Bacillus subtilis Contributes to Amylase Production in the Honey Sac of Apis mellifera
Simple Summary
Abstract
1. Introduction
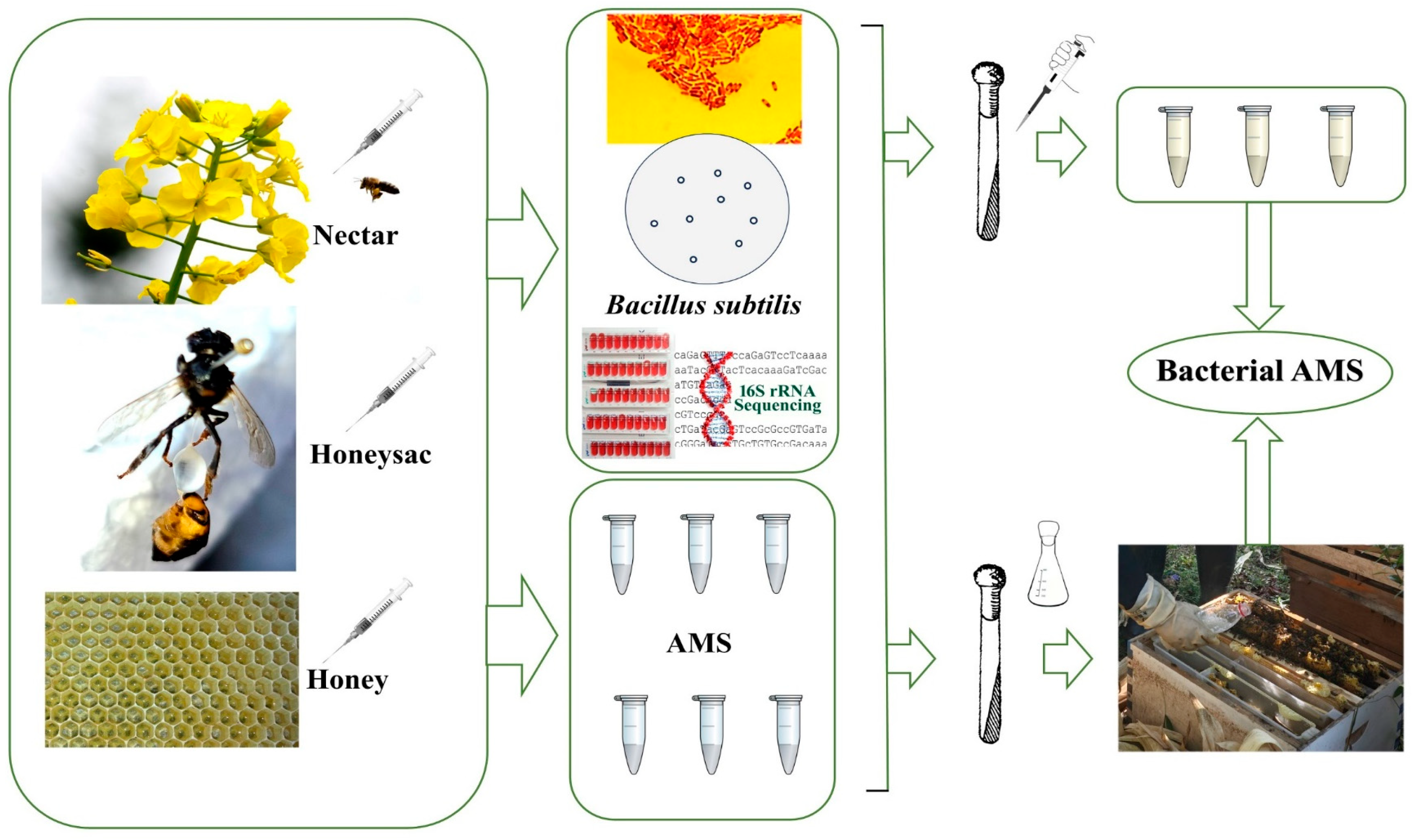
2. Materials and Methods
2.1. Sample Collection
2.1.1. Nectar Collection
2.1.2. Collection of Honey Sacs
2.1.3. Honey Sac Fluid Collection
2.1.4. Fresh Honey Collection
2.1.5. Sample Storage
2.2. Bacterial Culture and Identification Assessment
2.2.1. Methods for Culturing Bacteria
2.2.2. Identification Assessment for B. subtilis
16S rRNA Identification Methods for Bacillus Species
Determination of Biochemical Identification for Bacillus Species
2.3. Analyses of Amylase Activity
2.4. In Vitro Simulation Experiment
2.5. Feeding Experiment
2.6. Statistics Analyses
3. Results
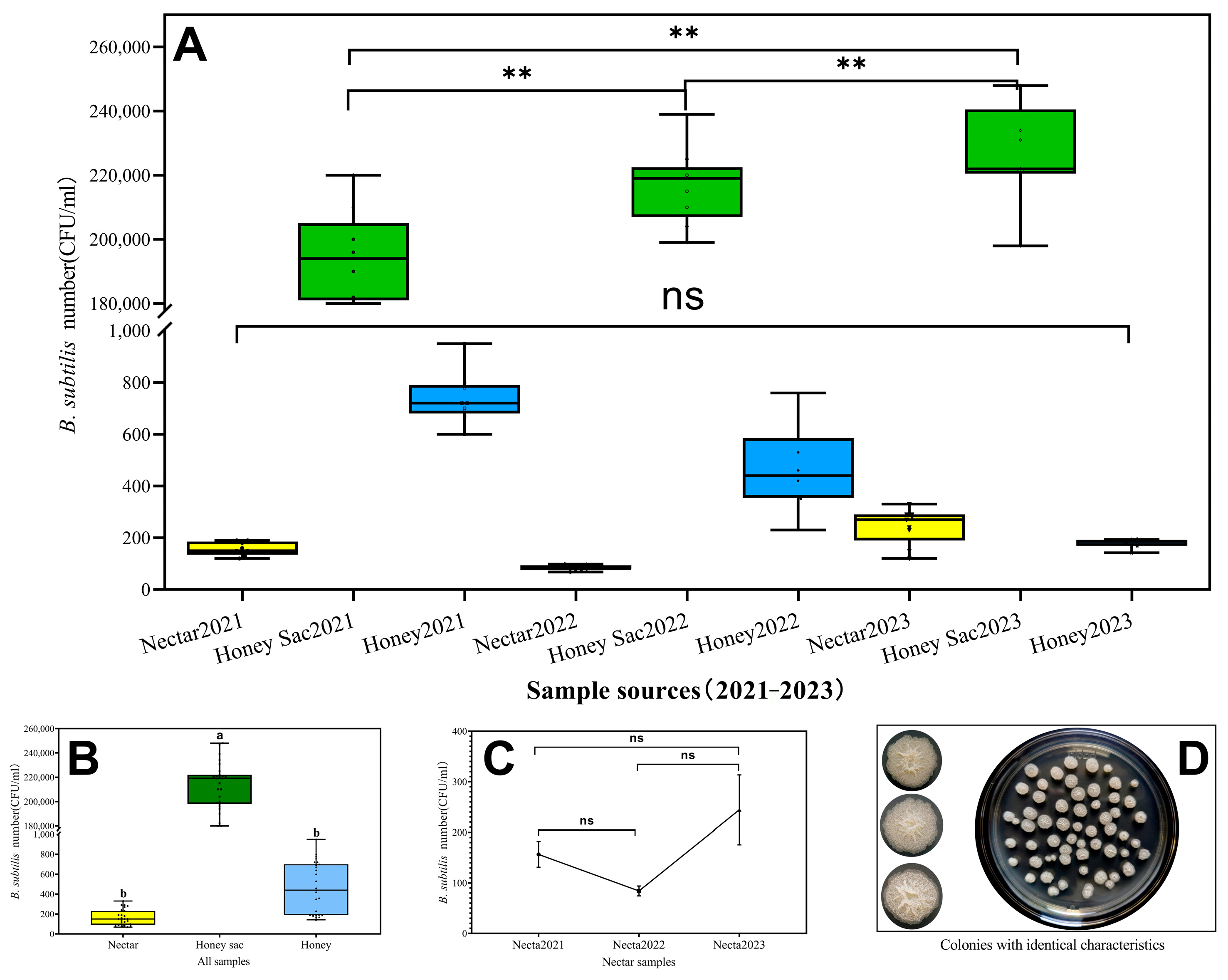
3.1. Sample Collection Results
3.2. Abundant B. subtilis Were Detected in Nectar, Honey Sac, and Fresh Honey
3.3. Molecular Bio-Identification of the Microbial Strains
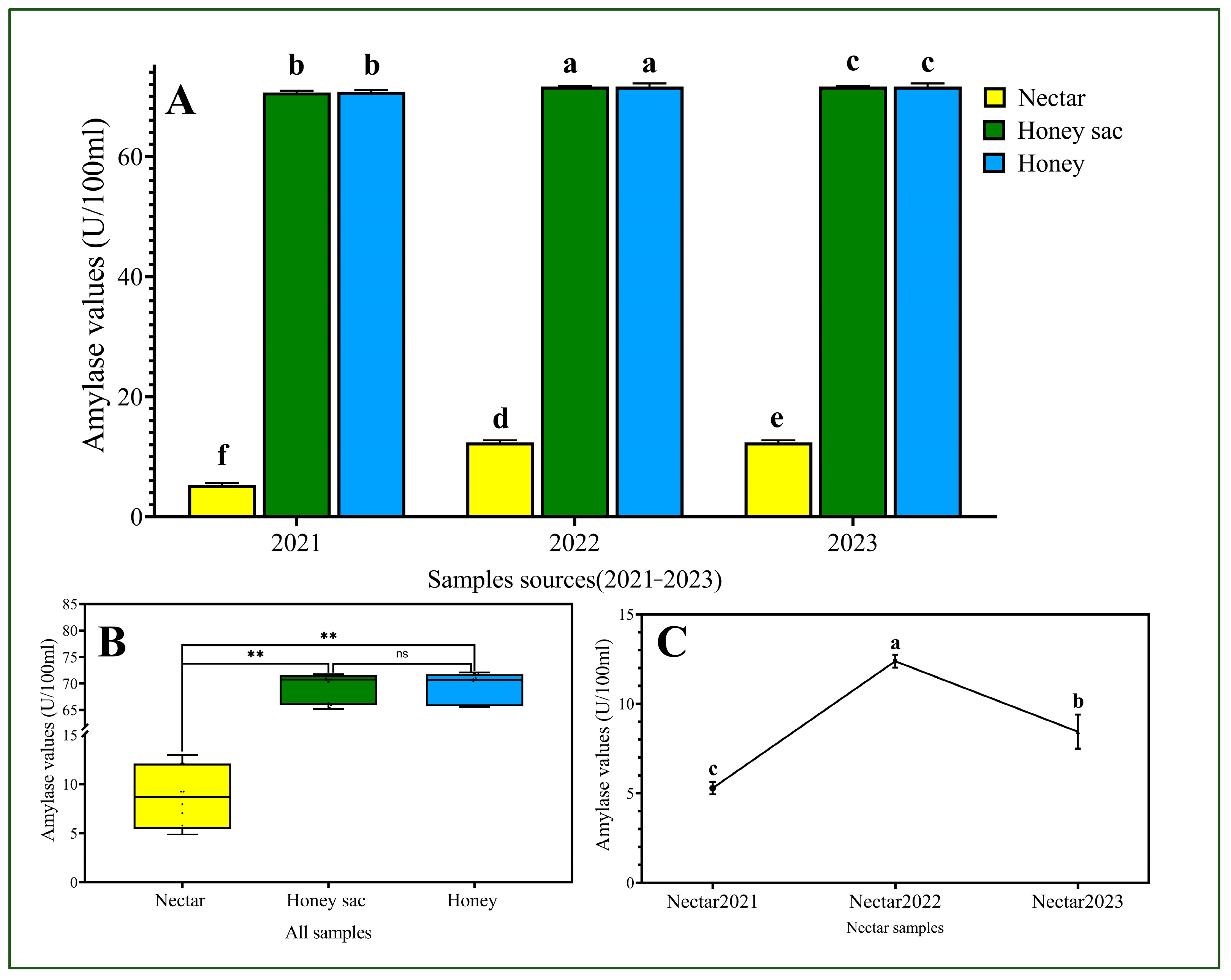
3.4. Higher Amylase Activities Were Detected in the Samples from Honey Sac and Honey Groups
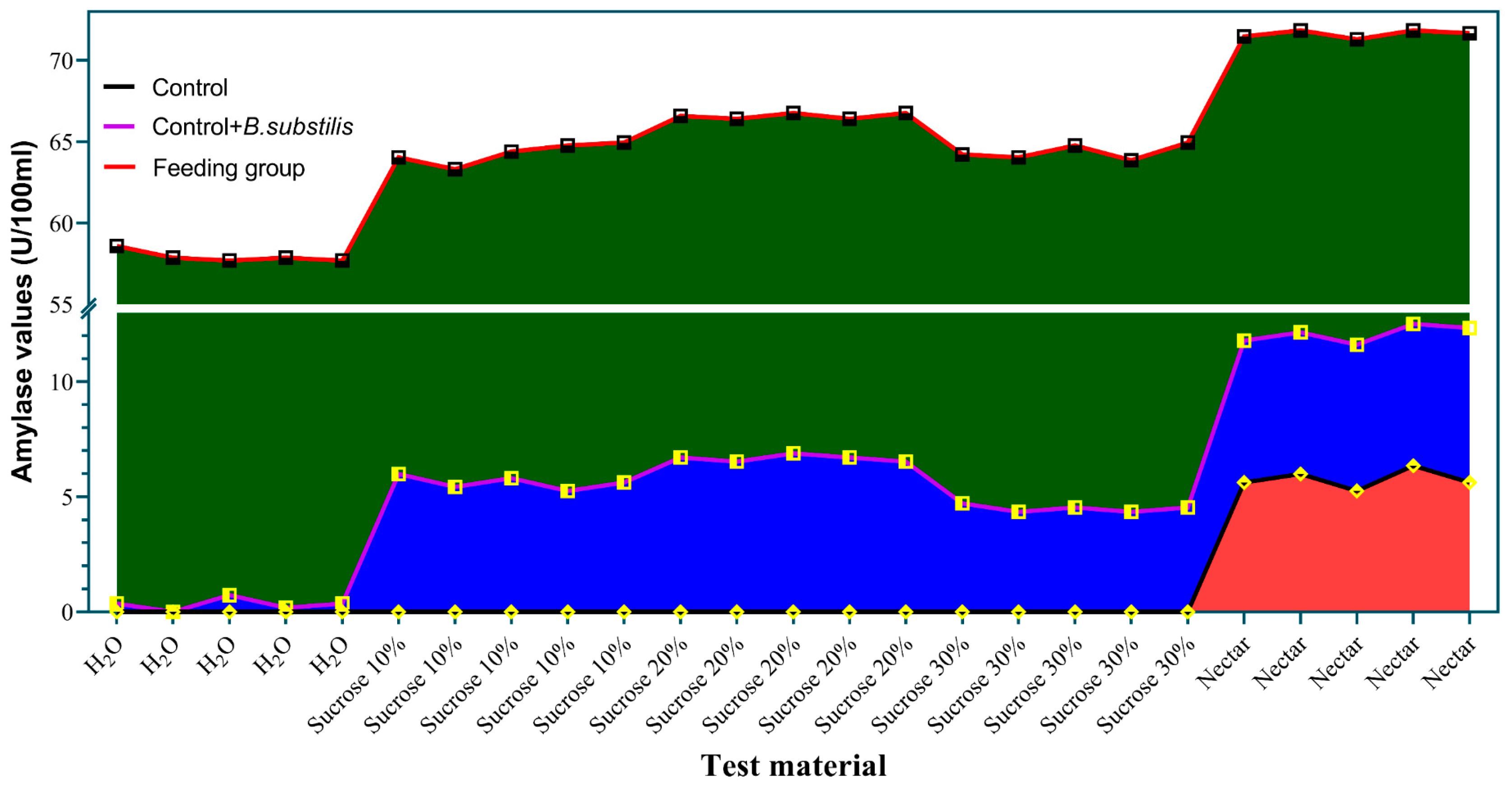
3.5. B. subtilis Can Produce Amylase in Culture When Provided with Sucrose or Nectar
3.6. Measurement of the Amylase Activity Produced by B. subtilis in the Honey Sacs of Bees
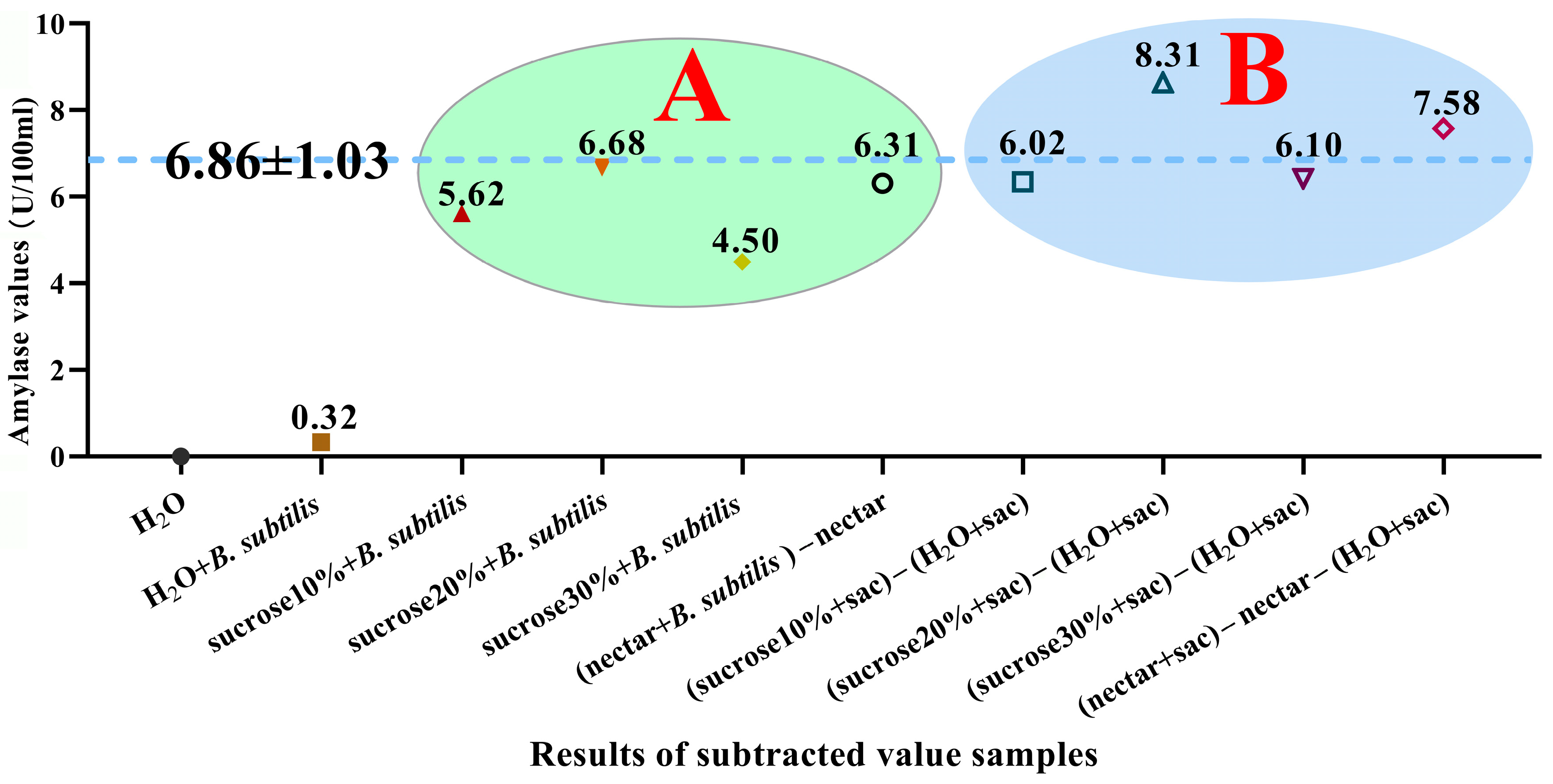
3.7. The Trend of Amylase Activity in Simulation and Feeding Experiments
3.8. The Amylase Activity Produced by B. subtilis in the Honey Sac Aligns with the Amylase Activity Observed in the Simulation Experiment
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Takahashi, Y.; Yoshida, I.; Yokozeki, T.; Igarashi, T.; Fujita, K. Investigation of Foreign Amylase Adulteration in Honey Distributed in Japan by Rapid and Improved Native PAGE Activity Staining Method. J. Appl. Glycosci. 2023, 70, 67–73. [Google Scholar] [CrossRef]
- Bong, J.; Middleditch, M.; Loomes, K.M.; Stephens, J.M. Proteomic analysis of honey. Identification of unique peptide markers for authentication of NZ mānuka (Leptospermum scoparium) honey. Food Chem. 2021, 350, 128442. [Google Scholar] [CrossRef] [PubMed]
- Babacan, S.; Rand, A.G. Characterization of Honey Amylase. J. Food Sci. 2007, 72, C050–C055. [Google Scholar] [CrossRef] [PubMed]
- Peláez-Acero, A.; Garrido-Islas, D.B.; Campos-Montiel, R.G.; González-Montiel, L.; Medina-Pérez, G.; Luna-Rodríguez, L.; González-Lemus, U.; Cenobio-Galindo, A.d.J. The Application of Ultrasound in Honey: Antioxidant Activity, Inhibitory Effect on α-amylase and α-glucosidase, and In Vitro Digestibility Assessment. Molecules 2022, 27, 5825. [Google Scholar] [CrossRef]
- Pasias, I.N.; Kiriakou, I.K.; Proestos, C. HMF and diastase activity in honeys: A fully validated approach and a chemometric analysis for identification of honey freshness and adulteration. Food Chem. 2017, 229, 425–431. [Google Scholar] [CrossRef]
- Babacan, S.; Pivarnik, L.F.; Rand, A.G. Honey amylase activity and food starch degradation. J. Food Sci. 2002, 5, 1625–1630. [Google Scholar] [CrossRef]
- Wang, J.; Li, Q.X. Chemical Composition, Characterization, and Differentiation of Honey Botanical and Geographical Origins. Adv. Food Nutr. Res. 2011, 62, 89–137. [Google Scholar] [CrossRef]
- Elyasi Far, B.; Dilmaghani, A.; Yari Khosroushahi, A. In Silico Study and Optimization of Bacillus megaterium alpha-Amylases Production Obtained from Honey Sources. Curr. Microbiol. 2020, 77, 2593–2601. [Google Scholar] [CrossRef] [PubMed]
- Habib, H.M.; Kheadr, E.; Ibrahim, W.H. Inhibitory effects of honey from arid land on some enzymes and protein damage. Food Chem. 2021, 364, 130415. [Google Scholar] [CrossRef] [PubMed]
- Ollerton, J.; Winfree, R.; Tarrant, S. How many flowering plants are pollinated by animals? Oikos 2011, 120, 321–326. [Google Scholar] [CrossRef]
- Jang, S.; Kikuchi, Y. Impact of the insect gut microbiota on ecology, evolution, and industry. Curr. Opin. Insect Sci. 2020, 41, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Nair, S. Dynamics of Insect–Microbiome Interaction Influence Host and Microbial Symbiont. Front. Microbiol. 2020, 11, 1357. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.F.; Rodriguez, V.; Brzoska, C.; Pfister, J.; Neumann, P.; Retschnig, G. Dream Team for Honey Bee Health: Pollen and Unmanipulated Gut Microbiota Promote Worker Longevity and Body Weight. Front. Sustain. Food Syst. 2022, 6, 864741. [Google Scholar] [CrossRef]
- Eleftherianos, I.; Zhang, W.; Tettamanti, G.; Daley, L.; Mohamed, A.; Stanley, D. Nutrition influences immunity: Diet and host-parasite interactions. Insect Biochem. Mol. Biol. 2024, 175, 104210. [Google Scholar] [CrossRef] [PubMed]
- Evans, J.D.; Armstrong, T.-N. Antagonistic interactions between honey bee bacterial symbionts and implications for disease. BMC Ecol. 2006, 6, 4. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Steele, M.I.; Leonard, S.P.; Motta, E.V.S.; Moran, N.A. Honey bees as models for gut microbiota research. Lab Anim. 2018, 47, 317–325. [Google Scholar] [CrossRef] [PubMed]
- Anee, I.J.; Alam, S.; Begum, R.A.; Shahjahan, R.M.; Khandaker, A.M. The role of probiotics on animal health and nutrition. J. Basic Appl. Zool. 2021, 82, 52. [Google Scholar] [CrossRef]
- Kwong, W.K.; Moran, N.A. Gut microbial communities of social bees. Nat. Rev. Microbiol. 2016, 14, 374–384. [Google Scholar] [CrossRef] [PubMed]
- Martinson, V.G.; Moy, J.; Moran, N.A. Establishment of Characteristic Gut Bacteria during Development of the Honeybee Worker. Appl. Environ. Microbiol. 2012, 78, 2830–2840. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Hong, X.; Yin, G.-F.; He, S.-Y. Diversity of bacteria in the honey stomach of Apis cerana and Apis mellifera during the rape blooming period. Chin. J. Appl. Entomol. 2014, 51, 1567–1575. [Google Scholar] [CrossRef]
- Su, Y.; Liu, C.; Fang, H.; Zhang, D. Bacillus subtilis: A universal cell factory for industry, agriculture, biomaterials and medicine. Microb. Cell Factories 2020, 19, 173. [Google Scholar] [CrossRef] [PubMed]
- Rinaudo, M.T.; Ponzetto, C.; Vidano, C.; Marletto, F. The origin of honey amylase. Comp. Biochem. Physiol. B 1973, 46, 253–256. [Google Scholar] [CrossRef]
- Wang, M.; Zhao, W.-Z.; Xu, H.; Wang, Z.-W.; He, S.-Y. Bacillus in the guts of honey bees (Apis mellifera; Hymenoptera: Apidae) mediate changes in amylase values. Eur. J. Entomol. 2015, 112, 619–624. [Google Scholar] [CrossRef]
- Klein, S.; Pasquaretta, C.; He, X.J.; Perry, C.; Søvik, E.; Devaud, J.-M.; Barron, A.B.; Lihoreau, M. Honey bees increase their foraging performance and frequency of pollen trips through experience. Sci. Rep. 2019, 9, 6778. [Google Scholar] [CrossRef]
- Nicolson, S.W.; Human, H.; Pirk, C.W.W. Honey bees save energy in honey processing by dehydrating nectar before returning to the nest. Sci. Rep. 2022, 12, 16224. [Google Scholar] [CrossRef]
- Sambo, F.; Finotello, F.; Lavezzo, E.; Baruzzo, G.; Masi, G.; Peta, E.; Falda, M.; Toppo, S.; Barzon, L.; Di Camillo, B. Optimizing PCR primers targeting the bacterial 16S ribosomal RNA gene. BMC Bioinform. 2018, 19, 343. [Google Scholar] [CrossRef] [PubMed]
- Foysal, M.J.; Lisa, A.K. Isolation and characterization of Bacillus sp. strain BC01 from soil displaying potent antagonistic activity against plant and fish pathogenic fungi and bacteria. J. Genet. Eng. Biotechnol. 2018, 16, 387–392. [Google Scholar] [CrossRef]
- Golnari, M.; Bahrami, N.; Milanian, Z.; Rabbani Khorasgani, M.; Asadollahi, M.A.; Shafiei, R.; Fatemi, S.S.-A. Isolation and characterization of novel Bacillus strains with superior probiotic potential: Comparative analysis and safety evaluation. Sci. Rep. 2024, 14, 1457. [Google Scholar] [CrossRef] [PubMed]
- Liao, C.; Xu, Y.; Sun, Y.; Lehnert, M.S.; Xiang, W.; Wu, J.; Wu, Z. Feeding behavior of honey bees on dry sugar. J. Insect Physiol. 2020, 124, 104059. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.; Gilet, T.; Bush, J.W. Optimal concentrations in nectar feeding. Proc. Natl. Acad. Sci. USA 2011, 108, 16618–16621. [Google Scholar] [CrossRef] [PubMed]
- Petanidou, T. Ecological and evolutionary aspects of floral nectars in Mediterranean habitats. Nectaries Nectar 2007, 8, 343–375. [Google Scholar]
- Kardas, M.; Staśkiewicz-Bartecka, W.; Sołtys, K.; Dul, L.; Sapała, A.-M.; Kiciak, A.; Bielaszka, A.; Kardas, J. The quality of selected raw and pasteurized honeys based on their sensory profiles and consumer preferences. Front. Nutr. 2024, 10, 1330307. [Google Scholar] [CrossRef]
- Davis, A.; Sawhney, V.; Fowke, L.; Low, N. Floral nectar secretion and ploidy in Brassica rapa and B napus (Brassicaceae). I. Nectary size and nectar carbohydrate production and composition. Apidologie 1994, 25, 602–614. [Google Scholar] [CrossRef]
- Hanadi Riyad Aljabi, E.P. Influence of weather conditions on the activity and properties of alpha-amylase in maize grains. J. Cereal Sci. 2022, 103, 103403. [Google Scholar] [CrossRef]
- Acharya, A.; Khanal, A.; Bajracharya, M.R.; Timalsina, A.; Bishwokarma, A.; Basnet, A. Production, purification and optimisation of amylase by submerged fermentation using Bacillus subtilis. Int. J. Sci. Res. Sci. Eng. Technol. 2019, 6, 265–275. [Google Scholar] [CrossRef]
- Abdella, M.; Rateb, S.H.; Khodairy, M.M.; Omar, E.M. Sucrose, glucose, and fructose preference in honeybees and their effects on food digestibility. Apidologie 2024, 55, 77. [Google Scholar] [CrossRef]
- Erban, T.; Shcherbachenko, E.; Talacko, P.; Harant, K. A single honey proteome dataset for identifying adulteration by foreign amylases and mining various protein markers natural to honey. J. Proteom. 2021, 239, 104157. [Google Scholar] [CrossRef] [PubMed]
| Year | Isolation and Accession Number of NCBI | Similar Bacterial Strain and Login Number of NCBI | Similar Interval | Sequence Lengths and Similarity | Concentration of B. subtilis Used in Simulation Experiment (Total Number 1000 µL) |
|---|---|---|---|---|---|
| 2021 | AMHS2039 (PQ288516) | B. subtilis strain SEM2H8 16S ribosomal RNA gene, partial sequence (MW380566.1) | 19–1355 | 1329/1338 (99%) | 10−5, 428 μL |
| 2022 | AMHS2785 (PQ288517) | B. subtilis strain 4I3 16S ribosomal RNA gene, partial sequence (MW380592.1) | 8–1449 | 1442/1442 (100%) | 10−5, 479 μL |
| 2023 | AMHS3076 (PQ288518) | B. subtilis strain FQ32 16S ribosomal RNA gene, partial sequence (MF144479.1) | 37–1336 | 1300/1300 (100%) | 10−5, 498 μL |
| 0~11 | Culture Time | 0 | GLY | ERY | DARA | LARA | RIB | DXYL | LXYL | ADO | MDX | GAL | GLU |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMHS2039 | 24 h | − | + | − | − | + | + | + | − | − | − | − | + |
| 48 h | − | − | − | − | + | + | + | − | − | − | − | − | |
| AMHS2785 | 24 h | − | + | − | − | + | + | + | − | − | − | − | + |
| 48 h | − | − | − | − | − | − | − | − | − | − | + | − | |
| AMHS3076 | 24 h | − | + | − | − | + | + | − | − | − | − | − | + |
| 48 h | − | − | − | − | − | − | + | + | + | + | − | − |
| Groups | Test Material | Amylase Values (U/100 mL) (Mean ± SD) Significance |
|---|---|---|
| Control groups | H2O | 0 (a) |
| 10% Sucrose | 0 (a) | |
| 20% Sucrose | 0 (a) | |
| 30% Sucrose | 0 (a) | |
| Nectar (Control) | 5.77 ± 0.41 (b) | |
| Experimental groups | H2O + B. subtilis | 0.32 ± 0.26 (a) |
| Sucrose 10% + B. subtilis | 5.62 ± 0.28 (b) | |
| Sucrose 20% + B. subtilis | 6.68 ± 0.15 (c) | |
| Sucrose 30% + B. subtilis | 4.49 ± 0.15 (d) | |
| Nectar + B. subtilis | 12.08 ± 0.38 (e) |
| Groups | Feeding Materials | Amylase Values (U/100 mL) (Mean ± SD) Significance |
|---|---|---|
| Control group | H2O | 57.94 ± 0.37 (a) |
| Experimental groups | 10% sucrose | 64.29 ± 0.64 (b) |
| 20% sucrose | 66.57 ± 0.18 (c) | |
| 30% sucrose | 64.36 ± 0.46 (b) | |
| Nectar | 71.61 ± 0.23 (d) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, M.; Zhao, W.; Zhou, D.; Huang, J. Bacillus subtilis Contributes to Amylase Production in the Honey Sac of Apis mellifera. Insects 2025, 16, 221. https://doi.org/10.3390/insects16020221
Wang M, Zhao W, Zhou D, Huang J. Bacillus subtilis Contributes to Amylase Production in the Honey Sac of Apis mellifera. Insects. 2025; 16(2):221. https://doi.org/10.3390/insects16020221
Chicago/Turabian StyleWang, Miao, Wenzheng Zhao, Danyin Zhou, and Jian Huang. 2025. "Bacillus subtilis Contributes to Amylase Production in the Honey Sac of Apis mellifera" Insects 16, no. 2: 221. https://doi.org/10.3390/insects16020221
APA StyleWang, M., Zhao, W., Zhou, D., & Huang, J. (2025). Bacillus subtilis Contributes to Amylase Production in the Honey Sac of Apis mellifera. Insects, 16(2), 221. https://doi.org/10.3390/insects16020221






