72 °C for 5 min.
2.4. Molecular Phylogenetic Analysis
All amplicons were sequenced using an ABI 3730XL automated sequencer. The raw sequences were initially manually corrected with Chromas v2.6.6 software (Technelysium Pty Ltd., Brisbane, Australia), followed by assembly of the bidirectional sequences in Geneious v9.0.2 (https://www.geneious.com, accessed on 27 October 2024). Phylogenetic analysis was conducted with sequences obtained in this study along with additional Limacodidae sequences downloaded from the GenBank database (https://www.ncbi.nlm.nih.gov/nucleotide, accessed on 23 July 2024) and BOLD database (Barcode of Life Data Systems, Guelph, Canada, https://v4.boldsystems.org/, accessed on 23 July 2024). PhyloSuite 1.2.3 (Zhang et al. (2020) [20]; Xiang et al. (2023) [21]) was used for the analysis, and 20 sequences were aligned with MAFFT v7.505 (Katoh and Standley (2013) [22]) using ‘--auto’ strategy and normal alignment mode. Ambiguously aligned fragments of 1 alignment was removed using Gblocks 0.91b (Talavera and Castresana (2007) [23]) with the following parameter settings: minimum number of sequences for a conserved/flank position (11/11), maximum number of contiguous non-conserved positions (8), minimum length of a block (10), allowed gap positions (with half).
ModelFinder v2.2.0 (Kalyaanamoorthy et al. (2017) [24]) was used to select the best-fit model using the BIC criterion. Phylogenetic analyses were performed using the Bayesian inference (BI) approach. Bayesian posterior probabilities were estimated in MrBayes v3.2.7a (Ronquist et al. (2012) [25]), with the best-fit model according to ModelFinder’s BIC recommendation: GTR+G+F. Sampling was conducted every 1000 generations. We conducted Bayesian inference using MCMC with four chains (one cold and three hot) for each run. The analysis was performed for 5 million generations in two independent runs to ensure convergence and robustness of the results, discarding the first 25% of sampled trees as burn-in. Tree nodes with BI posterior probabilities (PPs) > 0.95 were considered well-supported. The final phylogenetic tree was visualized using ChiPlot (Xie et al. (2023) [26]) (https://www.chiplot.online/index.html, accessed on 20 October 2024) and edited with Adobe Illustrator 2023.
Simultaneously, we aligned sequences using Clustal W in MEGA 11 (Tamura et al. (2021) [27]) and checked for stop codons after translating them to amino acid sequences. Uncorrected pairwise genetic distances of the COI gene were calculated among all individuals of the new species and the type species of each related genus. The analysis, comprising 8 nucleotide sequences, was conducted using the Kimura 2-parameter model (Kimura (1980) [28]), with codon positions set to 1st+2nd+3rd+Noncoding. All ambiguous positions were removed for each sequence pair (pairwise deletion option), yielding 658 positions in the final dataset. Evolutionary analyses were conducted in MEGA 11.
3. Result
3.1. Taxonomic Treatment
- Guium Wu & Han, gen. nov.
- Zoobank: urn:lsid:zoobank.org:act:C66396B0-701A-4CBD-BA49-CEBF3974A9BD
- Type species: Guium nebulum Wu & Han, sp. nov.
- Gender: neutral
- Chinese name: 桂刺蛾属
Diagnosis: The genus Guium (Figure 1a–d) shares the characteristic of a noticeably extended labial palpus with related genera such as Tanvia, Scopelodes, Hyphorma, Monema, and Mahanta (Figure 1e–i), but exhibits several distinct features that clearly differentiate it from them. Guium is characterized by its pale-yellow coloration and a large, diffuse ochre blotch on the forewing. In contrast, these related genera have a darker overall coloration, with predominant shades of ochre, brown, or dark brown. In the male antennae, Guium is similar to those of Tanvia, with bipectinate from base to apex, gradually narrowing; Scopelodes and Hyphorma are broadly bipectinate in the basal 1/2 to 2/3; those of Monema are filiform; and in Mahanta, only short rami are present.

Figure 1.
Male adults. (a) Guium nebulum sp. nov., holotype (Guangxi, China); (b,c) ditto, paratype, dorsal and ventral side (Guangxi, China); (d) ditto, paratype (Jiangxi, China); (e) Tanvia zolotuhini Witt & Solovyev, 2009 (Yunnan, China); (f) Scopelodes unicolor Westwood, 1841 (Borneo); (g) Hyphorma minax Walker, 1865 (Chongqing, China); (h) Monema flavescens Walker, 1855 (Heilongjiang, China); (i) Mahanta tanyae Solovyev, 2005 (Guizhou, China). Scale bars: 10 mm, all in NEFU.
Etymology: The genus name is derived from “Gui”, an abbreviation for Guangxi Autonomous Region, China, the type locality of the new genus.
The forewing venation of Guium (Figure 2a) is distinct: vein R1 has a pronounced outward curve and approaches Sc proximally, with the upper angle of the discal cell nearly a right angle and the lower angle extending noticeably outward. In case of the related genera (Figure 2b–f), R1 is straight, and does not approach Sc, while the upper angle forms a sharp point, and the lower angle is only slightly outward, level with, or shorter than the upper angle. Additionally, Guium resembles Scopelodes (Figure 2c) and Mahanta (Figure 2f) in having R2 branch after the upper angle of the discal cell and share a stem with R3+4+5. By contrast, in Tanvia (Figure 2b) and Hyphorma (Figure 2d), R2 branches near the upper angle, and in Monema (Figure 2e), R2 originates before this angle.
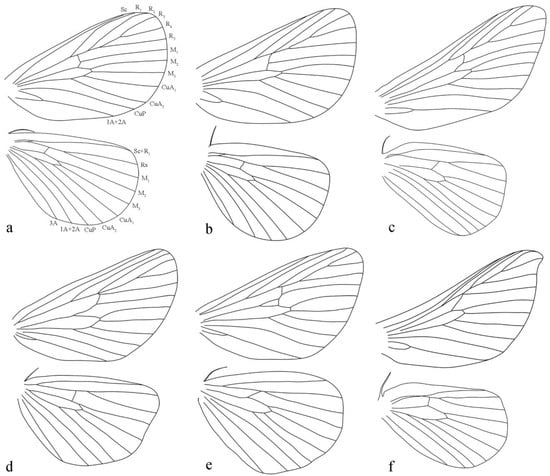
Figure 2.
Wing venations. (a) Guium nebulum sp. nov.; (b) Tanvia zolotuhini Witt & Solovyev, 2009; (c) Scopelodes unicolor Westwood, 1841; (d) Hyphorma minax Walker, 1865; (e) Monema flavescens Walker, 1855; (f) Mahanta tanyae Solovyev, 2005.
The labial palpus of Guium (Figure 3a) resembles that of Tanvia (Figure 3b) and Scopelodes (Figure 3c) in being elongated with a tuft of scales at the end of the third segment. However, the tuft in Guium is loosely arranged and not distinctly globular. Genera Hyphorma, and Monema, (Figure 3d,e) also have elongated palpi, but the third segment is covered with short scales and lacks a tufted appearance. The labial palpus in Mahanta (Figure 3f) is short.

Figure 3.
Labial palpi. (a) Guium nebulum sp. nov.; (b) Tanvia zolotuhini Witt & Solovyev, 2009; (c) Scopelodes unicolor Westwood, 1841; (d) Hyphorma minax Walker, 1865; (e) Monema flavescens Walker, 1855; (f) Mahanta tanyae Solovyev, 2005.
The male genitalia of Guium (Figure 4a,b) shows distinctive modifications: the uncus is broadened without an apical spine; the gnathos is wide and band-shaped; and the valva is divided into upper and lower lobes. Additionally, the eighth abdominal segment (Figure 4c–e) bears a pair of robust, sclerotized, tooth-like projections, each with one or more small spines at the base. These unique features in the male genitalia and abdomen are absent in related genera with elongated labial palpus mentioned herein, which exhibit the typical Limacodidae structure (Figure 5a–d). The male genitalia of Mahanta species (Figure 5e) share some similarities with those of the new genus, including the blunt uncus, the bifurcated valva, and a strongly curved phallus. However, the two genera can be distinguished by their general appearance.
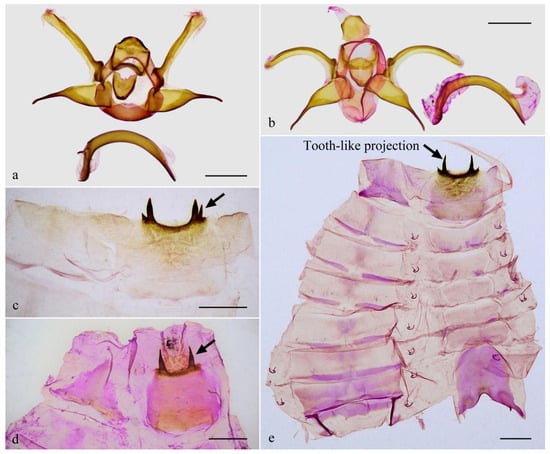
Figure 4.
Male genitalia (a,b), abdominal segment VIII (c,d), and abdomen (e) of Guium nebulum sp. nov. (a,c) holotype, slide WuJ-740-1 (Guangxi, China); (b,d) paratype, slide WuJ-1137-1 (Jiangxi, China); (e) paratype, slide WuJ-709-1 (Guangxi, China). In (c–e) the dorsum is shown on the left and the ventrum on the right, with the tooth-like projections indicated by arrows. Scale bars: 1 mm, all in NEFU.
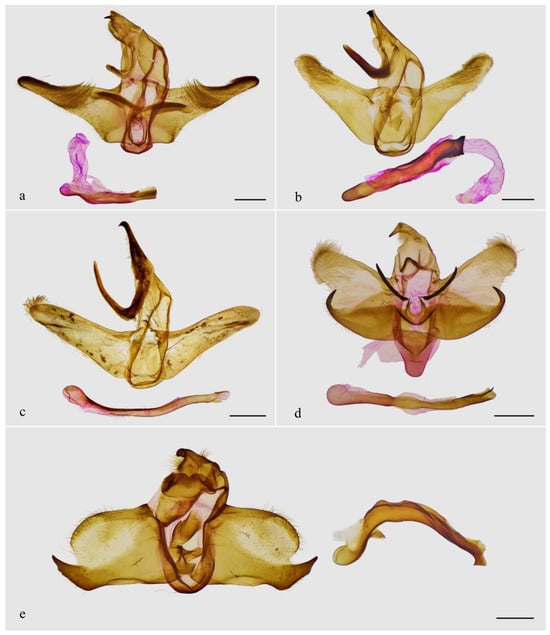
Figure 5.
Male genitalia of related genera. (a) Tanvia zolotuhini Witt & Solovyev, 2009, slide WuJ-1037-1; (b) Scopelodes unicolor Westwood, 1841, slide WuJ-1142-1; (c) Hyphorma minax Walker, 1865, slide WuJ-116-1; (d) Monema flavescens Walker, 1855, slide WuJ-976-1; (e) Mahanta tanyae Solovyev, 2005, slide WuJ-1043-1. Scale bars: 1 mm, all in NEFU.
Description: Male antennae bipectinate from base to apex, gradually narrowing. Labial palpus grayish-brown, extending prominently; first segment very short, second segment thicker, third segment tapering, ending with a tuft of long, pale yellow scales. Thorax dorsally grayish-white with a faint brownish tinge. Forewing broad with a pale-yellow base color, featuring a large, ochreous misty patch. Hindwing grayish-white. Forewing venation: vein R1 strongly curved distally, closely approaching vein Sc; R2 shares a stalk with veins R3 + R4, and R5; R5 originates after R2; M3 emerges from the lower angle of discal cell. Hindwing: Rs and M1 veins share a common stalk. Legs grayish-white; tibial spurs in formula 0-2-4. Sternite VIII bears a pair of hardened, tooth-like projections, each with one or more small spines at base.
Male genitalia. Highly modified compared to the typical male genitalia structure of Limacodidae, such as those of the related genera Tanvia and Scopelodes. Uncus wide, without apical spine; gnathos broad, band-shaped; tegumen broad. Valva divided into upper and lower lobes: upper lobe narrow, elongated; lower lobe sclerotized, with the basal half expanded. Juxta shield-like. Saccus indistinct. Phallus tubular, strongly sclerotized, down curved into a bow shape, with oblique slits near the caecum and apex.
Female. Unknown.
Remark: This genus is known to be a monotypic genus and is distributed in the Guangxi Autonomous Region and Jiangxi Province, China.
- Guium nebulum Wu & Han, sp. nov.
- Zoobank: urn:lsid:zoobank.org:act:7E92DB49-1E2A-465B-89A0-2B1D3BE3C1D4
- Chinese name. 桂刺蛾
HOLOTYPE: ♂, Guangxi Yinzhulaoshan Ziyuan Fir National Nature Reserve, Ziyuan County, Guilin City, Guangxi Autonomous Region, China, 26°16′15″ N, 110°35′44″ E, 2021-VII-15–18, Jun-Jie Fan, Biao Gao leg., genit. prep. WuJ-740-1 [NEFU]. PARATYPES: 4 ♂, same data as for holotype, genit. prep. WuJ-709-1, WuJ-739-1 [NEFU]. 1 ♂, Dazhangshan Township, Wuyuan County, Shangrao City, Jiangxi Province, China, 29°27′07″ N, 117°44′02″ E, 2024-VI-30, Kai Wu, Xin-Yu Cheng leg., genit. prep. WuJ-1137-1 [NEFU].
Diagnosis: The new species is similar to members of Tanvia and Scopelodes, in having a tuft of long scales at the end of the third labial palpus segment, but easily distinguished by appearance, male genitalia, and sternite VIII, as described for the genus.
Description: Male. Forewing length 14–15 mm, wingspan 30–34 mm (N = 6). Head pale grayish-white; antennae bipectinate till to tip. Labial palpus grayish-brown, extending prominently; first segment very short, second segment thicker, third segment tapering, approximately 4/5 the length of the second, ending with a tuft of long, pale yellow, loosely arranged scales. Thorax dorsally grayish-white with a faint brownish tinge. Forewing broad, with a pale-yellow base color, featuring a large, ochreous misty patch extending from approximately two-thirds along costa to halfway down the inner margin, scattered with dark brown scales; outer region without distinct markings. Terminal line distinct, thin, brown; fringes grayish-white. Hindwing grayish-white, with a faint brownish tinge along anal margin; fringes as in the forewing. Abdomen grayish-white with a faint brown; sternite VIII bears a pair of hardened, tooth-like projections, each with one or more small spines at base.
Male genitalia. Uncus wide, without apical spine; gnathos broad, band-shaped; tegumen broad. Valva divided into upper and lower lobes: upper lobe narrow, elongated, with a prominent, strongly sclerotized basal “pedestal”, with its upper edge weakly sclerotized, lower edge membranous; lower lobe sclerotized, with the basal half expanded, tapering sharply at midsection, ending in a strongly sclerotized, blunt rod-like process. Juxta shield-like, widening at top, narrowing below, edges strongly sclerotized with a notch at the mid-dorsal margin. Saccus indistinct. Phallus tubular, strongly sclerotized, down curved into a bow shape; caecum and apex each with long oblique slits, approximately 1/4 and 1/3 of the total phallus length, respectively.
Distribution: China (Guangxi, Jiangxi).
Bionomics: The new species was collected from montane regions in Guangxi Autonomous Region and Jiangxi Province, China. Specimens were captured in middle July in Guangxi and late June in Jiangxi, indicating adult activity during summer in these regions. The collection sites are characterized by mixed forest ecosystems, suggesting that the species may prefer forested habitats at low to mid elevations.
Etymology: The specific epithet is deriving from the Latin word nebula, meaning “mist” or “fog”, in reference to the ochre mist-like patch on the forewings of the new species.
3.2. Molecular Phylogenetic Analysis
Bayesian phylogenetic analysis showed good convergence, indicated by an average standard deviation of split frequencies close to 0 (0.004307) and potential scale reduction factors (PSRF) equal to 1 (maximum = 1.001). The topology generated by Bayesian inference is consistent with the results of Liang et al. (2024) [4], which used 13 mitochondrial markers. The new genus in this study formed an independent clade, supporting the monophyly of the Guium species (Figure 6).
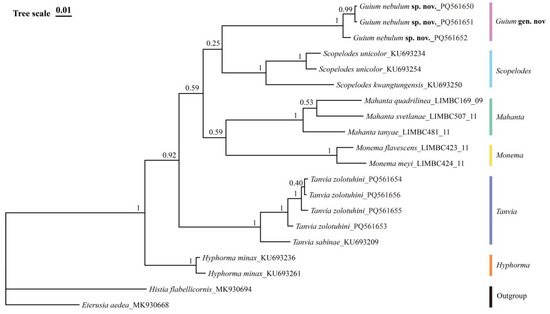
Figure 6.
Bayesian inference based on the DNA barcode sequences (COI) and rooted on two Zygaenidae species as outgroups. Nodal numbers are BI posterior probability values.
In the molecular analysis, interspecific distance values in Lepidoptera are generally greater than 3% (Hebert et al. 2003 [29]). In this study, the K2P distances of COI between G. nebulum sp. nov. and the type species of each related genus ranged from 9.7% to 12.4%, while the maximum intraspecific distance for G. nebulum is only 1.0% (Table 3).

Table 3.
Pairwise K2P distances of COI sequences for new species and the type species of each related genus used in this study.
Morphologically, Guium species share certain traits with neighboring genera, such as extended labial palpus and similar wing venation, likely indicating a shared evolutionary history. However, molecular analyses reveal phylogenetic divergence between Guium gen. nov. and these related genera. Bayesian inference indicate that Guium gen. nov. forms a distinct clade. Additionally, unique bifurcated valva and the highly modified sternite VIII in the male genitalia demonstrate marked evolutionary differentiation. These morphological differences may reflect functional adaptations, while molecular data indicate deeper phylogenetic distinctions from related genera.
4. Discussion
In this study, we conducted an integrated analysis of molecular data and morphological characteristics, providing evidence for the establishment of the new genus. Our molecular and morphological data support the validity of Guium gen. nov. Although this genus currently includes only the type species, G. nebulum sp. nov., found in Guangxi and Jiangxi (Figure 7), China, it may potentially have a broader distribution. Given similar ecological conditions in neighboring regions, such as Hunan and Fujian provinces, and in parts of Southeast Asia with comparable habitats, future surveys may uncover additional species within this genus or expand its known distribution.
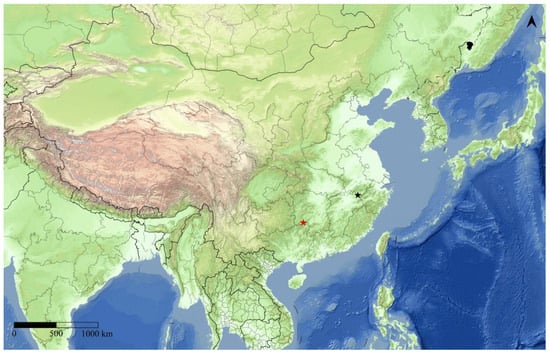
Figure 7.
Two collecting sites of Guium nebulum sp. nov. The red star indicates the type locality.
Among the three specimens, the two individuals from Guangxi showed no genetic divergence (0%), while the genetic distance between the Guangxi and Jiangxi individuals was 1.0%. Minor genetic divergence within the species may reflect population-level differences; ecological or climatic differences between Guangxi and Jiangxi might be affected by such divergence. In addition to these minor genetic variations, a slight difference in the male genitalia was observed: in the Jiangxi specimen, the upper lobe of the valva is curved, with small spines around the terminal tooth-like projection on sternite VIII, whereas the Guangxi specimen has a relatively straight upper lobe of the valva and more developed spines around the terminal projection of sternite VIII. We hypothesize that these differences might result from localized ecological or genetic factors, but further sampling and analysis across a broader geographic range are required to clarify the reasons behind their occurrence.
Due to the limited molecular marker used in this study (only a 658 bp COI sequence), we were able to confirm the monophyly of the new genus but could not fully resolve its phylogenetic relationships with closely related genera. Future studies should aim to incorporate additional genomic data, particularly large-scale nuclear markers, to clarify the phylogenetic position of Guium gen. nov. and its relatives, as well as to provide insights into their evolutionary history. The lack of information on juvenile stages and adult females also limits our ability to compare non-adult stages with closely related genera. Therefore, rearing larvae and documenting life histories are important, as these efforts will not only enhance our understanding of the group’s biology but also help obtain missing female specimens for further taxonomic and phylogenetic analyses.
Author Contributions
Conceptualization, J.W. and T.-T.Z.; investigation, J.W., T.-T.Z. and H.G.; writing—original draft, J.W. and T.-T.Z.; writing—review and editing, H.-L.H., H.G. and G.-Z.J.; funding acquisition, H.-L.H. and J.W. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the project of National Nature Science Foundation of China (grant number 31572294 to H.-L.H.); the financial assistance under Heilongjiang Postdoctoral Fund (grant number 415486 to J.W.); Full-time Postdoctoral Support Program (grant number 415895 to J.W.); and Northeast Asia Biodiversity Research Center (grant number NABRI202303 to J.W., and 2572022DS09 to H.-L.H.).
Data Availability Statement
Data are contained within the article.
Acknowledgments
We sincerely thank to Marc E. Epstein (Plant Pest Diagnostics Branch, California Department of Food and Agriculture, U.S.A.) and two anonymous reviewers for their invaluable suggestions, constructive comments, and meticulous linguistic refinements, which have greatly improved this paper. We are grateful to Shannon Wu, Assistant Editor, for her professional support and kind assistance during the manuscript revision and publication process. We thank Jia-Xin Wang (School of Forestry, Northeast Forestry University, China) for his valuable guidance in processing the COI sequences of the new species and uploading the data to GenBank. Special thanks to Kai Wu (College of Life Science, Shangrao Normal University, China) and Xin-Yu Cheng (Jiangxi Province Key Laboratory of Applied Optical Technology, China) for collecting and donating a paratype from Jiangxi, and to our colleagues Jun-Jie Fan and Biao Gao for collecting specimens from Guangxi. We are also grateful to Jun Yang (Jiangxi Wuyuan Forest Bird National Nature Reserve, China) and the staff of the Guangxi Yinzhulaoshan Ziyuan Fir National Nature Reserve for their assistance during fieldwork. We acknowledge the support of the Jiangxi Central Financial Forestry National Nature Reserve Subsidy project “Investigation and Monitoring of Insect Flora in Jiangxi Wuyuan Forest Bird National Nature Reserve”.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- van Nieukerken, E.J.; Kaila, L.; Kitching, I.J.; Kristensen, N.P.; Lees, D.C.; Minet, J.; Mitter, C.; Mutanen, M.; Regier, J.C.; Simonsen, T.J.; et al. Order Lepidoptera Linnaeus, 1758. In Animal Biodiversity: An Outline of Higher-Level Classification and Survey of Taxonomic Richness; Zhang, Z.Q., Ed.; Magnolia Press: Auckland, New Zealand, 2011; Volume 3148, pp. 212–221. [Google Scholar] [CrossRef]
- Wu, C.S. Analysis on the host plant diversity of slug caterpillar moths in China. For. Pest Dis. 2010, 29, 1–4. (In Chinese) [Google Scholar]
- Wu, C.S.; Fang, C.L. Fauna Sinica: Insecta, Volume 76: Limacodidae; Science Press: Beijing, China, 2023; 527p, ISBN 978-7-03-074634-4. (In Chinese) [Google Scholar]
- Liang, J.; Zhu, Y.; Solovyev, A.V.; He, M.; Lohman, D.J.; Wahlberg, N.; Li, W.; Li, J.; Wang, M.; Liang, D.; et al. A phylogenetic framework of Palaearctic and Indomalayan Limacodidae (Lepidoptera, Zygaenoidea) based on sequence capture data. Syst. Entomol. 2024, 49, 495–506. [Google Scholar] [CrossRef]
- Hering, M. Limacodidae (Cochliopodidae). In Die Gross-Schmetterlinge der Erde; Seitz, A., Ed.; Alfred Kerner Verlag: Stuttgart, Germany, 1931; Volume 10, pp. 665–728. [Google Scholar]
- Westwood, J.O. Scopelodes unicolor. In The Naturalist’s Library (Edn. 1) 33 (Ent. 7); Jardine, W., Ed.; W. H. Lizars: Edinburgh, UK, 1841; pp. 222–223. [Google Scholar]
- Walker, F. List of the Specimens of Lepidopterous Insects in the Collection of the British Museum, Volume 5: Lepidoptera Heterocera; The Trustees of the British Museum (Natural History): London, UK, 1855; pp. 977–1258. [Google Scholar]
- Turner, A.J. New Australian Lepidoptera. Trans. Proc. R. Soc. S. Aust. 1902, 26, 175–207. [Google Scholar]
- Turner, A.J. Revision of Australian Lepidoptera: Drepanidae, Limacodidae, Zygaenidae. Linn. Soc. N. S. W. 1926, 51, 437–445. [Google Scholar]
- Walker, F. List of the Specimens of Lepidopterous Insects in the Collection of the British Museum, Volume 32: Supplement—Part 2; The Trustees of the British Museum (Natural History): London, UK, 1865; pp. 323–706. [Google Scholar]
- Holloway, J.D. The moths of Borneo: Part I. Key to families; families Cossidae, Metarbelidae, Ratardidae, Dudgeoneidae, Epipyropidae and Limacodidae. Malay. Nat. J. 1986, 40, 1–165. [Google Scholar]
- Solovyev, A.V.; Witt, T.J. The Limacodidae of Vietnam. Entomofauna 2009, S16, 33–229. [Google Scholar]
- Solovyev, A.V.; Giusti, A. Revision of the genus Scopelodes Westwood, 1841(Lepidoptera, Limacodidae) with description of 16 new species. Insect Syst. Evol. 2017, 49, 520–581. [Google Scholar] [CrossRef]
- Bian, D.D.; Ye, W.T.; Dai, M.L.; Lu, Z.T.; Li, M.X.; Fang, Y.L.; Qu, J.W.; Su, W.J.; Li, F.C.; Sun, H.N.; et al. Phylogenetic relationships of Limacodidae and insights into the higher phylogeny of Lepidoptera. Int. J. Biol. Macromol. 2020, 159, 356–363. [Google Scholar] [CrossRef] [PubMed]
- Moore, F. Descriptions of New Indian Lepidopterous Insects from the Collection of the Late Mr. W.S. Atkinson, 1; Asiatic Society of Bengal: Calcutta, India, 1879; pp. 1–88. [Google Scholar]
- Kononenko, V.S.; Han, H.L. Atlas Genitalia of Noctuidae in Korea (Lepidoptera). In Insects of Korea. Series 11; Park, K.T., Ed.; Junhaeng-Sa: Seoul, Republic of Korea, 2007; 464p, ISBN 9788988154045. [Google Scholar]
- Epstein, M.E. Revision and phylogeny of the Limacodid-group families, with evolutionary studies on slug caterpillars (Lepidoptera: Zygaenoidea). Smithson. Contrib. Zool. 1996, 582, 1–102. [Google Scholar] [CrossRef]
- 4. Skeleton and muscles: Adults. In Vol. 4. Arthropoda: Insecta, Teilband/Part 36. Lepidoptera, Moths and Butterflies. Vol. 2. Morphology, Physiology, and Development. Handbook of Zoology/Handbuch der Zoologie. Part IV; Kristensen, N.P., Ed.; Walter de Gruyter: Berlin, Germany, 2003; pp. 39–131. [Google Scholar] [CrossRef]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochromec c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar] [PubMed]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Xiang, C.Y.; Gao, F.L.; Jakovlić, I.; Lei, H.P.; Hu, Y.; Zhang, H.; Zou, H.; Wang, G.T.; Zhang, D. Using PhyloSuite for molecular phylogeny and tree-based analyses. iMeta 2023, 2, e87. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Talavera, G.; Castresana, J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007, 56, 564–577. [Google Scholar] [CrossRef] [PubMed]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.M.; Chen, Y.R.; Cai, G.J.; Cai, R.L.; Hu, Z.; Wang, H. Tree Visualization by One Table (tvBOT): A web application for visualizing, modifying and annotating phylogenetic trees. Nucleic Acids Res. 2023, 51, W587–W592. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA 11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Hebert, P.D.N.; Cywinska, A.; Ball, S.L.; deWaard, J.R. Biological identifications through DNA barcodes. Proc. R. Soc. B 2003, 270, 313–321. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).