Oncogenic Roles of UHRF1 in Cancer
Abstract
:1. Introduction
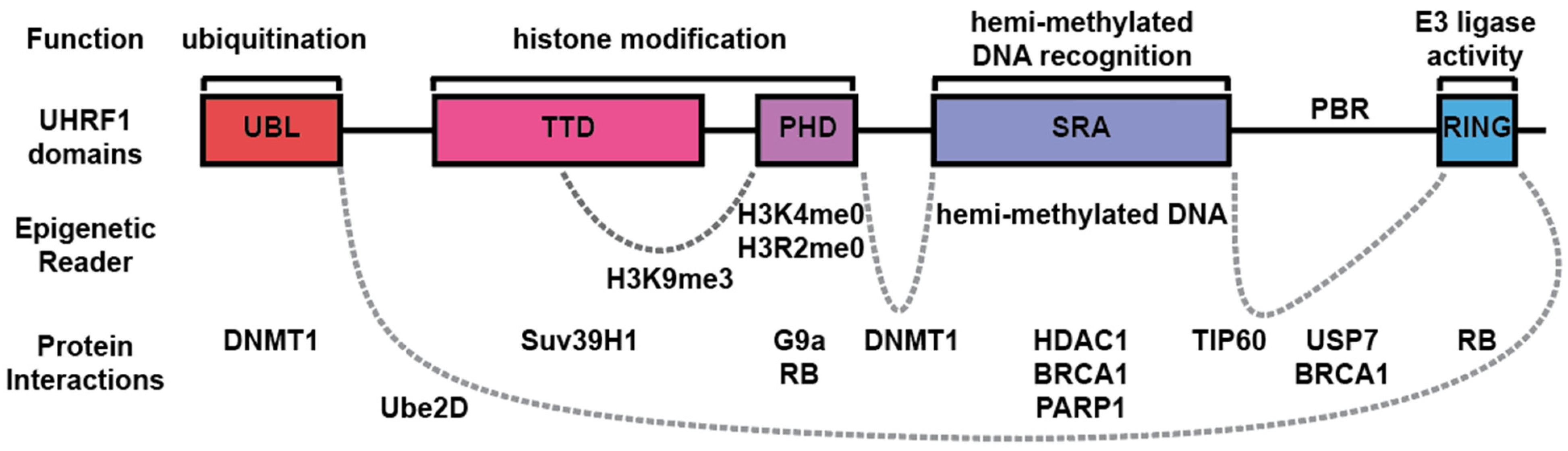
2. UHRF1 Functional Domains
2.1. UBL Domain
2.2. TTD and PHD
2.3. SRA Domain
2.4. RING Domain
3. UHRF1 Functional Protein Interactions
3.1. DNMT1
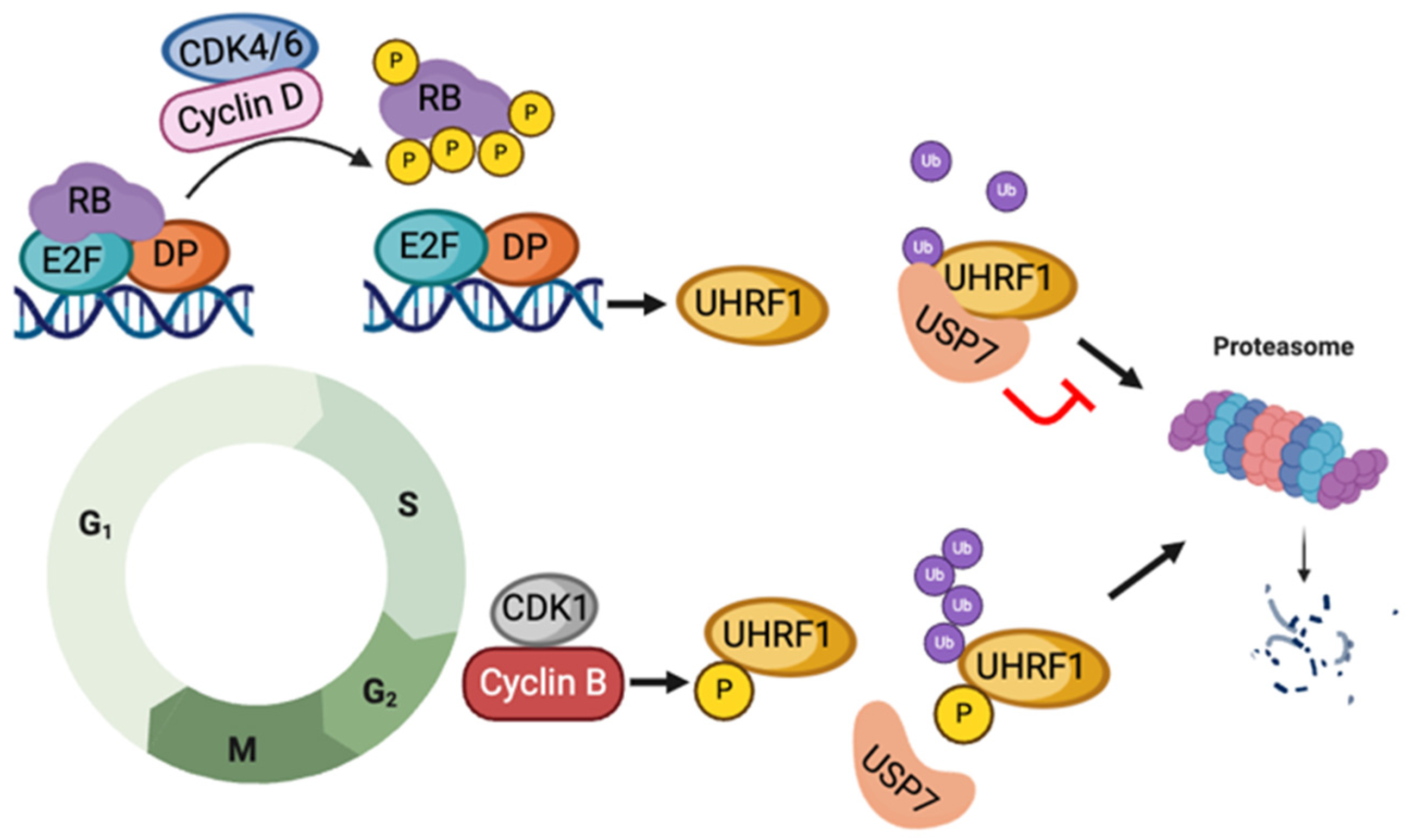
3.2. USP7
3.3. HDAC1
3.4. TIP60
3.5. BRCA1
3.6. PARP1
4. UHRF1 in Cancers
4.1. Sustaining Proliferation and Tumor Growth
4.2. Resisting Cell Death
4.3. Inducing Angiogenesis and Metastasis
4.4. Increasing Chemoresistance
4.5. Epigenetic Silencing
5. Current Potential UHRF1 Therapeutics
5.1. Targeting UHRF1 Domains
5.2. Alternative Therapeutic Approaches
6. Conclusions and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ashraf, W.; Ibrahim, A.; Alhosin, M.; Zaayter, L.; Ouararhni, K.; Papin, C.; Ahmad, T.; Hamiche, A.; Mely, Y.; Bronner, C.; et al. The Epigenetic Integrator UHRF1: On the Road to Become a Universal Biomarker for Cancer. Oncotarget 2017, 8, 51946–51962. [Google Scholar] [CrossRef]
- Hashimoto, H.; Horton, J.R.; Zhang, X.; Cheng, X. UHRF1, a Modular Multi-Domain Protein, Regulates Replication-Coupled Crosstalk between DNA Methylation and Histone Modifications. Epigenetics 2009, 4, 8–14. [Google Scholar] [CrossRef]
- Rottach, A.; Frauer, C.; Pichler, G.; Bonapace, I.M.; Spada, F.; Leonhardt, H. The Multi-Domain Protein Np95 Connects DNA Methylation and Histone Modification. Nucleic Acids Res. 2010, 38, 1796–1804. [Google Scholar] [CrossRef]
- Du, J.; Johnson, L.M.; Jacobsen, S.E.; Patel, D.J. DNA Methylation Pathways and Their Crosstalk with Histone Methylation. Nat. Rev. Mol. Cell Biol. 2015, 16, 519–532. [Google Scholar] [CrossRef]
- Fang, J.; Cheng, J.; Wang, J.; Zhang, Q.; Liu, M.; Gong, R.; Wang, P.; Zhang, X.; Feng, Y.; Lan, W.; et al. Hemi-Methylated DNA Opens a Closed Conformation of UHRF1 to Facilitate Its Histone Recognition. Nat. Commun. 2016, 7, 11197. [Google Scholar] [CrossRef]
- Li, T.; Wang, L.; Du, Y.; Xie, S.; Yang, X.; Lian, F.; Zhou, Z.; Qian, C. Structural and Mechanistic Insights into UHRF1-Mediated DNMT1 Activation in the Maintenance DNA Methylation. Nucleic Acids Res. 2018, 46, 3218–3231. [Google Scholar] [CrossRef]
- Foster, B.M.; Stolz, P.; Mulholland, C.B.; Montoya, A.; Kramer, H.; Bultmann, S.; Bartke, T. Critical Role of the UBL Domain in Stimulating the E3 Ubiquitin Ligase Activity of UHRF1 toward Chromatin. Mol. Cell 2018, 72, 739–752.e9. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Yang, Y.; Fang, J.; Xiao, J.; Zhu, T.; Chen, F.; Wang, P.; Li, Z.; Yang, H.; Xu, Y. Structural Insight into Coordinated Recognition of Trimethylated Histone H3 Lysine 9 (H3K9me3) by the Plant Homeodomain (PHD) and Tandem Tudor Domain (TTD) of UHRF1 (Ubiquitin-like, Containing PHD and RING Finger Domains, 1) Protein. J. Biol. Chem. 2013, 288, 1329–1339. [Google Scholar] [CrossRef] [PubMed]
- Rothbart, S.B.; Dickson, B.M.; Ong, M.S.; Krajewski, K.; Houliston, S.; Kireev, D.B.; Arrowsmith, C.H.; Strahl, B.D. Multivalent Histone Engagement by the Linked Tandem Tudor and PHD Domains of UHRF1 Is Required for the Epigenetic Inheritance of DNA Methylation. Genes. Dev. 2013, 27, 1288–1298. [Google Scholar] [CrossRef]
- Unoki, M.; Nishidate, T.; Nakamura, Y. ICBP90, an E2F-1 Target, Recruits HDAC1 and Binds to Methyl-CpG through Its SRA Domain. Oncogene 2004, 23, 7601–7610. [Google Scholar] [CrossRef]
- Bostick, M.; Kim, J.K.; Esteve, P.O.; Clark, A.; Pradhan, S.; Jacobsen, S.E. UHRF1 Plays a Role in Maintaining DNA Methylation in Mammalian Cells. Science 2007, 317, 1760–1764. [Google Scholar] [CrossRef] [PubMed]
- DaRosa, P.A.; Harrison, J.S.; Zelter, A.; Davis, T.N.; Brzovic, P.; Kuhlman, B.; Klevit, R.E. A Bifunctional Role for the UHRF1 UBL Domain in the Control of Hemi-Methylated DNA-Dependent Histone Ubiquitylation. Mol. Cell 2018, 72, 753–765.e6. [Google Scholar] [CrossRef]
- Benavente, C.A.; Finkelstein, D.; Johnson, D.A.; Marine, J.-C.; Ashery-Padan, R.; Dyer, M.A. Chromatin Remodelers HELLS and UHRF1 Mediate the Epigenetic Deregulation of Genes That Drive Retinoblastoma Tumor Progression. Oncotarget 2014, 5, 9594–9608. [Google Scholar] [CrossRef]
- Wu, S.C.; Kim, A.; Gu, Y.; Martinez, D.I.; Zocchi, L.; Chen, C.C.; Lopez, J.; Salcido, K.; Singh, S.; Wu, J.; et al. UHRF1 Overexpression Promotes Osteosarcoma Metastasis through Altered Exosome Production and AMPK/SEMA3E Suppression. Oncogenesis 2022, 11, 51. [Google Scholar] [CrossRef]
- Ma, H.; Chen, H.; Guo, X.; Wang, Z.; Sowa, M.E.; Zheng, L.; Hu, S.; Zeng, P.; Guo, R.; Diao, J.; et al. M Phase Phosphorylation of the Epigenetic Regulator UHRF1 Regulates Its Physical Association with the Deubiquitylase USP7 and Stability. Proc. Natl. Acad. Sci. USA 2012, 109, 4828–4833. [Google Scholar] [CrossRef]
- Zhang, Z.-M.; Rothbart, S.B.; Allison, D.F.; Cai, Q.; Harrison, J.S.; Li, L.; Wang, Y.; Strahl, B.D.; Wang, G.G.; Song, J. An Allosteric Interaction Links USP7 to Deubiquitination and Chromatin Targeting of UHRF1. Cell Rep. 2015, 12, 1400–1406. [Google Scholar] [CrossRef]
- Mousli, M.; Hopfner, R.; Abbady, A.Q.; Monte, D.; Jeanblanc, M.; Oudet, P.; Louis, B.; Bronner, C. ICBP90 Belongs to a New Family of Proteins with an Expression That Is Deregulated in Cancer Cells. Br. J. Cancer 2003, 89, 120–127. [Google Scholar] [CrossRef]
- Liu, Y.; Liang, G.; Zhou, T.; Liu, Z. Silencing UHRF1 Inhibits Cell Proliferation and Promotes Cell Apoptosis in Retinoblastoma Via the PI3K/Akt Signalling Pathway. Pathol. Oncol. Res. 2020, 26, 1079–1088. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Qiao, R.H.; Wang, D.M.; Huang, X.W.; Li, B.; Wang, D. UHRF1 Promotes Human Osteosarcoma Cell Invasion by Downregulating the Expression of E-cadherin in an Rb1-dependent Manner. Mol. Med. Rep. 2016, 13, 315–320. [Google Scholar] [CrossRef]
- Li, X.-L.; Xu, J.-H.; Nie, J.-H.; Fan, S.-J. Exogenous Expression of UHRF1 Promotes Proliferation and Metastasis of Breast Cancer Cells. Oncol. Rep. 2012, 28, 375–383. [Google Scholar] [CrossRef]
- Gao, S.-P.; Sun, H.-F.; Li, L.-D.; Fu, W.-Y.; Jin, W. UHRF1 Promotes Breast Cancer Progression by Suppressing KLF17 Expression by Hypermethylating Its Promoter. Am. J. Cancer Res. 2017, 7, 1554–1565. [Google Scholar]
- Unoki, M.; Daigo, Y.; Koinuma, J.; Tsuchiya, E.; Hamamoto, R.; Nakamura, Y. UHRF1 Is a Novel Diagnostic Marker of Lung Cancer. Br. J. Cancer 2010, 103, 217–222. [Google Scholar] [CrossRef]
- Cui, L.; Chen, J.; Zhang, Q.; Wang, X.; Qu, J.; Zhang, J.; Dang, S. Up-Regulation of UHRF1 by Oncogenic Ras Promoted the Growth, Migration, and Metastasis of Pancreatic Cancer Cells. Mol. Cell Biochem. 2015, 400, 223–232. [Google Scholar] [CrossRef]
- Hu, Q.; Qin, Y.; Ji, S.; Xu, W.; Liu, W.; Sun, Q.; Zhang, Z.; Liu, M.; Ni, Q.; Yu, X.; et al. UHRF1 Promotes Aerobic Glycolysis and Proliferation via Suppression of SIRT4 in Pancreatic Cancer. Cancer Lett. 2019, 452, 226–236. [Google Scholar] [CrossRef] [PubMed]
- Beck, A.; Trippel, F.; Wagner, A.; Joppien, S.; Felle, M.; Vokuhl, C.; Schwarzmayr, T.; Strom, T.M.; von Schweinitz, D.; Längst, G.; et al. Overexpression of UHRF1 Promotes Silencing of Tumor Suppressor Genes and Predicts Outcome in Hepatoblastoma. Clin. Epigenetics 2018, 10, 27. [Google Scholar] [CrossRef]
- Zhou, L.; Shang, Y.; Jin, Z.; Zhang, W.; Lv, C.; Zhao, X.; Liu, Y.; Li, N.; Liang, J. UHRF1 Promotes Proliferation of Gastric Cancer via Mediating Tumor Suppressor Gene Hypermethylation. Cancer Biol. Ther. 2015, 16, 1241–1251. [Google Scholar] [CrossRef] [PubMed]
- Mudbhary, R.; Hoshida, Y.; Chernyavskaya, Y.; Jacob, V.; Villanueva, A.; Fiel, M.I.; Chen, X.; Kojima, K.; Thung, S.; Bronson, R.T.; et al. UHRF1 Overexpression Drives DNA Hypomethylation and Hepatocellular Carcinoma. Cancer Cell 2014, 25, 196–209. [Google Scholar] [CrossRef]
- Xie, S.; Qian, C. The Growing Complexity of UHRF1-Mediated Maintenance DNA Methylation. Genes 2018, 9, 600. [Google Scholar] [CrossRef]
- Hu, L.; Li, Z.; Wang, P.; Lin, Y.; Xu, Y. Crystal Structure of PHD Domain of UHRF1 and Insights into Recognition of Unmodified Histone H3 Arginine Residue 2. Cell Res. 2011, 21, 1374–1378. [Google Scholar] [CrossRef]
- Nady, N.; Lemak, A.; Walker, J.R.; Avvakumov, G.V.; Kareta, M.S.; Achour, M.; Xue, S.; Duan, S.; Allali-Hassani, A.; Zuo, X.; et al. Recognition of Multivalent Histone States Associated with Heterochromatin by UHRF1 Protein. J. Biol. Chem. 2011, 286, 24300–24311. [Google Scholar] [CrossRef]
- Alhosin, M.; Omran, Z.; Zamzami, M.A.; Al-Malki, A.L.; Choudhry, H.; Mousli, M.; Bronner, C. Signalling Pathways in UHRF1-Dependent Regulation of Tumor Suppressor Genes in Cancer. J. Exp. Clin. Cancer Res. 2016, 35, 174. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Tan, X.-F.; Zhang, S.; Wu, T.; Zhang, Z.-M.; Ai, H.; Song, J. An Intramolecular Interaction of UHRF1 Reveals Dual Control for Its Histone Association. Structure 2018, 26, 304–311.e3. [Google Scholar] [CrossRef] [PubMed]
- Avvakumov, G.V.; Walker, J.R.; Xue, S.; Li, Y.; Duan, S.; Bronner, C.; Arrowsmith, C.H.; Dhe-Paganon, S. Structural Basis for Recognition of Hemi-Methylated DNA by the SRA Domain of Human UHRF1. Nature 2008, 455, 822–825. [Google Scholar] [CrossRef] [PubMed]
- Arita, K.; Ariyoshi, M.; Tochio, H.; Nakamura, Y.; Shirakawa, M. Recognition of Hemi-Methylated DNA by the SRA Protein UHRF1 by a Base-Flipping Mechanism. Nature 2008, 455, 818–821. [Google Scholar] [CrossRef] [PubMed]
- Qian, C.; Li, S.; Jakoncic, J.; Zeng, L.; Walsh, M.J.; Zhou, M.-M. Structure and Hemimethylated CpG Binding of the SRA Domain from Human UHRF1. J. Biol. Chem. 2008, 283, 34490–34494. [Google Scholar] [CrossRef] [PubMed]
- Bashtrykov, P.; Jankevicius, G.; Jurkowska, R.Z.; Ragozin, S.; Jeltsch, A. The UHRF1 Protein Stimulates the Activity and Specificity of the Maintenance DNA Methyltransferase DNMT1 by an Allosteric Mechanism*. J. Biol. Chem. 2014, 289, 4106–4115. [Google Scholar] [CrossRef] [PubMed]
- Berkyurek, A.C.; Suetake, I.; Arita, K.; Takeshita, K.; Nakagawa, A.; Shirakawa, M.; Tajima, S. The DNA Methyltransferase Dnmt1 Directly Interacts with the SET and RING Finger-Associated (SRA) Domain of the Multifunctional Protein Uhrf1 to Facilitate Accession of the Catalytic Center to Hemi-Methylated DNA. J. Biol. Chem. 2014, 289, 379–386. [Google Scholar] [CrossRef] [PubMed]
- Nishiyama, A.; Yamaguchi, L.; Sharif, J.; Johmura, Y.; Kawamura, T.; Nakanishi, K.; Shimamura, S.; Arita, K.; Kodama, T.; Ishikawa, F.; et al. Uhrf1-Dependent H3K23 Ubiquitylation Couples Maintenance DNA Methylation and Replication. Nature 2013, 502, 249–253. [Google Scholar] [CrossRef] [PubMed]
- Qin, W.; Wolf, P.; Liu, N.; Link, S.; Smets, M.; Mastra, F.L.; Forné, I.; Pichler, G.; Hörl, D.; Fellinger, K.; et al. DNA Methylation Requires a DNMT1 Ubiquitin Interacting Motif (UIM) and Histone Ubiquitination. Cell Res. 2015, 25, 911–929. [Google Scholar] [CrossRef]
- Ibrahim, A.; Alhosin, M.; Papin, C.; Ouararhni, K.; Omran, Z.; Zamzami, M.A.; Al-Malki, A.L.; Choudhry, H.; Mély, Y.; Hamiche, A.; et al. Thymoquinone Challenges UHRF1 to Commit Auto-Ubiquitination: A Key Event for Apoptosis Induction in Cancer Cells. Oncotarget 2018, 9, 28599–28611. [Google Scholar] [CrossRef]
- Jenkins, Y.; Markovtsov, V.; Lang, W.; Sharma, P.; Pearsall, D.; Warner, J.; Franci, C.; Huang, B.; Huang, J.; Yam, G.C.; et al. Critical Role of the Ubiquitin Ligase Activity of UHRF1, a Nuclear RING Finger Protein, in Tumor Cell Growth. MBoC 2005, 16, 5621–5629. [Google Scholar] [CrossRef] [PubMed]
- Harrison, J.S.; Cornett, E.M.; Goldfarb, D.; DaRosa, P.A.; Li, Z.M.; Yan, F.; Dickson, B.M.; Guo, A.H.; Cantu, D.V.; Kaustov, L.; et al. Hemi-Methylated DNA Regulates DNA Methylation Inheritance through Allosteric Activation of H3 Ubiquitylation by UHRF1. eLife 2016, 5, e17101. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Gao, Q.; Li, P.; Zhao, Q.; Zhang, J.; Li, J.; Koseki, H.; Wong, J. UHRF1 Targets DNMT1 for DNA Methylation through Cooperative Binding of Hemi-Methylated DNA and Methylated H3K9. Nat. Commun. 2013, 4, 1563. [Google Scholar] [CrossRef] [PubMed]
- Qin, W.; Leonhardt, H.; Spada, F. Usp7 and Uhrf1 Control Ubiquitination and Stability of the Maintenance DNA Methyltransferase Dnmt1. J. Cell. Biochem. 2011, 112, 439–444. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, R.; Jin, J.; Han, M.; Chen, Z.; Gao, Y.; Hu, X.; Zhu, H.; Gao, H.; Lu, K.; et al. USP7 Negatively Controls Global DNA Methylation by Attenuating Ubiquitinated Histone-Dependent DNMT1 Recruitment. Cell Discov. 2020, 6, 58. [Google Scholar] [CrossRef] [PubMed]
- Hervouet, E.; Peixoto, P.; Delage-Mourroux, R.; Boyer-Guittaut, M.; Cartron, P.-F. Specific or Not Specific Recruitment of DNMTs for DNA Methylation, an Epigenetic Dilemma. Clin. Epigenetics 2018, 10, 17. [Google Scholar] [CrossRef] [PubMed]
- Niinuma, T.; Kitajima, H.; Kai, M.; Yamamoto, E.; Yorozu, A.; Ishiguro, K.; Sasaki, H.; Sudo, G.; Toyota, M.; Hatahira, T.; et al. UHRF1 Depletion and HDAC Inhibition Reactivate Epigenetically Silenced Genes in Colorectal Cancer Cells. Clin. Epigenetics 2019, 11, 70. [Google Scholar] [CrossRef] [PubMed]
- Squatrito, M.; Gorrini, C.; Amati, B. Tip60 in DNA Damage Response and Growth Control: Many Tricks in One HAT. Trends Cell Biol. 2006, 16, 433–442. [Google Scholar] [CrossRef] [PubMed]
- Ikura, T.; Ogryzko, V.V.; Grigoriev, M.; Groisman, R.; Wang, J.; Horikoshi, M.; Scully, R.; Qin, J.; Nakatani, Y. Involvement of the TIP60 Histone Acetylase Complex in DNA Repair and Apoptosis. Cell 2000, 102, 463–473. [Google Scholar] [CrossRef]
- Achour, M.; Fuhrmann, G.; Alhosin, M.; Rondé, P.; Chataigneau, T.; Mousli, M.; Schini-Kerth, V.B.; Bronner, C. UHRF1 Recruits the Histone Acetyltransferase Tip60 and Controls Its Expression and Activity. Biochem. Biophys. Res. Commun. 2009, 390, 523–528. [Google Scholar] [CrossRef]
- Ashraf, W.; Bronner, C.; Zaayter, L.; Ahmad, T.; Richert, L.; Alhosin, M.; Ibrahim, A.; Hamiche, A.; Mely, Y.; Mousli, M. Interaction of the Epigenetic Integrator UHRF1 with the MYST Domain of TIP60 inside the Cell. J. Exp. Clin. Cancer Res. 2017, 36, 188. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, T.; Ashraf, W.; Ibrahim, A.; Zaayter, L.; Muller, C.D.; Hamiche, A.; Mély, Y.; Bronner, C.; Mousli, M. TIP60 Governs the Auto-ubiquitination of UHRF1 through USP7 Dissociation from the UHRF1/USP7 Complex. Int. J. Oncol. 2021, 59, 89. [Google Scholar] [CrossRef] [PubMed]
- Haber, J.E. Partners and Pathways: Repairing a Double-Strand Break. Trends Genet. 2000, 16, 259–264. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, H.; Chen, Y.; Yang, X.; Wang, P.; Liu, T.; Deng, M.; Qin, B.; Correia, C.; Lee, S.; et al. A Cell Cycle-Dependent BRCA1–UHRF1 Cascade Regulates DNA Double-Strand Break Repair Pathway Choice. Nat. Commun. 2016, 7, 10201. [Google Scholar] [CrossRef]
- Jin, W.; Chen, L.; Chen, Y.; Xu, S.; Di, G.; Yin, W.; Wu, J.; Shao, Z. UHRF1 Is Associated with Epigenetic Silencing of BRCA1 in Sporadic Breast Cancer. Breast Cancer Res. Treat. 2010, 123, 359–373. [Google Scholar] [CrossRef]
- Vos, M.D.; Ramy, R.E.; Quénet, D.; Wolf, P.; Spada, F.; Magroun, N.; Babbio, F.; Schreiber, V.; Leonhardt, H.; Bonapace, I.M.; et al. Poly(ADP-Ribose) Polymerase 1 (PARP1) Associates with E3 Ubiquitin-Protein Ligase UHRF1 and Modulates UHRF1 Biological Functions. J. Biol. Chem. 2014, 289, 16223–16238. [Google Scholar] [CrossRef] [PubMed]
- Hahm, J.Y.; Kang, J.-Y.; Park, J.W.; Jung, H.; Seo, S.-B. Methylated-UHRF1 and PARP1 Interaction Is Critical for Homologous Recombination. BMB Rep. 2020, 53, 112–117. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Petit, S.A.; Ficarro, S.B.; Toomire, K.J.; Xie, A.; Lim, E.; Cao, S.A.; Park, E.; Eck, M.J.; Scully, R.; et al. PARP1-Driven Poly-ADP-Ribosylation Regulates BRCA1 Function in Homologous Recombination Mediated DNA Repair. Cancer Discov. 2014, 4, 1430–1447. [Google Scholar] [CrossRef]
- Role of BRCA Mutations in Cancer Treatment with Poly(ADP-Ribose) Polymerase (PARP) Inhibitors—PMC. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6316750/ (accessed on 27 February 2024).
- Wang, F.; Yang, Y.-Z.; Shi, C.-Z.; Zhang, P.; Moyer, M.P.; Zhang, H.-Z.; Zou, Y.; Qin, H.-L. UHRF1 Promotes Cell Growth and Metastasis Through Repression of P16ink4a in Colorectal Cancer. Ann. Surg. Oncol. 2012, 19, 2753–2762. [Google Scholar] [CrossRef]
- Zhang, Z.-Y.; Cai, J.-J.; Hong, J.; Li, K.K.-W.; Ping, Z.; Wang, Y.; Ng, H.-K.; Yao, Y.; Mao, Y. Clinicopathological Analysis of UHRF1 Expression in Medulloblastoma Tissues and Its Regulation on Tumor Cell Proliferation. Med. Oncol. 2016, 33, 99. [Google Scholar] [CrossRef]
- Hou, J.; Li, W.; Zhang, S.; Tan, D.; Lv, K.; Zhu, Y.; Hou, Y.; Guo, H.; Jiang, L. UHRF1 Plays an Oncogenic Role in Small Cell Lung Cancer. Mol. Carcinog. 2023, 62, 385–397. [Google Scholar] [CrossRef]
- Wang, H.; Cao, D.; Wu, F. Long Noncoding RNA UPAT Promoted Cell Proliferation via Increasing UHRF1 Expression in Non-Small Cell Lung Cancer. Oncol. Lett. 2018, 16, 1491–1498. [Google Scholar] [CrossRef]
- Wei, C.; Lu, N.; Wang, L.; Zhang, Y.; Feng, Z.; Yang, Y.; Qi, F.; Gu, J. Upregulation of UHRF1 Promotes the Progression of Melanoma by Inducing Cell Proliferation. Oncol. Rep. 2018, 39, 2553–2562. [Google Scholar] [CrossRef]
- Kostyrko, K.; Román, M.; Lee, A.G.; Simpson, D.R.; Dinh, P.T.; Leung, S.G.; Marini, K.D.; Kelly, M.R.; Broyde, J.; Califano, A.; et al. UHRF1 Is a Mediator of KRAS Driven Oncogenesis in Lung Adenocarcinoma. Nat. Commun. 2023, 14, 3966. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Lee, C.; Kim, J.K. UHRF1 Depletion Sensitizes Retinoblastoma Cells to Chemotherapeutic Drugs via Downregulation of XRCC4. Cell Death Dis. 2018, 9, 164. [Google Scholar] [CrossRef]
- Liu, X.; Ou, H.; Xiang, L.; Li, X.; Huang, Y.; Yang, D. Elevated UHRF1 Expression Contributes to Poor Prognosis by Promoting Cell Proliferation and Metastasis in Hepatocellular Carcinoma. Oncotarget 2017, 8, 10510–10522. [Google Scholar] [CrossRef] [PubMed]
- Tian, D.; Tang, J.; Geng, X.; Li, Q.; Wang, F.; Zhao, H.; Narla, G.; Yao, X.; Zhang, Y. Targeting UHRF1-Dependent DNA Repair Selectively Sensitizes KRAS Mutant Lung Cancer to Chemotherapy. Cancer Lett. 2020, 493, 80–90. [Google Scholar] [CrossRef]
- Daskalos, A.; Oleksiewicz, U.; Filia, A.; Nikolaidis, G.; Xinarianos, G.; Gosney, J.R.; Malliri, A.; Field, J.K.; Liloglou, T. UHRF1-Mediated Tumor Suppressor Gene Inactivation in Nonsmall Cell Lung Cancer. Cancer 2011, 117, 1027–1037. [Google Scholar] [CrossRef]
- Babbio, F.; Pistore, C.; Curti, L.; Castiglioni, I.; Kunderfranco, P.; Brino, L.; Oudet, P.; Seiler, R.; Thalman, G.N.; Roggero, E.; et al. The SRA Protein UHRF1 Promotes Epigenetic Crosstalks and Is Involved in Prostate Cancer Progression. Oncogene 2012, 31, 4878–4887. [Google Scholar] [CrossRef]
- Ramos, A.; Sadeghi, S.; Tabatabaeian, H. Battling Chemoresistance in Cancer: Root Causes and Strategies to Uproot Them. Int. J. Mol. Sci. 2021, 22, 9451. [Google Scholar] [CrossRef]
- Kazanets, A.; Shorstova, T.; Hilmi, K.; Marques, M.; Witcher, M. Epigenetic Silencing of Tumor Suppressor Genes: Paradigms, Puzzles, and Potential. Biochim. Biophys. Acta (BBA) Rev. Cancer 2016, 1865, 275–288. [Google Scholar] [CrossRef] [PubMed]
- Senisterra, G.; Zhu, H.Y.; Luo, X.; Zhang, H.; Xun, G.; Lu, C.; Xiao, W.; Hajian, T.; Loppnau, P.; Chau, I.; et al. Discovery of Small-Molecule Antagonists of the H3K9me3 Binding to UHRF1 Tandem Tudor Domain. SLAS Discov. 2018, 23, 930–940. [Google Scholar] [CrossRef]
- Chang, L.; Campbell, J.; Raji, I.O.; Guduru, S.K.R.; Kandel, P.; Nguyen, M.; Liu, S.; Tran, K.; Venugopal, N.K.; Taylor, B.C.; et al. Discovery of Small Molecules Targeting the Tandem Tudor Domain of the Epigenetic Factor UHRF1 Using Fragment-Based Ligand Discovery. Sci. Rep. 2021, 11, 1121. [Google Scholar] [CrossRef]
- Liu, W.H.; Miner, R.E.I.; Albaugh, B.N.; Ananiev, G.E.; Wildman, S.A.; Denu, J.M. Discovery and Mechanism of Small Molecule Inhibitors Selective for the Chromatin-Binding Domains of Oncogenic UHRF1. Biochemistry 2022, 61, 354–366. [Google Scholar] [CrossRef]
- Myrianthopoulos, V.; Cartron, P.F.; Liutkevičiūtė, Z.; Klimašauskas, S.; Matulis, D.; Bronner, C.; Martinet, N.; Mikros, E. Tandem Virtual Screening Targeting the SRA Domain of UHRF1 Identifies a Novel Chemical Tool Modulating DNA Methylation. Eur. J. Med. Chem. 2016, 114, 390–396. [Google Scholar] [CrossRef]
- Giovinazzo, H.; Walker, D.; Wyhs, N.; Liu, J.; Esopi, D.M.; Vaghasia, A.M.; Jain, Y.; Bhamidipati, A.; Zhou, J.; Nelson, W.G.; et al. A High-Throughput Screen of Pharmacologically Active Compounds for Inhibitors of UHRF1 Reveals Epigenetic Activity of Anthracycline Derivative Chemotherapeutic Drugs. Oncotarget 2019, 10, 3040–3050. [Google Scholar] [CrossRef]
- Yin, L.; Liu, Y.; Peng, Y.; Peng, Y.; Yu, X.; Gao, Y.; Yuan, B.; Zhu, Q.; Cao, T.; He, L.; et al. PARP Inhibitor Veliparib and HDAC Inhibitor SAHA Synergistically Co-Target the UHRF1/BRCA1 DNA Damage Repair Complex in Prostate Cancer Cells. J. Exp. Clin. Cancer Res. 2018, 37, 153. [Google Scholar] [CrossRef]
- Fu, Y.; Cao, T.; Zou, X.; Ye, Y.; Liu, Y.; Peng, Y.; Deng, T.; Yin, L.; Li, X. AKT1 Regulates UHRF1 Protein Stability and Promotes the Resistance to Abiraterone in Prostate Cancer. Oncogenesis 2023, 12, 1. [Google Scholar] [CrossRef] [PubMed]
- Ding, G.; Chen, P.; Zhang, H.; Huang, X.; Zang, Y.; Li, J.; Li, J.; Wong, J. Regulation of Ubiquitin-like with Plant Homeodomain and RING Finger Domain 1 (UHRF1) Protein Stability by Heat Shock Protein 90 Chaperone Machinery. J. Biol. Chem. 2016, 291, 20125–20135. [Google Scholar] [CrossRef]
- Kong, X.; Chen, J.; Xie, W.; Brown, S.M.; Cai, Y.; Wu, K.; Fan, D.; Nie, Y.; Yegnasubramanian, S.; Tiedemann, R.L.; et al. Defining UHRF1 Domains That Support Maintenance of Human Colon Cancer DNA Methylation and Oncogenic Properties. Cancer Cell 2019, 35, 633–648.e7. [Google Scholar] [CrossRef]
- Zhou, X.; Wang, X.; Huang, Z.; Wang, J.; Zhu, W.; Shu, Y.; Liu, P. Prognostic Value of miR-21 in Various Cancers: An Updating Meta-Analysis. PLoS ONE 2014, 9, e102413. [Google Scholar] [CrossRef] [PubMed]
- Matsushita, R.; Yoshino, H.; Enokida, H.; Goto, Y.; Miyamoto, K.; Yonemori, M.; Inoguchi, S.; Nakagawa, M.; Seki, N. Regulation of UHRF1 by Dual-Strand Tumor-Suppressor microRNA-145 (miR-145-5p and miR-145-3p): Inhibition of Bladder Cancer Cell Aggressiveness. Oncotarget 2016, 7, 28460–28487. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Zhao, X.; Han, Y.; Lu, Y.; Shang, Y.; Liu, C.; Li, T.; Jin, Z.; Fan, D.; Wu, K. Regulation of UHRF1 by miR-146a/b Modulates Gastric Cancer Invasion and Metastasis. FASEB J. 2013, 27, 4929–4939. [Google Scholar] [CrossRef] [PubMed]
- Wotschofsky, Z.; Gummlich, L.; Liep, J.; Stephan, C.; Kilic, E.; Jung, K.; Billaud, J.-N.; Meyer, H.-A. Integrated microRNA and mRNA Signature Associated with the Transition from the Locally Confined to the Metastasized Clear Cell Renal Cell Carcinoma Exemplified by miR-146-5p. PLoS ONE 2016, 11, e0148746. [Google Scholar] [CrossRef] [PubMed]
| Role in Cancer | Cancer | UHRF1 | Reference |
|---|---|---|---|
| Sustaining proliferation and tumor growth | SCLC | YAP1 stability promotes proliferation | [62] |
| NSCLC | Increases proliferation, enhances G1/S transition | [63] | |
| Melanoma | Elevates levels of Ki67, facilitates cell division | [64] | |
| Medulloblastoma | UHRF1 downregulation activates p16 and limits CDK4 | [61] | |
| KRAS-driven lung cancer | Associated with tumor growth and poor prognosis | [65] | |
| TNBC | Reduces G1 cell population, enhances tumor growth | [20] | |
| Resisting cell death | Retinoblastoma | UHRF1 depletion increases apoptotic markers | [66] |
| TNBC | Inhibits apoptosis | [20] | |
| HCC | UHRF1 downregulation does not trigger apoptosis | [67] | |
| Inducing angiogenesis and metastasis | Osteosarcoma | Downregulates E-cadherin and promotes EMT | [19] |
| Suppresses AMPK and decreases SEMA3E | [14] | ||
| Increasing chemoresistance | Retinoblastoma | UHRF1 depletion increases sensitivity to etoposide, etc. | [66] |
| SCLC | UHRF1 reduction increases cisplatin chemosensitivity | [62] | |
| KRAS mutant lung cancer | Target for cardiac glycosides to increase sensitivity | [68] | |
| Epigenetic silencing | NSCLC | UHRF1 downregulation is linked to reactivation of TSGs | [69] |
| Breast cancer | Transcriptional complex silences BRCA1 gene | [55] | |
| Prostate cancer | Modulates H3K9 methylation via EZH2 | [70] |
| Compound | Target | Function | Reference |
|---|---|---|---|
| NV01 | UHRF1 TTD–PHD | Bind H3K9me3 | [73] |
| 2,4-lutidine | UHRF1 TTD | Bind critical site involved in H3K9me3 and PBR | [74] |
| MLD3-5 | UHRF1 PHD | Disrupt histone-TTD interaction | [75] |
| NSC232003 | UHRF1 SRA | Target 5-methylcytosine; reduce DNMT1–UHRF1 interaction | [76] |
| Mitoxantrone, idarubicin | UHRF1 SRA | Inhibit UHRF1-hemi-methylated DNA interaction | [77] |
| Veliparib with SAHA | BRCA1 | Decrease cell viability; induce apoptosis and DNA damage; decrease in BRCA1, which degrades UHRF1 | [78] |
| MK2206 | AKT1/USP7 | Inhibit AKT1 and induce phosphorylated UHRF1; reduce UHRF1–USP7 binding | [79] |
| 17-AAG/17-DMAG | HSP90 | Degrade UHRF1 via ubiquitin–proteasome system | [80] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, A.; Benavente, C.A. Oncogenic Roles of UHRF1 in Cancer. Epigenomes 2024, 8, 26. https://doi.org/10.3390/epigenomes8030026
Kim A, Benavente CA. Oncogenic Roles of UHRF1 in Cancer. Epigenomes. 2024; 8(3):26. https://doi.org/10.3390/epigenomes8030026
Chicago/Turabian StyleKim, Ahhyun, and Claudia A. Benavente. 2024. "Oncogenic Roles of UHRF1 in Cancer" Epigenomes 8, no. 3: 26. https://doi.org/10.3390/epigenomes8030026
APA StyleKim, A., & Benavente, C. A. (2024). Oncogenic Roles of UHRF1 in Cancer. Epigenomes, 8(3), 26. https://doi.org/10.3390/epigenomes8030026






