Comparative Analysis of the Mitochondrial Genomes of Three Species of Elmidae (Coleoptera: Dryopoidea)
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection, Identification, and DNA Extraction
2.2. Mitogenome Sequencing, Assembly, and Annotation
2.3. Sequence Analyses
2.4. Phylogenetic Analysis
3. Results
3.1. Mitogenome Features of the Three Elmidae Species
3.2. Protein-Coding Genes (PCGs) and Codon Usage
3.3. Transfer RNAs and Ribosomal RNA Genes
3.4. Control Region
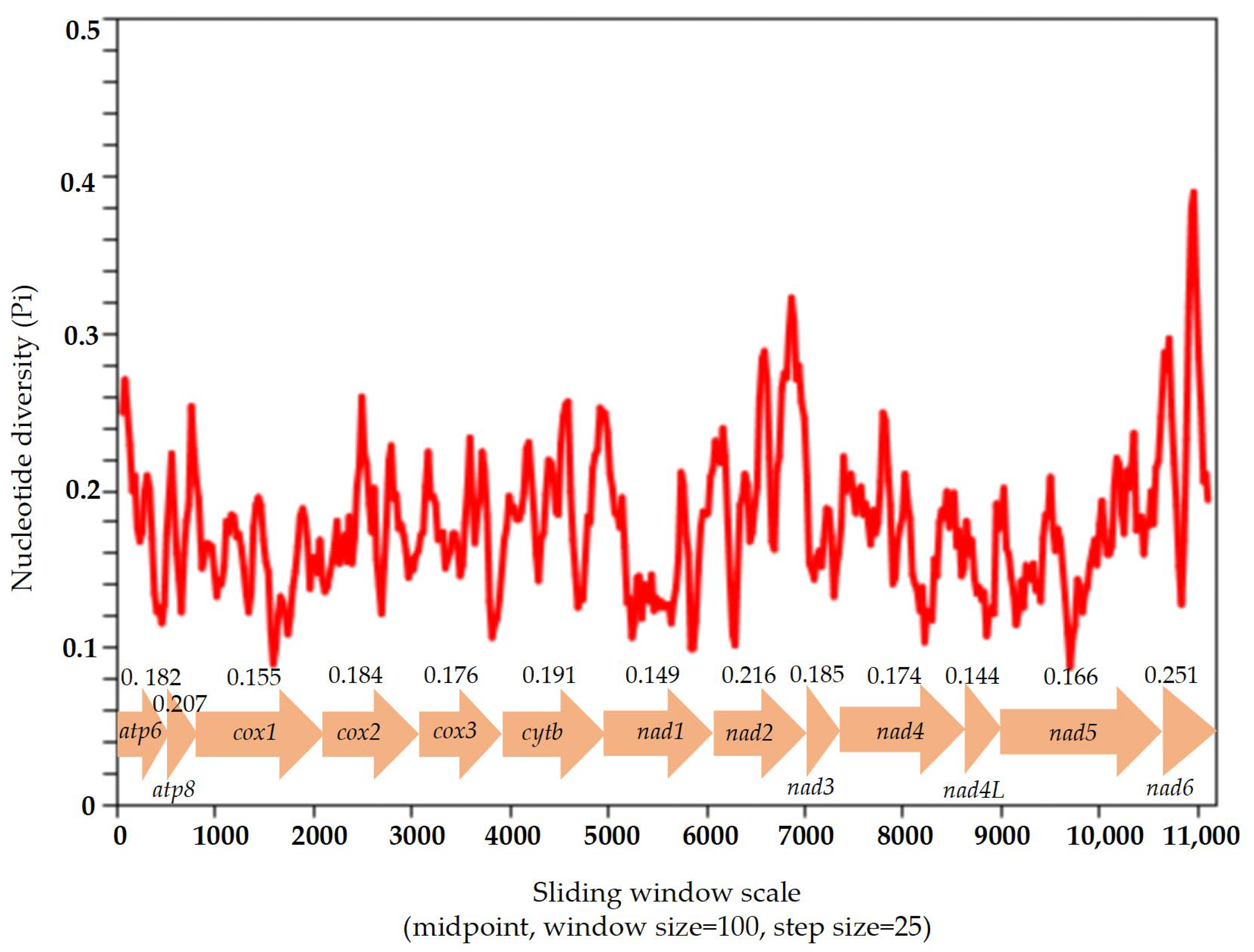
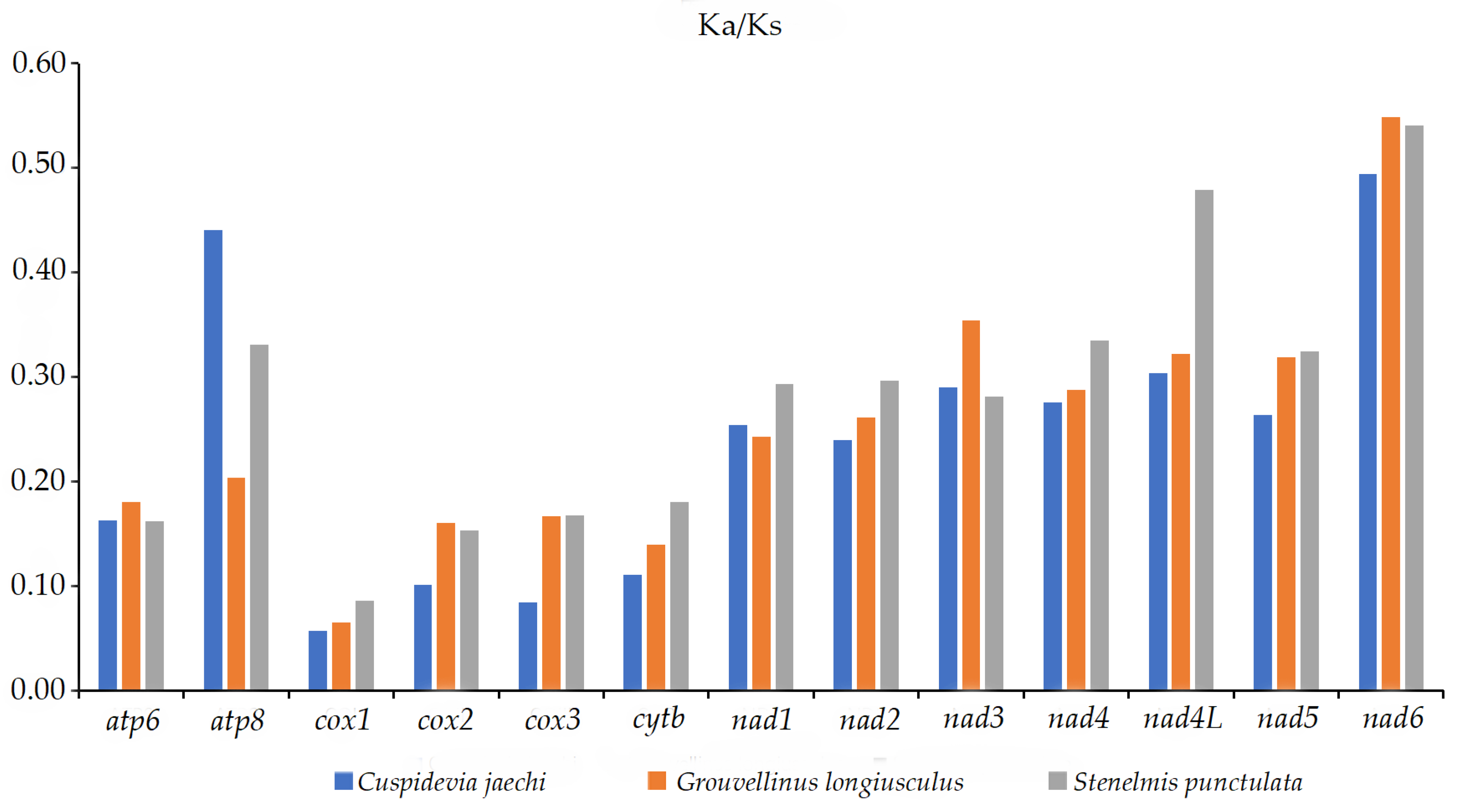
3.5. Nucleotide Diversity and Evolutionary Rate Analysis
3.6. Heterogeneity and Substitution Saturation Tests
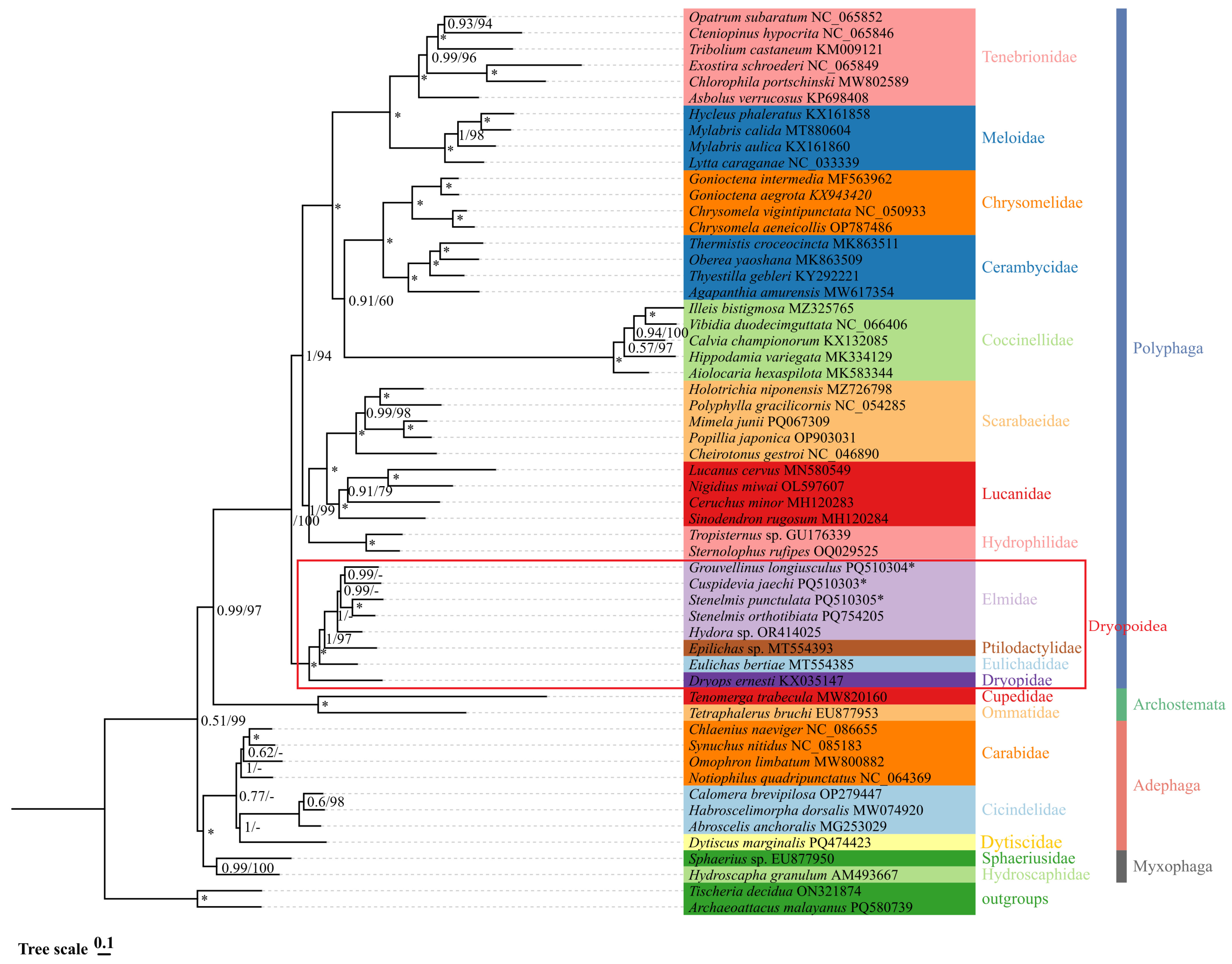
3.7. Phylogenetic Relationships
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kodada, K.; Jäch, M.A.; Ciampor, F.J. Elmidae. In Handbook of Zoology. Coleoptera, Vol. 1: Morphology & Systematics, 2nd ed.; Beutel, R.G., Leschen, R.A.B., Eds.; De Gruyter: Vienna, Austria, 2016; pp. 561–598. [Google Scholar]
- Kobayashi, T.; Hayashi, M.; Kamite, Y.; Sota, T. Molecular phylogeny of Elmidae (Coleoptera: Byrrhoidea) with a focus on Japanese species: Implications for intrafamilial classification. Syst. Entomol. 2021, 46, 870–886. [Google Scholar] [CrossRef]
- Compin, A.; Céréghino, R. Sensitivity of aquatic insect species richness to disturbance in the Adour–Garonne stream system (France). Ecol. Indic. 2003, 3, 135–142. [Google Scholar] [CrossRef]
- Garcia-Criado, F.; Fernandez-Aláez, M. Hydraenidae and Elmidae assemblages (Coleoptera) from a Spanish river basin: Good indicators of coal mining pollution? Arch. Hydrobiol. 2001, 150, 641–660. [Google Scholar] [CrossRef]
- Ribera, I.; Foster, G.N. Uso de coleópteros acuáticos como indicadores biológicos (Coleoptera). Elytron 1992, 6, 61–75. [Google Scholar]
- Ribera, I. Biogeography and conservation of Iberian Water Beetles. Biol. Conserv. 2000, 92, 131–150. [Google Scholar] [CrossRef]
- Bouchard, P.; Bousquet, Y.; Davies, A.E.; Alonso-Zarazaga, M.A.; Lawrence, J.F.; Lyal, C.H.; Newton, A.F.; Reid, C.A.; Schmitt, M.; Slipiński, S.A.; et al. Family-group names in Coleoptera (Insecta). Zookeys 2011, 4, 1–972. [Google Scholar]
- Jäch, M.A.; Kodada, J.; Brojer, M.; Shepard, W.D.; Ciampor, F. Coleoptera: Elmidae and Protelmidae. World Catalogue of Insects; Brill: Leiden, The Netherlands, 2016; Volume 14. [Google Scholar]
- Hayashi, M.; Yoshitomi, H.; Kamite, Y.; Kobayashi, T.; Sota, T. Description of adults and larvae of Orientelmis parvula (Nomura & Baba, 1961) (Coleoptera: Elmidae) with their molecular phylogenetic analysis. Zootaxa 2019, 4568, 483–500. [Google Scholar]
- Wolstenholme, D.R. Animal mitochondrial DNA: Structure and evolution. Int. Rev. Cytol. 1992, 141, 173–216. [Google Scholar]
- Boore, J.L. Animal mitochondrial genomes. Nucleic Acids Res. 1999, 27, 1767–1780. [Google Scholar] [CrossRef]
- Shadel, G.S.; Clayton, D.A. Mitochondrial transcription initiation: Variation and conservation. J. Biol. Chem. 1993, 268, 16083–16086. [Google Scholar] [CrossRef]
- Zhang, D.X.; Hewitt, G.M. Insect mitochondrial control region: A review of its structure, evolution and usefulness in evolutionary studies. Biochem. Syst. Ecol. 1997, 25, 99–120. [Google Scholar] [CrossRef]
- Cameron, S.L. Insect mitochondrial genomics: Implications for evolution and phylogeny. Annu. Rev. Entomol. 2014, 59, 95–117. [Google Scholar] [CrossRef] [PubMed]
- Macino, G.; Scazzocchio, C.; Waring, R.B.; Berks, M.M.P.; Wayne Davies, R. Conservation and rearrangement of mitochondrial structural gene sequences. Nature 1980, 288, 404–406. [Google Scholar] [CrossRef]
- Johnston, N.P.; Piwczy’nski, M.; Trzeciak, P.; Walczak, K.; Szpila, K. Integration of mitogenomic and morphological data disentangles the systematics of Pollenia and establishes a revised phylogenetic hypothesis for the Polleniidae. Syst. Entomol. 2022, 48, 296–315. [Google Scholar] [CrossRef]
- Bian, D.; Ji, L. Two new species of Cuspidevia Jäch & Boukal, 1995 from China (Coleoptera: Elmidae). Zootaxa 2010, 2663, 53–58. [Google Scholar]
- Bian, D.; Jäch, M.A. Revision of the species Grouvellinus Champion, 1923 (Coleoptera: Elmidae) with long median pronotal carina, including descriptions of four new species from China. Zootaxa 2019, 4586, 127–140. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenetics Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef]
- Meng, G.; Li, Y.; Yang, C.; Liu, S. MitoZ: A toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 2019, 47, e63. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Chan, P.P.; Lowe, T.M. tRNAscan-SE: Searching for tRNA Genes in Genomic Sequences. Methods Mol. Biol. 2019, 1962, 1–14. [Google Scholar] [PubMed]
- Grant, J.R.; Stothard, P. The CGView Server: A comparative genomics tool for circular genomes. Nucleic Acids Res. 2008, 36, W181–W184. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Perna, N.T.; Kocher, T.D. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 1995, 41, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Laslett, D.; Canbäck, B. ARWEN: A program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics 2008, 24, 172–175. [Google Scholar] [CrossRef]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Talavera, G.; Castresana, J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007, 56, 564–577. [Google Scholar] [CrossRef]
- Xiang, C.-Y.; Gao, F.; Jakovlić, I.; Lei, H.-P.; Hu, Y.; Zhang, H.; Zou, H.; Wang, G.-T.; Zhang, D. Using PhyloSuite for molecular phylogeny and tree-based analyses. iMeta 2023, 2, e87. [Google Scholar] [CrossRef]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.; Lemey, P. Assessing substitution saturation with DAMBE. In The Phylogenetic Handbook: A Practical Approach to DNA and Protein Phylogeny; Cambridge University Press: Cambridge, UK, 2009; Volume 2, pp. 587–589. [Google Scholar]
- Xia, X.; Xie, Z. DAMBE: Software package for data analysis in molecular biology and evolution. J. Hered. 2001, 92, 371–373. [Google Scholar] [CrossRef] [PubMed]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New Methods for Selecting Partitioned Models of Evolution for Molecular and Morphological Phylogenetic Analyses. Mol. Biol. Evol. 2017, 34, 772–773. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Minh, B.Q.; Nguyen, M.A.T.; von Haeseler, A. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 2013, 30, 1188–1195. [Google Scholar] [CrossRef]
- Lartillot, N.; Lepage, T.; Blanquart, S. PhyloBayes 3: A Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics 2009, 25, 2286–2288. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef]
- Ojala, D.; Montoya, J.; Attardi, G. tRNA punctuation model of RNA processing in human mitochondria. Nature 1981, 290, 470–474. [Google Scholar] [CrossRef]
- Sheffield, N.C.; Song, H.; Cameron, S.L.; Whiting, M.F. A comparative analysis of mitochondrial genomes in Coleoptera (Arthropoda: Insecta) and genome descriptions of six new beetles. Mol. Biol. Evol. 2008, 25, 2499–2509. [Google Scholar] [CrossRef]
- Zhang, H.L.; Ye, F. Comparative Mitogenomic Analyses of Praying Mantises (Dictyoptera, Mantodea): Origin and Evolution of Unusual Intergenic Gaps. Int. J. Biol. Sci. 2017, 13, 367–382. [Google Scholar] [CrossRef][Green Version]
- Shepard, W.D.; Sites, R.W. Aquatic beetles of the families Dryopidae and Elmidae (Insecta: Coleoptera: Byrrhoidea) of Thailand: Annotated list and illustrated key to genera. Nat. Hist. Bull. Siam Soc. 2016, 61, 89–126. [Google Scholar]
- Nie, R.-E.; Yang, X.-K. Research progress in mitochondrial genemes of Coleoptera. Acta Entomol. Sin. 2014, 57, 860–868. [Google Scholar]
- Timmermans, M.J.T.N.; Vogler, A.P. Phylogenetically informative rearrangements in mitochondrial genomes of Coleoptera, and monophyly of aquatic elateriform beetles (Dryopoidea). Mol. Phylogenetics Evol. 2012, 63, 229–304. [Google Scholar] [CrossRef] [PubMed]
- McKenna, D.D.; Shin, S.; Ahrens, D.; Balke, M.; Beza-Beza, C.; Clarke, D.J.; Donath, A.; Escalona, H.E.; Friedrich, F.; Letsch, H.; et al. The evolution and genomic basis of beetle diversity. Proc. Natl. Acad. Sci. USA 2019, 116, 24729–24737. [Google Scholar] [CrossRef] [PubMed]
- Cai, C.; Tihelka, E.; Giacomelli, M.; Lawrence, J.F.; Ślipiński, A.; Kundrata, R.; Yamamoto, S.; Thayer, M.K.; Newton, A.F.; Leschen Richard, A.B.; et al. Integrated phylogenomics and fossil data illuminate the evolution of beetles. R. Soc. Open Sci. 2022, 9, 211771. [Google Scholar] [CrossRef]
- Bocak, L.; Barton, C.; Alexcrampton-Platt, D.C.; Ahrens, D.; Alfried, P.V. Building the Coleoptera tree-of-life for >8000 species: Composition of public DNA data and fit with Linnaean classification. Syst. Entomol. 2014, 39, 97–110. [Google Scholar] [CrossRef]


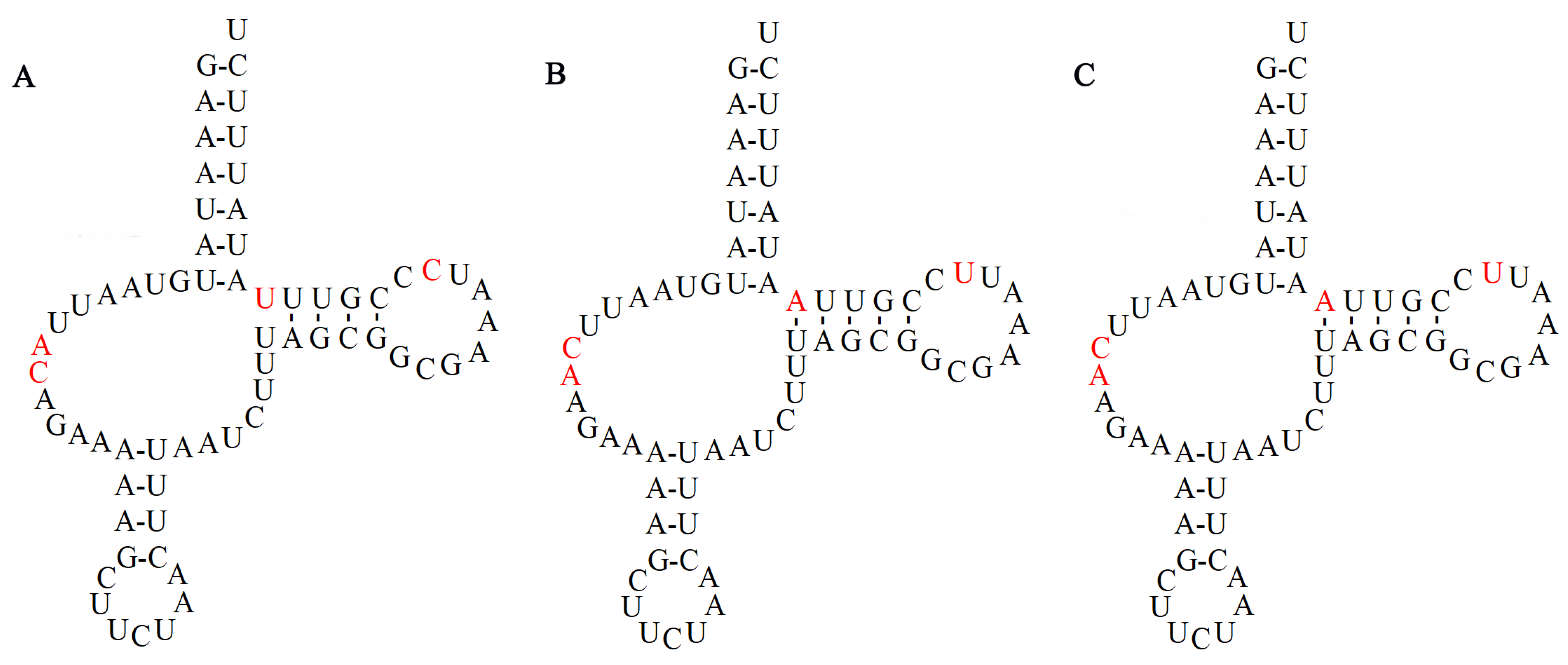


| Specimen | Date of Collection | Collection Site | Longitude (E) | Latitude (N) | GB Number |
|---|---|---|---|---|---|
| C. jaechi | 15 October 2023 | Jiulianshan Natural Reserve, Jiangxi | 114.504167 | 24.591389 | PQ510303 |
| G. longiusculus | 9 October 2023 | Yunju Mountains, Jiangxi | 115.583611 | 29.093889 | PQ510304 |
| S. punctulata | 25 September 2023 | Mount Li, Shanxi | 111.993611 | 35.355000 | PQ510305 |
| Species | Type | Length (bp) | A% | T% | G% | C% | A + T% | G + C% | AT-Skew | GC-Skew |
|---|---|---|---|---|---|---|---|---|---|---|
| C. jaechi | PCGs | 11,184 (68.6%) | 31.6 | 41.7 | 12.6 | 14.2 | 73.2 | 26.8 | −0.1382 | −0.0601 |
| tRNAs | 1452 (8.9%) | 38.4 | 37.9 | 13.2 | 10.5 | 76.2 | 23.8 | 0.0063 | 0.1130 | |
| rRNAs | 2076 (12.7%) | 32.5 | 46.0 | 14.9 | 6.6 | 78.5 | 21.5 | −0.1713 | 0.3870 | |
| l-rRNA | 1298 (8.0%) | 46.6 | 32.6 | 6.2 | 14.6 | 79.2 | 20.8 | 0.1770 | −0.4000 | |
| s-rRNA | 778 (4.8%) | 44.9 | 32.4 | 7.2 | 15.6 | 77.2 | 22.8 | 0.1614 | −0.3672 | |
| control region | 1553 (9.5%) | 45.0 | 35.9 | 6.2 | 12.9 | 80.9 | 19.1 | 0.1122 | −0.3514 | |
| J-strand | 7816 (47.9%) | 35.7 | 36.2 | 10.6 | 17.5 | 71.9 | 28.1 | −0.0073 | −0.2476 | |
| N-strand | 6896 (42.3%) | 28.6 | 48.3 | 15.7 | 7.4 | 76.9 | 23.1 | −0.2570 | 0.3610 | |
| full genome | 16,309 | 42.0 | 33.0 | 8.8 | 16.3 | 74.9 | 25.1 | 0.1202 | −0.3009 | |
| G. longiusculus | PCGs | 11,178 (68.6%) | 32.5 | 42.0 | 12.5 | 13.0 | 74.5 | 25.5 | −0.1273 | −0.0196 |
| tRNAs | 1458 (8.9%) | 39.4 | 37.9 | 13.0 | 9.7 | 77.3 | 22.7 | 0.0204 | 0.1420 | |
| rRNAs | 2082 (12.8%) | 32.9 | 46.1 | 14.1 | 6.9 | 79.0 | 21.0 | −0.1679 | 0.3425 | |
| l-rRNA | 1306 (8.0%) | 46.7 | 32.9 | 6.4 | 13.9 | 79.6 | 20.4 | 0.1731 | −0.3684 | |
| s-rRNA | 776 (4.8%) | 45.1 | 32.7 | 7.7 | 14.4 | 77.8 | 22.2 | 0.1589 | −0.3023 | |
| control region | 1543 (9.5%) | 44.8 | 38.0 | 7.3 | 9.9 | 82.8 | 17.2 | 0.0829 | −0.1472 | |
| J-strand | 7809 (47.9%) | 36.1 | 37.3 | 11.4 | 15.2 | 73.4 | 26.6 | −0.0154 | −0.1419 | |
| N-strand | 6909 (42.4%) | 30.0 | 47.7 | 14.3 | 8.0 | 77.7 | 22.3 | −0.2283 | 0.2824 | |
| full genome | 16,291 | 41.9 | 34.2 | 9.6 | 14.3 | 76.1 | 23.9 | 0.1005 | −0.2001 | |
| S. punctulata | PCGs | 11,188 (72.3%) | 32.5 | 42.2 | 12.3 | 13.0 | 74.7 | 25.3 | −0.1306 | −0.0258 |
| tRNAs | 1479 (9.6%) | 38.5 | 38.7 | 13.3 | 9.5 | 77.2 | 22.8 | −0.0035 | 0.1632 | |
| rRNAs | 2071 (13.4%) | 33.5 | 45.9 | 14.1 | 6.5 | 79.4 | 20.6 | −0.1557 | 0.3677 | |
| l-rRNA | 1296 (8.4%) | 46.4 | 33.8 | 6.3 | 13.6 | 80.2 | 19.8 | 0.1569 | −0.3696 | |
| s-rRNA | 777 (5.0%) | 45.0 | 33.1 | 6.9 | 14.9 | 78.1 | 21.9 | 0.1532 | −0.3647 | |
| control region | 753 (4.9%) | 47.4 | 39.6 | 4.9 | 8.1 | 87.0 | 13.0 | 0.0901 | −0.2449 | |
| J-strand | 7827 (50.6%) | 36.1 | 37.2 | 11.1 | 15.5 | 73.3 | 26.7 | −0.0159 | −0.1648 | |
| N-strand | 6911 (44.6%) | 30.0 | 48.2 | 14.4 | 7.4 | 78.2 | 21.8 | −0.2332 | 0.3201 | |
| full genome | 15,480 | 42.0 | 34.1 | 9.2 | 14.7 | 76.1 | 23.9 | 0.1037 | −0.2308 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qin, Z.; Li, N.; Mo, Y.; Wang, J.; Peng, Y.; Song, F. Comparative Analysis of the Mitochondrial Genomes of Three Species of Elmidae (Coleoptera: Dryopoidea). Insects 2025, 16, 247. https://doi.org/10.3390/insects16030247
Qin Z, Li N, Mo Y, Wang J, Peng Y, Song F. Comparative Analysis of the Mitochondrial Genomes of Three Species of Elmidae (Coleoptera: Dryopoidea). Insects. 2025; 16(3):247. https://doi.org/10.3390/insects16030247
Chicago/Turabian StyleQin, Zeliang, Na Li, Yaqi Mo, Juping Wang, Yunfei Peng, and Fan Song. 2025. "Comparative Analysis of the Mitochondrial Genomes of Three Species of Elmidae (Coleoptera: Dryopoidea)" Insects 16, no. 3: 247. https://doi.org/10.3390/insects16030247
APA StyleQin, Z., Li, N., Mo, Y., Wang, J., Peng, Y., & Song, F. (2025). Comparative Analysis of the Mitochondrial Genomes of Three Species of Elmidae (Coleoptera: Dryopoidea). Insects, 16(3), 247. https://doi.org/10.3390/insects16030247






