Genome-Wide Identification and Expression Profiling of Glycosidases, Lipases, and Proteases from Invasive Asian Palm Weevil, Rhynchophorus ferrugineus
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Insect Collection and Tissue Dissection
2.2. Total RNA Extraction, cDNA Library Construction, and Sequencing
2.3. Data Processing, Assembly and Annotation, and Differential Expression Analysis
2.4. Phylogenetic and Genome-Wide Analysis of the Candidate Digestive Enzymes
2.5. Structural Modeling and Molecular Docking
3. Results
3.1. Digestive Enzyme (DE) Transcriptome Profiling and Abundance
3.1.1. Glycosidase Gene Expression
3.1.2. Lipase Gene Expression
3.1.3. Protease Gene Expression
3.2. DE Phylogenetic Analysis
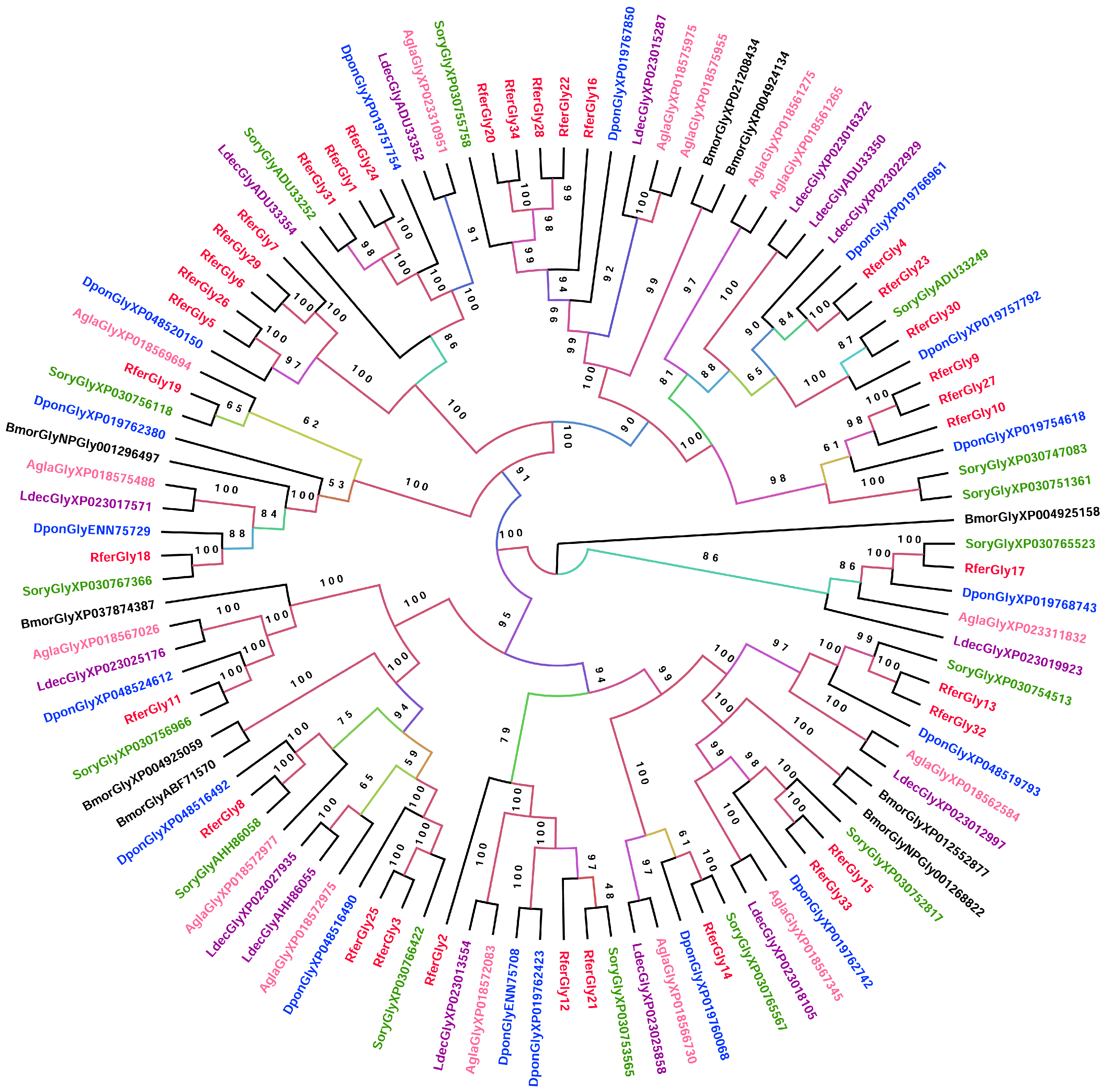
3.2.1. Glycosidase
3.2.2. Lipase
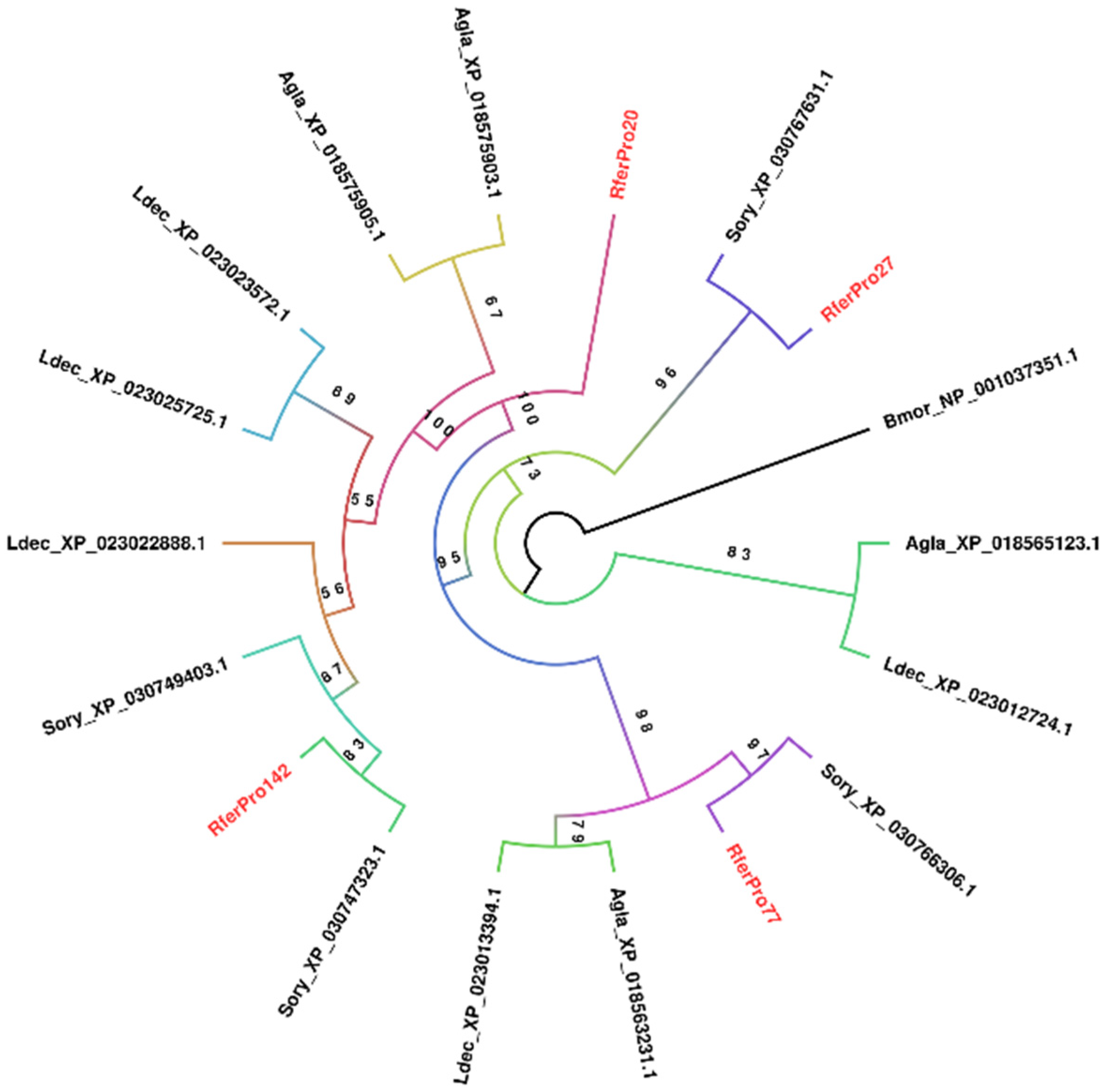
3.2.3. Aspartic Protease
3.3. Genome-Wide Analysis of the Candidate Digestive Enzymes
3.4. Modeling Evaluation and Molecular Docking Analysis
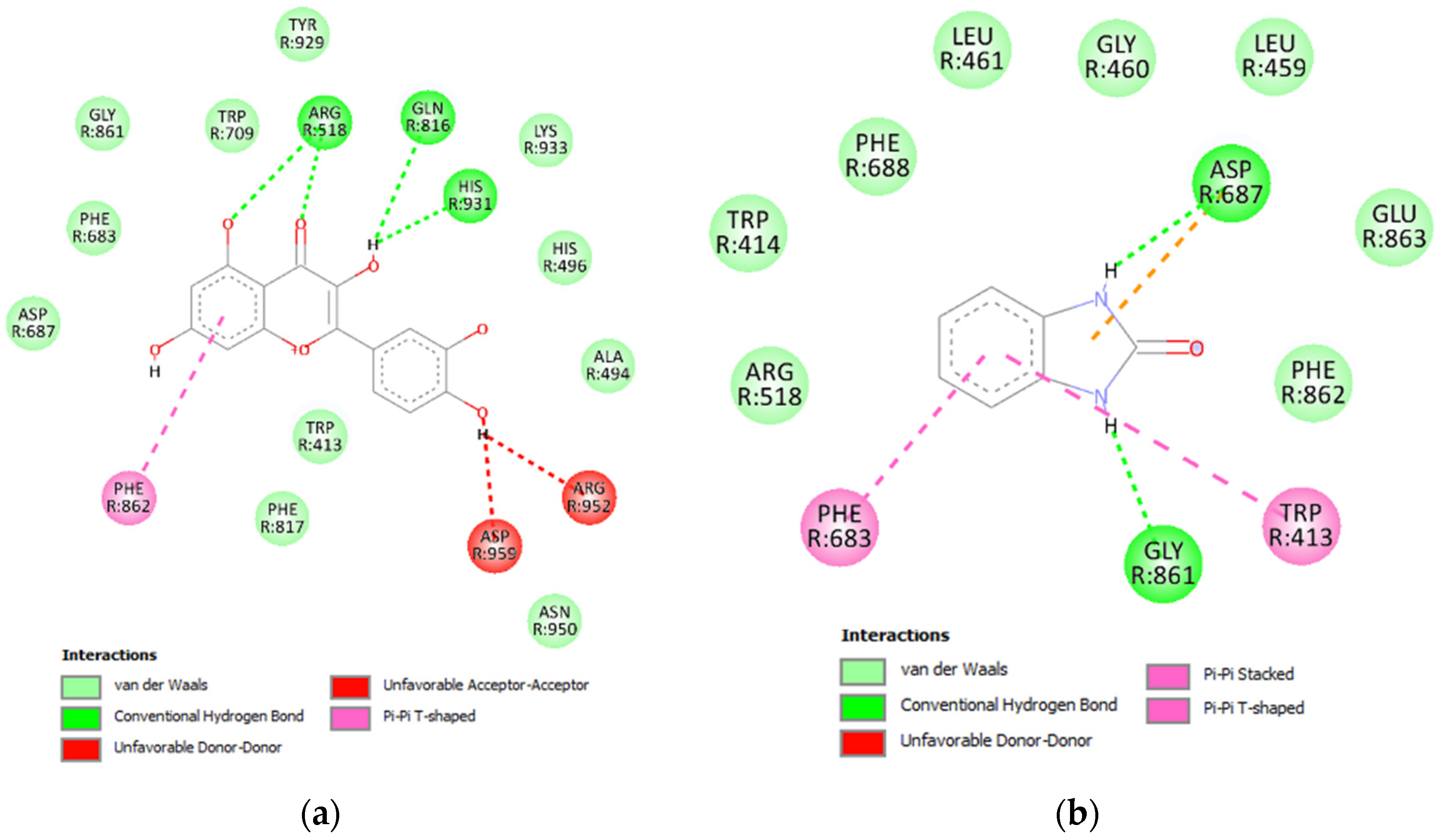
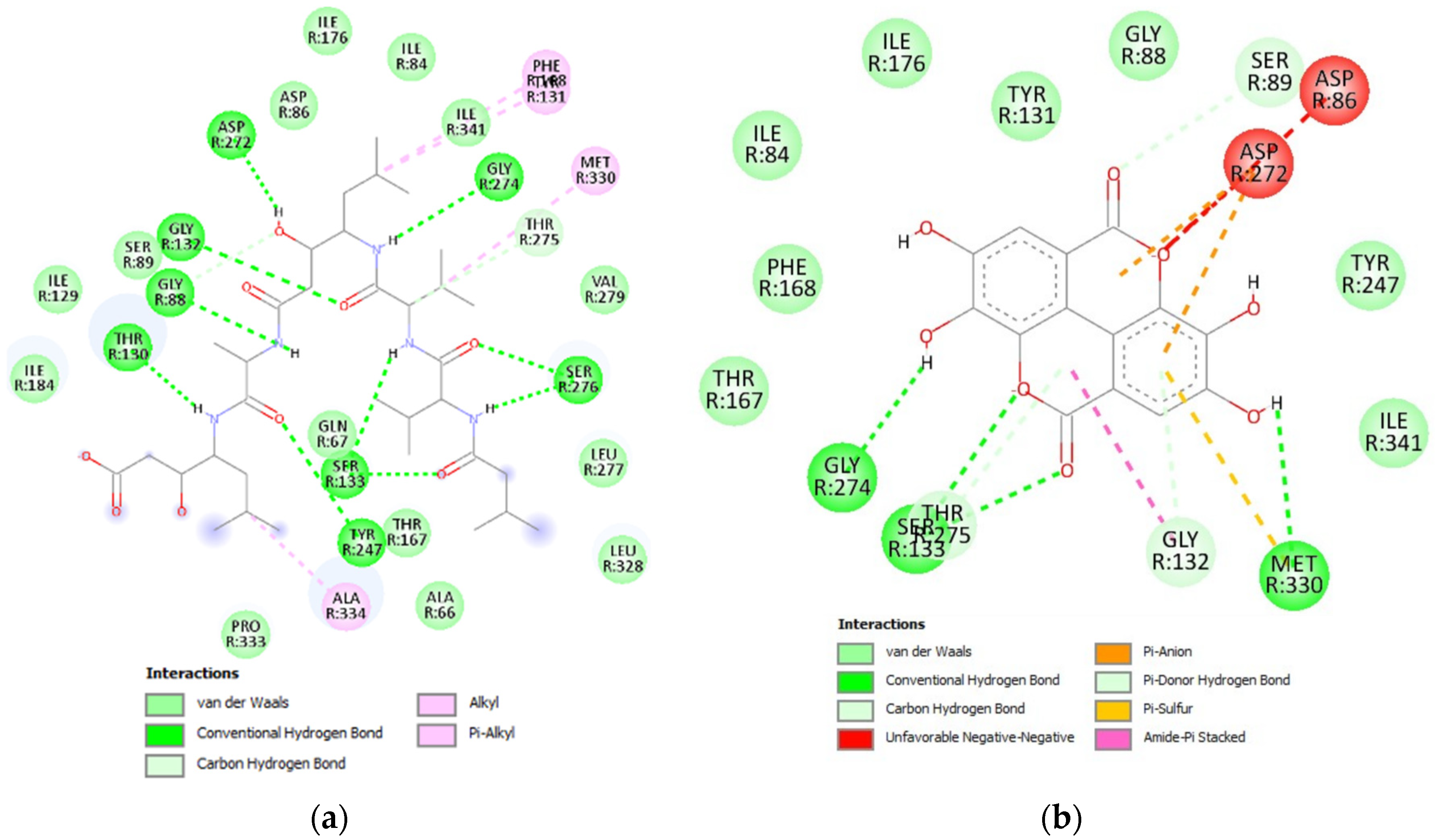
3.4.1. Glycosidase Candidate Model and Putative Inhibitor Docking
3.4.2. Lipase Candidate Model and Putative Inhibitor Docking
3.4.3. Protease Candidate Model and Putative Inhibitor Docking
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Salem, H.; Kaltenpoth, M. Beetle-Bacterial Symbioses: Endless Forms Most Functional. Annu. Rev. Entomol. 2022, 67, 201–219. [Google Scholar] [CrossRef]
- McKenna, D.D.; Shin, S.; Ahrens, D.; Balke, M.; Beza-Beza, C.; Clarke, D.J.; Donath, A.; Escalona, H.E.; Friedrich, F.; Letsch, H.; et al. The Evolution and Genomic Basis of Beetle Diversity. Proc. Natl. Acad. Sci. USA 2019, 116, 24729–24737. [Google Scholar] [CrossRef]
- Terra, W.R.; Ferreira, C.; Jordão, B.P.; Dillon, R.J. Digestive enzymes. In Biology of the Insect Midgut; Springer: Dordrecht, The Netherlands, 1996; Volume 85, pp. 153–194. [Google Scholar]
- Zhang, H.G.; Tan, J.; Reynolds, E.; Kuebler, D.; Faulhaber, S.; Tanouye, M. The Drosophila Slamdance Gene: A Mutation in an Aminopeptidase Can Cause Seizure, Paralysis and Neuronal Failure. Genetics 2002, 162, 1283–1299. [Google Scholar] [CrossRef]
- Park, Y.; Kumar, S.; Kanumuri, R.; Stanley, D.; Kim, Y. A Novel Calcium-Independent Cellular PLA2 Acts in Insect Immunity and Larval Growth. Insect Biochem. Mol. Biol. 2015, 66, 13–23. [Google Scholar] [CrossRef]
- Rugman-Jones, P.F.; Hoddle, C.D.; Hoddle, M.S.; Stouthamer, R. The Lesser of Two Weevils: Molecular-Genetics of Pest Palm Weevil Populations Confirm Rhynchophorus vulneratus (Panzer 1798) as a Valid Species Distinct from R. ferrugineus (Olivier 1790), and Reveal the Global Extent of Both. PLoS ONE 2013, 8, e78379. [Google Scholar] [CrossRef]
- Hoddle, M.S.; Antony, B.; El-Shafie, H.A.F.; Chamorro, M.L.; Milosavljević, I.; Löhr, B.; Faleiro, J.R. Taxonomy, Biology, Symbionts, Omics, and Management of Rhynchophorus Palm Weevils (Coleoptera: Curculionidae: Dryophthorinae). Annu. Rev. Entomol. 2024, 69, 455–479. [Google Scholar] [CrossRef]
- FAO. Red Palm Weevil: Guidelines on Management Practices; Elkahky, M., Faleiro, J.R., Eds.; Food and Agriculture Organizations of United Nations: Rome, Italy, 2020; ISBN 9789251321898. [Google Scholar]
- CABI Invasive Species Compedium. Available online: https://www.cabidigitallibrary.org/doi/10.1079/cabicompendium.47472 (accessed on 10 February 2023).
- Rochat, D.; Dembilio, O.; Jaques, J.A.; Suma, P.; La Pergola, A.; Hamidi, R.; Kontodimas, D.; Soroker, V. Rhynchophorus ferrugineus: Taxonomy, distribution, biology, and life cycle. In Handbook of Major Palm Pests: Biology and Management; Soroker, V., Colazza, S., Eds.; John Wiley & Sons: Hoboken, NJ, USA, 2017; pp. 69–101. ISBN 9781119057482. [Google Scholar]
- Antony, B.; Johny, J.; Abdelazim, M.M.; Jakše, J.; Al-Saleh, M.A.; Pain, A. Global Transcriptome Profiling and Functional Analysis Reveal That Tissue-Specific Constitutive Overexpression of Cytochrome P450s Confers Tolerance to Imidacloprid in Palm Weevils in Date Palm Fields. BMC Genom. 2019, 20, 440. [Google Scholar] [CrossRef]
- Borzoui, E.; Nouri-Ganbalani, G.; Naseri, B. In Vitro and In Vivo Effects of α-Amylase Inhibitor from Avena sativa Seeds on Life History and Physiological Characteristics of Sitotroga cerealella (Lepidoptera: Gelechiidae). J. Insect Sci. 2017, 17, 108. [Google Scholar] [CrossRef]
- Mehrabadi, M.; Bandani, A.R.; Mehrabadi, R.; Alizadeh, H. Inhibitory Activity of Proteinaceous α-Amylase Inhibitors from Triticale Seeds against Eurygaster integriceps Salivary α-Amylases: Interaction of the Inhibitors and the Insect Digestive Enzymes. Pestic. Biochem. Physiol. 2012, 102, 220–228. [Google Scholar] [CrossRef]
- Rodrigues Macedo, M.; Das Graças, M.; Freire, M. Insect Digestive Enzymes as a Target for Pest Control. ISJ-Invertebr. Surviv. J. 2011, 8, 190–198. [Google Scholar]
- Jurat-Fuentes, J.L.; Heckel, D.G.; Ferré, J. Mechanisms of Resistance to Insecticidal Proteins from Bacillus thuringiensis. Annu. Rev. Entomol. 2021, 66, 121–140. [Google Scholar] [CrossRef]
- Antony, B.; Soffan, A.; Jakše, J.; Abdelazim, M.M.; Aldosari, S.A.; Aldawood, A.S.; Pain, A. Identification of the Genes Involved in Odorant Reception and Detection in the Palm Weevil Rhynchophorus ferrugineus, an Important Quarantine Pest, by Antennal Transcriptome Analysis. BMC Genom. 2016, 17, 69. [Google Scholar] [CrossRef]
- Antony, B.; Johny, J.; Montagné, N.; Jacquin-Joly, E.; Capoduro, R.; Cali, K.; Persaud, K.; Al-Saleh, M.A.; Pain, A. Pheromone Receptor of the Globally Invasive Quarantine Pest of the Palm Tree, the Red Palm Weevil (Rhynchophorus ferrugineus). Mol. Ecol. 2021, 30, 2025–2039. [Google Scholar] [CrossRef]
- Antony, B.; Montagné, N.; Comte, A.; Mfarrej, S.; Jakše, J.; Capoduro, R.; Shelke, R.; Cali, K.; AlSaleh, M.A.; Persaud, K.; et al. Deorphanizing an Odorant Receptor Tuned to Palm Tree Volatile Esters in the Asian Palm Weevil Sheds Light on the Mechanisms of Palm Tree Selection. Insect Biochem. Mol. Biol. 2024, 169, 104129. [Google Scholar] [CrossRef]
- DeLeo, D.M.; Pérez-Moreno, J.L.; Vázquez-Miranda, H.; Bracken-Grissom, H.D. RNA Profile Diversity across Arthropoda: Guidelines, Methodological Artifacts, and Expected Outcomes. Biol. Methods Protoc. 2018, 3, bpy012. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT Online Service: Multiple Sequence Alignment, Interactive Sequence Choice and Visualization. Brief. Bioinform. 2018, 20, 1160–1166. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast Model Selection for Accurate Phylogenetic Estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Trifinopoulos, J.; Nguyen, L.T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A Fast Online Phylogenetic Tool for Maximum Likelihood Analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef]
- Dias, G.B.; Altammami, M.A.; El-Shafie, H.A.F.; Alhoshani, F.M.; Al-Fageeh, M.B.; Bergman, C.M.; Manee, M.M. Haplotype-Resolved Genome Assembly Enables Gene Discovery in the Red Palm Weevil Rhynchophorus ferrugineus. Sci. Rep. 2021, 11, 9987. [Google Scholar] [CrossRef]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate Structure Prediction of Biomolecular Interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef]
- Williams, C.J.; Headd, J.J.; Moriarty, N.W.; Prisant, M.G.; Videau, L.L.; Deis, L.N.; Verma, V.; Keedy, D.A.; Hintze, B.J.; Chen, V.B.; et al. MolProbity: More and Better Reference Data for Improved All-Atom Structure Validation. Protein Sci. 2018, 27, 293–315. [Google Scholar] [CrossRef]
- Schrödinger, L. The Pymol Molecular Graphics System Version 2.0. Available online: https://www.pubcompare.ai/product-search/ (accessed on 10 February 2023).
- Colovos, C.; Yeates, T.O. Verification of Protein Structures: Patterns of Nonbonded Atomic Interactions. Protein Sci. 1993, 2, 1511–1519. [Google Scholar] [CrossRef]
- Lüthy, R.; Bowie, J.U.; Eisenberg, D. Assessment of Protein Models with Three-Dimensional Profiles. Nature 1992, 356, 83–85. [Google Scholar] [CrossRef]
- Benkert, P.; Biasini, M.; Schwede, T. Toward the Estimation of the Absolute Quality of Individual Protein Structure Models. Bioinformatics 2011, 27, 343–350. [Google Scholar] [CrossRef]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent Updates, New Developments and Status in 2020. Nucleic Acids Res. 2021, 49, D458–D460. [Google Scholar] [CrossRef]
- Holm, L. Dali Server: Structural Unification of Protein Families. Nucleic Acids Res. 2022, 50, W210–W215. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera-A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Miyazaki, T.; Park, E.Y. Structure-Function Analysis of Silkworm Sucrose Hydrolase Uncovers the Mechanism of Substrate Specificity in GH13 Subfamily 17 Exo-a-Glucosidases. J. Biol. Chem. 2020, 295, 8784–8797. [Google Scholar] [CrossRef]
- Bruntz, R.C.; Lindsley, C.W.; Brown, H.A. Phospholipase D Signaling Pathways and Phosphatidic Acid as Therapeutic Targets in Cancer. Pharmacol. Rev. 2014, 66, 1033–1079. [Google Scholar] [CrossRef]
- Yang, X.; Li, Z.; Zhao, L.; She, Z.; Gao, Z.; Sui, S.F.; Dong, Y.; Li, Y. Structural Insights into PA3488-Mediated Inactivation of Pseudomonas aeruginosa PldA. Nat. Commun. 2022, 13, 5979. [Google Scholar] [CrossRef]
- Park, M.H.; Min, D.S. Quercetin-Induced Downregulation of Phospholipase D1 Inhibits Proliferation and Invasion in U87 Glioma Cells. Biochem. Biophys. Res. Commun. 2011, 412, 710–715. [Google Scholar] [CrossRef] [PubMed]
- May-Dracka, T.L.; Gao, F.; Hopkins, B.T.; Hronowski, X.; Chen, T.; Chodaparambil, J.V.; Metrick, C.M.; Cullivan, M.; Enyedy, I.; Kaliszczak, M.; et al. Discovery of Phospholipase D Inhibitors with Improved Drug-like Properties and Central Nervous System Penetrance. ACS Med. Chem. Lett. 2022, 13, 665–673. [Google Scholar] [CrossRef] [PubMed]
- Hánová, I.; Brynda, J.; Houštecká, R.; Alam, N.; Sojka, D.; Kopáček, P.; Marešová, L.; Vondrášek, J.; Horn, M.; Schueler-Furman, O.; et al. Novel Structural Mechanism of Allosteric Regulation of Aspartic Peptidases via an Evolutionarily Conserved Exosite. Cell Chem. Biol. 2018, 25, 318–329.e4. [Google Scholar] [CrossRef] [PubMed]
- Punia, A.; Singh Chauhan, N.; Kaur, S.; Kaur Sohal, S. Effect of Ellagic Acid on the Larvae of Spodoptera litura (Lepidoptera:Noctuidae) and Its Parasitoid Bracon hebetor (Hymenoptera:Braconidae). J. Asia Pac. Entomol. 2020, 23, 660–665. [Google Scholar] [CrossRef]
- Bothe, I.; Baylies, M.K.; Sloan, M.; Cancer, K. Primer: Drosophila Myogenesis. Curr. Biol. 2017, 26, R786–R791. [Google Scholar] [CrossRef]
- da Costa-Latgé, S.G.; Bates, P.; Dillon, R.; Genta, F.A. Characterization of Glycoside Hydrolase Families 13 and 31 Reveals Expansion and Diversification of α-Amylase Genes in the Phlebotomine Lutzomyia longipalpis and Modulation of Sandfly Glycosidase Activities by Leishmania Infection. Front. Physiol. 2021, 12, 635633. [Google Scholar] [CrossRef]
- CAZY. Carbohydrate-Binding Module Family Classification. Available online: https://www.cazy.org/ (accessed on 13 July 2023).
- Sin, J.; Andres, A.M.; Taylor, D.J.; Weston, T.; Hiraumi, Y.; Stotland, A.; Kim, B.J.; Huang, C.; Doran, K.S.; Gottlieb, R.A. Mitophagy Is Required for Mitochondrial Biogenesis and Myogenic Differentiation of C2C12 Myoblasts. Autophagy 2016, 12, 369–380. [Google Scholar] [CrossRef]
- Nicolas, G.; Pottier, C.; Charbonnier, C.; Guyant-Maréchal, L.; Le Ber, I.; Pariente, J.; Labauge, P.; Ayrignac, X.; Defebvre, L.; Maltête, D.; et al. Phenotypic Spectrum of Probable and Genetically-Confirmed Idiopathic Basal Ganglia Calcification. Brain 2013, 136, 3395–3407. [Google Scholar] [CrossRef]
- Meek, R.W.; Brockerman, J.; Fordwour, O.B.; Zandberg, W.F.; Davies, G.J.; Vocadlo, D.J. The Primary Familial Brain Calcification-Associated Protein MYORG Is an α-Galactosidase with Restricted Substrate Specificity. PLoS Biol. 2022, 20, e3001764. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, J.; Guo, H.; Zhao, A.; Shao, D.; Dong, Z.; Sun, Y.; Fan, Y.; Yang, F.; Li, P.; et al. Effects of Mulberry Leaf and White Kidney Bean Extract Mix on Postprandial Glycaemic Control in Pre-Diabetic Subjects Aged 45–65 Years: A Randomized Controlled Trial. J. Funct. Foods 2020, 73, 104117. [Google Scholar] [CrossRef]
- Konno, K.; Ono, H.; Nakamura, M.; Tateishi, K.; Hirayama, C.; Tamura, Y.; Hattori, M.; Koyama, A.; Kohno, K. Mulberry Latex Rich in Antidiabetic Sugar-Mimic Alkaloids Forces Dieting on Caterpillars. Proc. Natl. Acad. Sci. USA 2006, 103, 1337–1341. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Wheeler, M.M.; Oi, F.M.; Scharf, M.E. Inhibition of Termite Cellulases by Carbohydrate-Based Cellulase Inhibitors: Evidence from in Vitro Biochemistry and in Vivo Feeding Studies. Pestic. Biochem. Physiol. 2008, 90, 31–41. [Google Scholar] [CrossRef]
- Oguri, E.; Steele, J.E. Use of the Glucosidase Inhibitor 1,4-Dideoxy-1,4-Imino-D-Arabinitol as a Probe to Investigate Hypertrehalosemic Hormone-Mediated Change in Carbohydrate and Lipid Metabolism in the Cockroach, Periplaneta americana. Arch. Insect Biochem. Physiol. 2006, 63, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Cipolla, L.; Sgambato, A.; Forcella, M.; Fusi, P.; Parenti, P.; Cardona, F.; Bini, D. N-Bridged 1-Deoxynojirimycin Dimers as Selective Insect Trehalase Inhibitors. Carbohydr. Res. 2014, 389, 46–49. [Google Scholar] [CrossRef]
- Joensuu, M.; Wallis, T.P.; Saber, S.H.; Meunier, F.A. Phospholipases in Neuronal Function: A Role in Learning and Memory? J. Neurochem. 2020, 153, 300–333. [Google Scholar] [CrossRef]
- Burkhardt, U.; Stegner, D.; Hattingen, E.; Beyer, S.; Nieswandt, B.; Klein, J. Impaired Brain Development and Reduced Cognitive Function in Phospholipase D-Deficient Mice. Neurosci. Lett. 2014, 572, 48–52. [Google Scholar] [CrossRef]
- Foster, D.A.; Xu, L. Phospholipase D in Cell Proliferation and Cancer. Mol. Cancer Res. 2003, 1, 789–800. [Google Scholar]
- Richmond, G.S.; Smith, T.K. Phospholipases A 1. Int. J. Mol. Sci. 2011, 12, 588–612. [Google Scholar] [CrossRef]
- Herrera-Mayorga, V.; Guerrero-Sánchez, J.A.; Méndez-álvarez, D.; Paredes-Sánchez, F.A.; Rodríguez-Duran, L.V.; Niño-García, N.; Paz-González, A.D.; Rivera, G. Insecticidal Activity of Organic Extracts of Solidago graminifolia and Its Main Metabolites (Quercetin and Chlorogenic Acid) against Spodoptera frugiperda: An In Vitro and In Silico Approach. Molecules 2022, 27, 3325. [Google Scholar] [CrossRef]
- Riddick, E.W. Potential of Quercetin to Reduce Herbivory without Disrupting Natural Enemies and Pollinators. Agriculture 2021, 11, 476. [Google Scholar] [CrossRef]
- Nakagawa, Y.; Kuwano, E.; Fujita, T. Effects of Insect-Growth-Regulatory Benzimidazole Derivatives on Cultured Integument of the Rice Stem Borer and Mitochondria from Rat Liver. Agric. Biol. Chem. 1985, 49, 3569–3573. [Google Scholar] [CrossRef]
- Moreau, S.J.M.; Asgari, S. Venom Proteins from Parasitoid Wasps and Their Biological Functions. Toxins 2015, 7, 2385–2412. [Google Scholar] [CrossRef] [PubMed]
- Roh, K.B.; Kim, C.H.; Lee, H.; Kwon, H.M.; Park, J.W.; Ryu, J.H.; Kurokawa, K.; Ha, N.C.; Lee, W.J.; Lemaitre, B.; et al. Proteolytic Cascade for the Activation of the Insect Toll Pathway Induced by the Fungal Cell Wall Component. J. Biol. Chem. 2009, 284, 19474–19481. [Google Scholar] [CrossRef] [PubMed]
- Figueiredo, L.; Santos, R.B.; Figueiredo, A. Defense and Offense Strategies: The Role of Aspartic Proteases in Plant–Pathogen Interactions. Biology 2021, 10, 75. [Google Scholar] [CrossRef]
- Rabossi, A.; Stoka, V.; Puizdar, V.; Turk, V.; Quesada-Allué, L.A. Novel Aspartyl Proteinase Associated to Fat Body Histolysis during Ceratitis Capitata Early Metamorphosis. Arch. Insect Biochem. Physiol. 2004, 57, 51–67. [Google Scholar] [CrossRef]
- Zhong, Z.G.; Kwang, S.L.; Bo, Y.K.; Yong, S.C.; Ya, D.W.; Young, M.C.; Pil, D.K.; Hyung, J.Y.; Kim, I.; Yeon, H.J.; et al. Functional Role of Aspartic Proteinase Cathepsin D in Insect Metamorphosis. BMC Dev. Biol. 2006, 6, 49. [Google Scholar] [CrossRef]
- Yang, L.; Cheng, Y.; Wang, Q.; Hou, J.; Rong, Q.; Xiao, C.; Zhang, Y.; Yan, J.; Xia, Q.; Hou, Y. Insights into the Activation Mechanism of Bm-CPA: Implications for Insect Molting Regulation. Insect Biochem. Mol. Biol. 2024, 173, 104175. [Google Scholar] [CrossRef]
- Riddiford, L.M. Chapter 170-Molting. In Encyclopedia of Insects, 2nd ed.; Resh, V.H., Cardé, R.T., Eds.; Academic Press: San Diego, CA, USA, 2009; pp. 649–654. ISBN 978-0-12-374144-8. [Google Scholar]
- Umezawa, H.; Aoyagi, T.; Morishima, H.; Matsuzaki, M.; Hamada, M.; Takeuchi, T. Pepstatin, a New Pepsin Inhibitor Produced by Actinomycetes. J. Antibiot. 1970, 23, 259–262. [Google Scholar] [CrossRef]
- Marks, N.; Grynbaum, A.; Lajtha, A. Pentapeptide (Pepstatin) Inhibition of Brain Acid Proteinase. Science 1973, 181, 949–951. [Google Scholar] [CrossRef]
- Maynadier, M.; Vezenkov, L.L.; Amblard, M.; Martin, V.; Gandreuil, C.; Vaillant, O.; Gary-Bobo, M.; Basile, I.; Hernandez, J.-F.; Garcia, M.; et al. Dipeptide Mimic Oligomer Transporter Mediates Intracellular Delivery of Cathepsin D Inhibitors: A Potential Target for Cancer Therapy. J. Control Release 2013, 171, 251–257. [Google Scholar] [CrossRef]
- Matúz, K.; Mótyán, J.; Li, M.; Wlodawer, A.; Tőzsér, J. Inhibition of XMRV and HIV-1 Proteases by Pepstatin A and Acetyl-pepstatin. FEBS J. 2012, 279, 3276–3286. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.E.; Zhu-Salzman, K. CmCatD, a Cathepsin D-like Protease Has a Potential Role in Insect Defense against a Phytocystatin. J. Insect Physiol. 2009, 55, 678–685. [Google Scholar] [CrossRef] [PubMed]
- Landete, J.M. Ellagitannins, Ellagic Acid and Their Derived Metabolites: A Review about Source, Metabolism, Functions and Health. Food Res. Int. 2011, 44, 1150–1160. [Google Scholar] [CrossRef]
- Zhang, H.M.; Zhao, L.; Li, H.; Xu, H.; Chen, W.W.; Tao, L. Research Progress on the Anticarcinogenic Actions and Mechanisms of Ellagic Acid. Cancer Biol. Med. 2014, 11, 92–100. [Google Scholar] [CrossRef] [PubMed]
- De, R.; Sarkar, A.; Ghosh, P.; Ganguly, M.; Karmakar, B.C.; Saha, D.R.; Halder, A.; Chowdhury, A.; Mukhopadhyay, A.K. Antimicrobial Activity of Ellagic Acid against Helicobacter pylori Isolates from India and during Infections in Mice. J. Antimicrob. Chemother. 2018, 73, 1595–1603. [Google Scholar] [CrossRef]
- Busch, A.; Danchin, E.G.J.; Pauchet, Y. Functional Diversification of Horizontally Acquired Glycoside Hydrolase Family 45 (GH45) Proteins in Phytophaga Beetles. BMC Evol. Biol. 2019, 19, 100. [Google Scholar] [CrossRef]
- Kirsch, R.; Okamura, Y.; García-Lozano, M.; Weiss, B.; Keller, J.; Vogel, H.; Fukumori, K.; Fukatsu, T.; Konstantinov, A.S.; Montagna, M.; et al. Symbiosis and Horizontal Gene Transfer Promote Herbivory in the Megadiverse Leaf Beetles. Curr. Biol. 2025, 35, 640–654. [Google Scholar] [CrossRef]
- Lu, H.; Lu, J.; Li, L.; Zhang, Z.; Chen, J.; Li, J.; Zhang, C.; Huang, H. Functional Analysis of Neutral Lipases in Bug Feeding and Reproduction. Pest Manag. Sci. 2023, 79, 4809–4818. [Google Scholar] [CrossRef]

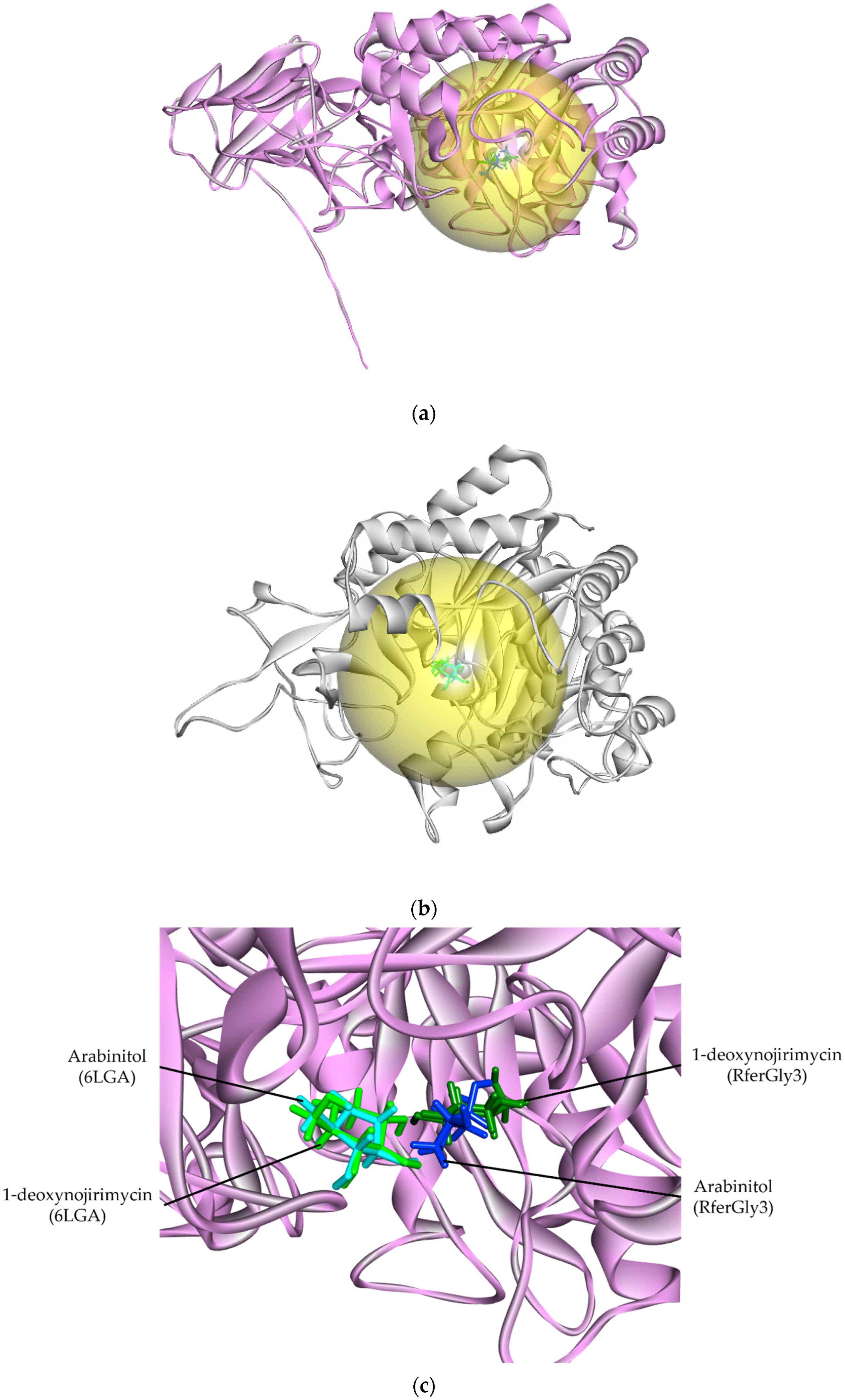

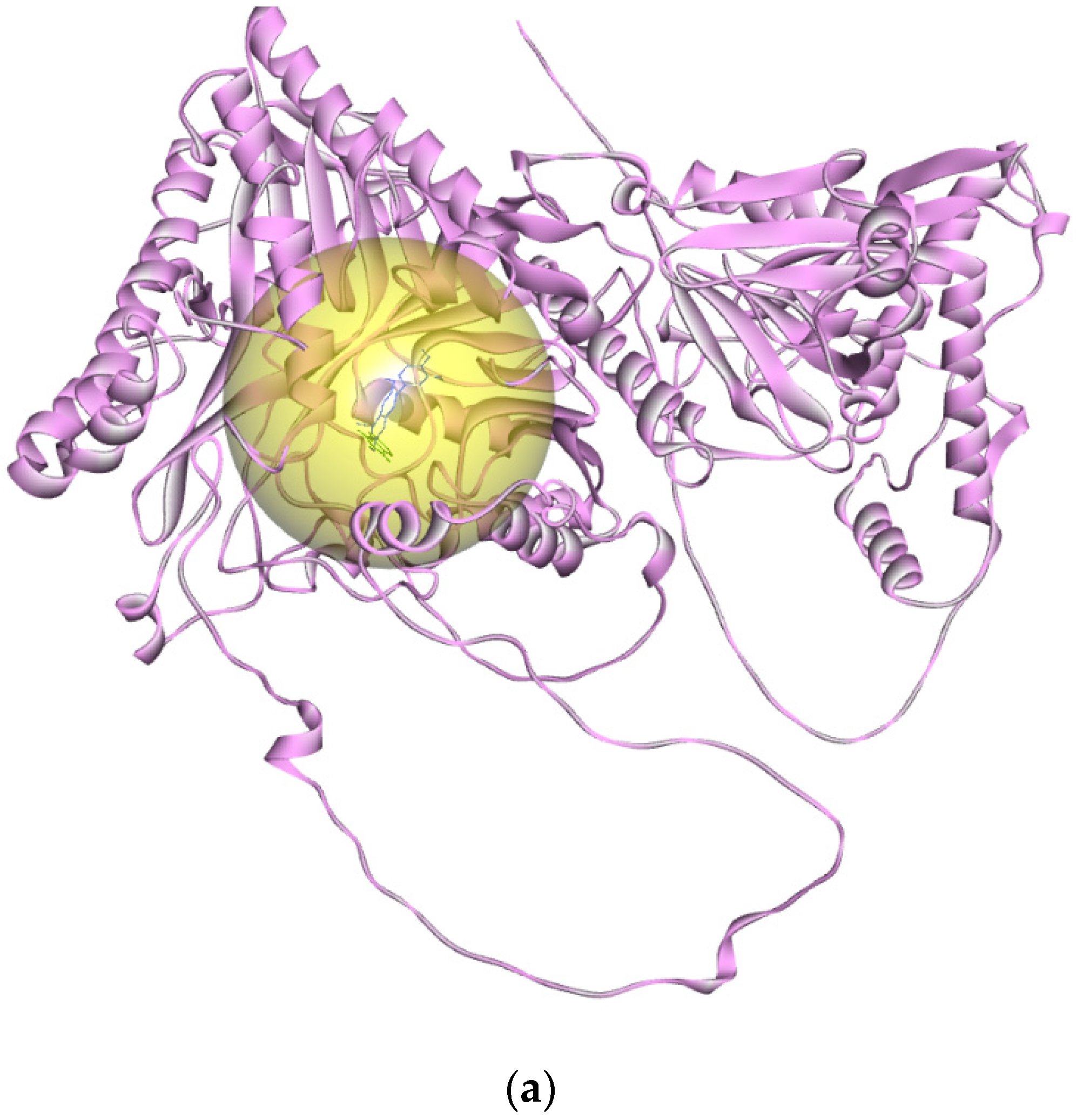


| Digestive Enzyme | pTM | ERRAT | Verify3D (%) | Ramachandran | MolProbity | QMean Disco Global | Root Mean Square Difference (Å) | |
|---|---|---|---|---|---|---|---|---|
| Glycosidase | RferGly3 | 0.94 | 98.36 | 88.15 | 94.77% favorable | 1.67 | 0.68 | 1.16 |
| 0.16% outliers | ||||||||
| 6LGA (reference) | n/a | 96.58 | 96.11 | 97.69% favorable | 0.99 | 0.94 | ||
| 0% outliers | ||||||||
| Lipase | RferLip3 | 0.82 | 91.56 | 73.35 | 93.88% favorable | 1.73 | 0.6 | 2.2 |
| 0.81% outliers | ||||||||
| 7V55 (reference) | n/a | 94.55 | 84.44 | 96.35% favorable | 1.45 | 0.9 | ||
| 0% outliers | ||||||||
| Protease | RferPro27 | 0.85 | 88.5196 | 76.42 | 95.64% favorable | 1.85 | 0.75 | 0.187 |
| 0.27% outliers | ||||||||
| 5N7Q (reference) | n/a | 90.7051 | 96.14 | 98.51% favorable | 0.97 | 0.92 | ||
| 0.30% outliers | ||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Harith-Fadzilah, N.; Nihad, M.; AlSaleh, M.A.; Bazeyad, A.Y.; Pandurangan, S.-B.; Munawar, K.; Vidyawan, A.; Alharbi, H.A.; Jakše, J.; Pain, A.; et al. Genome-Wide Identification and Expression Profiling of Glycosidases, Lipases, and Proteases from Invasive Asian Palm Weevil, Rhynchophorus ferrugineus. Insects 2025, 16, 421. https://doi.org/10.3390/insects16040421
Harith-Fadzilah N, Nihad M, AlSaleh MA, Bazeyad AY, Pandurangan S-B, Munawar K, Vidyawan A, Alharbi HA, Jakše J, Pain A, et al. Genome-Wide Identification and Expression Profiling of Glycosidases, Lipases, and Proteases from Invasive Asian Palm Weevil, Rhynchophorus ferrugineus. Insects. 2025; 16(4):421. https://doi.org/10.3390/insects16040421
Chicago/Turabian StyleHarith-Fadzilah, Nazmi, Mohammad Nihad, Mohammed Ali AlSaleh, Abdulqader Yaslam Bazeyad, Subash-Babu Pandurangan, Kashif Munawar, Arya Vidyawan, Hattan A. Alharbi, Jernej Jakše, Arnab Pain, and et al. 2025. "Genome-Wide Identification and Expression Profiling of Glycosidases, Lipases, and Proteases from Invasive Asian Palm Weevil, Rhynchophorus ferrugineus" Insects 16, no. 4: 421. https://doi.org/10.3390/insects16040421
APA StyleHarith-Fadzilah, N., Nihad, M., AlSaleh, M. A., Bazeyad, A. Y., Pandurangan, S.-B., Munawar, K., Vidyawan, A., Alharbi, H. A., Jakše, J., Pain, A., & Antony, B. (2025). Genome-Wide Identification and Expression Profiling of Glycosidases, Lipases, and Proteases from Invasive Asian Palm Weevil, Rhynchophorus ferrugineus. Insects, 16(4), 421. https://doi.org/10.3390/insects16040421









