α-Crystalline Domains and Intrinsically Disordered Regions Can Work in Parallel to Induce Accumulation of MBD6 at Chromocenters in Arabidopsis thaliana
Abstract
1. Introduction
2. Results
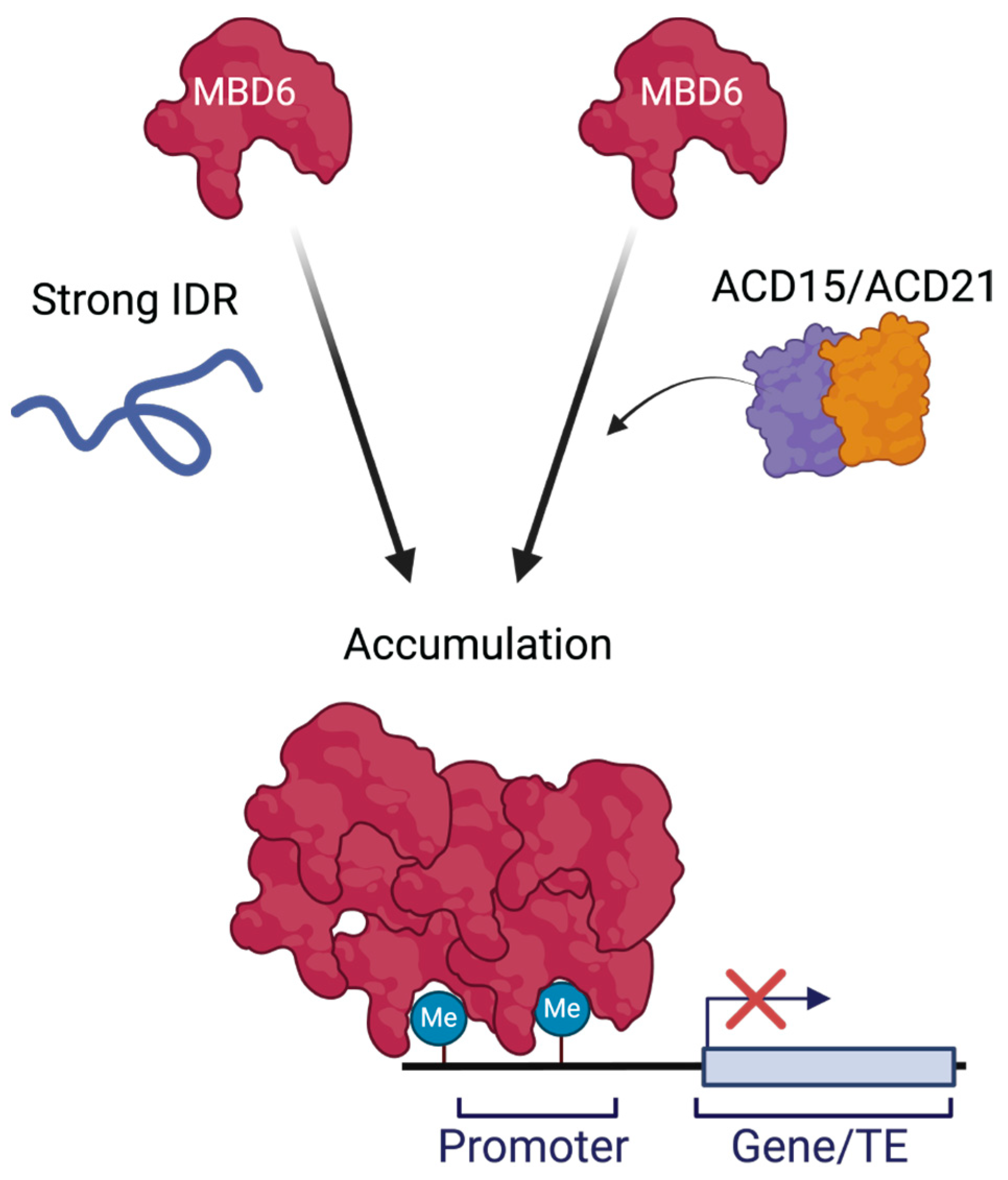
2.1. Human IDRs Can Stimulate the Accumulation of MBD6 Together with ACD15 and ACD21
2.2. IDR Accumulation of MBD6 Is Associated with Gene Silencing

3. Conclusions
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Generation of Transgenic Lines
4.3. Confocal Microscopy
4.4. Quantification of Foci Counts and Nuclear RFP Signal Distributions
4.5. RT-qPCR
- FWA RT-qPCR Forward: TTAGATCCAAAGGAGTATCAAAG
- FWA RT-qPCR Reverse: CTTTGGTACCAGCGGAGA
- IPP2 RT-qPCR Forward: GTATGAGTTGCTTCTCCAGCAAAG
- IPP2 RT-qPCR Reverse: GAGGATGGCTGCAACAAGTGT
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Boeynaems, S.; Alberti, S.; Fawzi, N.L.; Mittag, T.; Polymenidou, M.; Rousseau, F.; Schymkowitz, J.; Shorter, J.; Wolozin, B.; Bosch, L.V.D.; et al. Protein Phase Separation: A New Phase in Cell Biology. Trends Cell Biol. 2018, 28, 420–435. [Google Scholar] [CrossRef]
- Emenecker, R.J.; Holehouse, A.S.; Strader, L.C. Emerging Roles for Phase Separation in Plants. Dev. Cell 2020, 55, 69–83. [Google Scholar] [CrossRef] [PubMed]
- Chong, P.A.; Forman-Kay, J.D. Liquid–liquid phase separation in cellular signaling systems. Curr. Opin. Struct. Biol. 2016, 41, 180–186. [Google Scholar] [CrossRef]
- Buttress, T.; He, S.; Wang, L.; Zhou, S.; Saalbach, G.; Vickers, M.; Li, G.; Li, P.; Feng, X. Histone H2B.8 compacts flowering plant sperm through chromatin phase separation. Nature 2022, 611, 614–622. [Google Scholar] [CrossRef]
- Bergeron-Sandoval, L.-P.; Safaee, N.; Michnick, S.W. Mechanisms and Consequences of Macromolecular Phase Separation. Cell 2016, 165, 1067–1079. [Google Scholar] [CrossRef]
- Brangwynne, C.P.; Tompa, P.; Pappu, R.V. Polymer physics of intracellular phase transitions. Nat. Phys 2015, 11, 899–904. [Google Scholar] [CrossRef]
- Maiti, S.; De, S. Identification of potential short linear motifs (SLiMs) in intrinsically disordered sequences of proteins by fast time-scale backbone dynamics. J. Magn. Reson. Open 2022, 10–11, 100029. [Google Scholar] [CrossRef]
- Davey, N.E.; Roey, K.V.; Weatheritt, R.J.; Toedt, G.; Uyar, B.; Altenberg, B.; Budd, A.; Diella, F.; Dinkel, H.; Gibson, T.J. Attributes of short linear motifs. Mol. BioSyst. 2011, 8, 268–281. [Google Scholar] [CrossRef]
- Van Roey, K.; Uyar, B.; Weatheritt, R.J.; Dinkel, H.; Seiler, M.; Budd, A.; Gibson, T.J.; Davey, N.E. Short Linear Motifs: Ubiquitous and Functionally Diverse Protein Interaction Modules Directing Cell Regulation. Chem. Rev. 2014, 114, 6733–6778. [Google Scholar] [CrossRef] [PubMed]
- O’Shea, C.; Staby, L.; Bendsen, S.K.; Tidemand, F.G.; Redsted, A.; Willemoës, M.; Kragelund, B.B.; Skriver, K. Structures and Short Linear Motif of Disordered Transcription Factor Regions Provide Clues to the Interactome of the Cellular Hub Protein Radical-induced Cell Death1. J. Biol. Chem. 2017, 292, 512–527. [Google Scholar] [CrossRef] [PubMed]
- Boone, B.A.; Ichino, L.; Wang, S.; Gardiner, J.; Yun, J.; Jami-Alahmadi, Y.; Sha, J.; Mendoza, C.P.; Steelman, B.J.; Aardenne, A.v.; et al. ACD15, ACD21, and SLN regulate accumulation and mobility of MBD6 to silence genes and transposable elements. Sci. Adv. 2023, 9, eadi9036. [Google Scholar] [CrossRef] [PubMed]
- Zemach, A.; Grafi, G. Methyl-CpG-binding domain proteins in plants: Interpreters of DNA methylation. Trends Plant Sci. 2007, 12, 80–85. [Google Scholar] [CrossRef]
- Zemach, A.; Paul, L.K.; Stambolsky, P.; Efroni, I.; Rotter, V.; Grafi, G. The C-terminal domain of the Arabidopsis AtMBD7 protein confers strong chromatin binding activity. Exp. Cell Res. 2009, 315, 3554–3562. [Google Scholar] [CrossRef]
- Ichino, L.; Boone, B.A.; Strauskulage, L.; Harris, C.J.; Kaur, G.; Gladstone, M.A.; Tan, M.; Feng, S.; Jami-Alahmadi, Y.; Duttke, S.H.; et al. MBD5 and MBD6 couple DNA methylation to gene silencing through the J-domain protein SILENZIO. Science 2021, 372, 1434–1439. [Google Scholar] [CrossRef]
- Ren, Z.; Gou, R.; Zhuo, W.; Chen, Z.; Yin, X.; Cao, Y.; Wang, Y.; Mi, Y.; Liu, Y.; Wang, Y.; et al. The MBD–ACD DNA methylation reader complex recruits MICRORCHIDIA6 to regulate rRNA gene expression in Arabidopsis. Plant Cell 2023, 36, 1098–1118. [Google Scholar] [CrossRef]
- Haslbeck, M.; Weinkauf, S.; Buchner, J. Small heat shock proteins: Simplicity meets complexity. J. Biol. Chem. 2019, 294, 2121–2132. [Google Scholar] [CrossRef]
- Haslbeck, M.; Vierling, E. A First Line of Stress Defense: Small Heat Shock Proteins and Their Function in Protein Homeostasis. J. Mol. Biol. 2015, 427, 1537–1548. [Google Scholar] [CrossRef] [PubMed]
- Park, C.-J.; Seo, Y.-S. Heat Shock Proteins: A Review of the Molecular Chaperones for Plant Immunity. Plant Pathol. J. 2015, 31, 323–333. [Google Scholar] [CrossRef]
- Boelens, W.C. Structural aspects of the human small heat shock proteins related to their functional activities. Cell Stress Chaperones 2020, 25, 581–591. [Google Scholar] [CrossRef] [PubMed]
- Hochberg, G.K.A.; Shepherd, D.A.; Marklund, E.G.; Santhanagoplan, I.; Degiacomi, M.T.; Laganowsky, A.; Allison, T.M.; Basha, E.; Marty, M.T.; Galpin, M.R.; et al. Structural principles that enable oligomeric small heat-shock protein paralogs to evolve distinct functions. Science 2018, 359, 930–935. [Google Scholar] [CrossRef]
- Mymrikov, E.V.; Riedl, M.; Peters, C.; Weinkauf, S.; Haslbeck, M.; Buchner, J. Regulation of small heat-shock proteins by hetero-oligomer formation. J. Biol. Chem. 2020, 295, 158–169. [Google Scholar] [CrossRef]
- Waters, E.R.; Vierling, E. Plant small heat shock proteins—Evolutionary and functional diversity. New Phytol. 2020, 227, 24–37. [Google Scholar] [CrossRef]
- Simon, L.; Voisin, M.; Tatout, C.; Probst, A.V. Structure and Function of Centromeric and Pericentromeric Heterochromatin in Arabidopsis thaliana. Front. Plant Sci. 2015, 6, 1049. [Google Scholar] [CrossRef]
- Fransz, P.; de Jong, J.H.; Lysak, M.; Castiglione, M.R.; Schubert, I. Interphase chromosomes in Arabidopsis are organized as well defined chromocenters from which euchromatin loops emanate. Proc. Natl. Acad. Sci. USA 2002, 99, 14584–14589. [Google Scholar] [CrossRef]
- Larson, A.G.; Elnatan, D.; Keenen, M.M.; Trnka, M.J.; Johnston, J.B.; Burlingame, A.L.; Agard, D.A.; Redding, S.; Narlikar, G.J. Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature 2017, 547, 236–240. [Google Scholar] [CrossRef]
- Li, C.H.; Coffey, E.L.; Dall’Agnese, A.; Hannett, N.M.; Tang, X.; Henninger, J.E.; Platt, J.M.; Oksuz, O.; Zamudio, A.V.; Afeyan, L.K.; et al. MeCP2 links heterochromatin condensates and neurodevelopmental disease. Nature 2020, 586, 440–444. [Google Scholar] [CrossRef]
- Cheutin, T.; McNairn, A.J.; Jenuwein, T.; Gilbert, D.M.; Singh, P.B.; Misteli, T. Maintenance of Stable Heterochromatin Domains by Dynamic HP1 Binding. Science 2003, 299, 721–725. [Google Scholar] [CrossRef]
- Schmidt, A.; Zhang, H.; Cardoso, M.C. MeCP2 and Chromatin Compartmentalization. Cells 2020, 9, 878. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Yu, Y.; Wang, L.; Zhang, J.; Bai, Z.; Li, G.; Li, P.; Feng, X. Linker histone H1 drives heterochromatin condensation via phase separation in Arabidopsis. Plant Cell 2024, 36, 1829–1843. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Zhang, S.; Gu, J.; Tong, Y.; Li, Y.; Gui, X.; Long, H.; Wang, C.; Zhao, C.; Lu, J.; et al. Hsp27 chaperones FUS phase separation under the modulation of stress-induced phosphorylation. Nat. Struct. Mol. Biol. 2020, 27, 363–372. [Google Scholar] [CrossRef] [PubMed]
- Wallace, E.W.J.; Kear-Scott, J.L.; Pilipenko, E.V.; Schwartz, M.H.; Laskowski, P.R.; Rojek, A.E.; Katanski, C.D.; Riback, J.A.; Dion, M.F.; Franks, A.M.; et al. Reversible, Specific, Active Aggregates of Endogenous Proteins Assemble upon Heat Stress. Cell 2015, 162, 1286–1298. [Google Scholar] [CrossRef] [PubMed]
- Yoo, H.; Triandafillou, C.; Drummond, D.A. Cellular sensing by phase separation: Using the process, not just the products. J. Biol. Chem. 2019, 294, 7151–7159. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boone, B.A.; Mendoza, C.P.; Behrendt, N.J.; Jacobsen, S.E. α-Crystalline Domains and Intrinsically Disordered Regions Can Work in Parallel to Induce Accumulation of MBD6 at Chromocenters in Arabidopsis thaliana. Epigenomes 2024, 8, 33. https://doi.org/10.3390/epigenomes8030033
Boone BA, Mendoza CP, Behrendt NJ, Jacobsen SE. α-Crystalline Domains and Intrinsically Disordered Regions Can Work in Parallel to Induce Accumulation of MBD6 at Chromocenters in Arabidopsis thaliana. Epigenomes. 2024; 8(3):33. https://doi.org/10.3390/epigenomes8030033
Chicago/Turabian StyleBoone, Brandon A., Cristy P. Mendoza, Noah J. Behrendt, and Steven E. Jacobsen. 2024. "α-Crystalline Domains and Intrinsically Disordered Regions Can Work in Parallel to Induce Accumulation of MBD6 at Chromocenters in Arabidopsis thaliana" Epigenomes 8, no. 3: 33. https://doi.org/10.3390/epigenomes8030033
APA StyleBoone, B. A., Mendoza, C. P., Behrendt, N. J., & Jacobsen, S. E. (2024). α-Crystalline Domains and Intrinsically Disordered Regions Can Work in Parallel to Induce Accumulation of MBD6 at Chromocenters in Arabidopsis thaliana. Epigenomes, 8(3), 33. https://doi.org/10.3390/epigenomes8030033






