Swine Conjunctivitis Associated with a Novel Mycoplasma Species Closely Related to Mycoplasma hyorhinis
Abstract
1. Introduction
2. Results
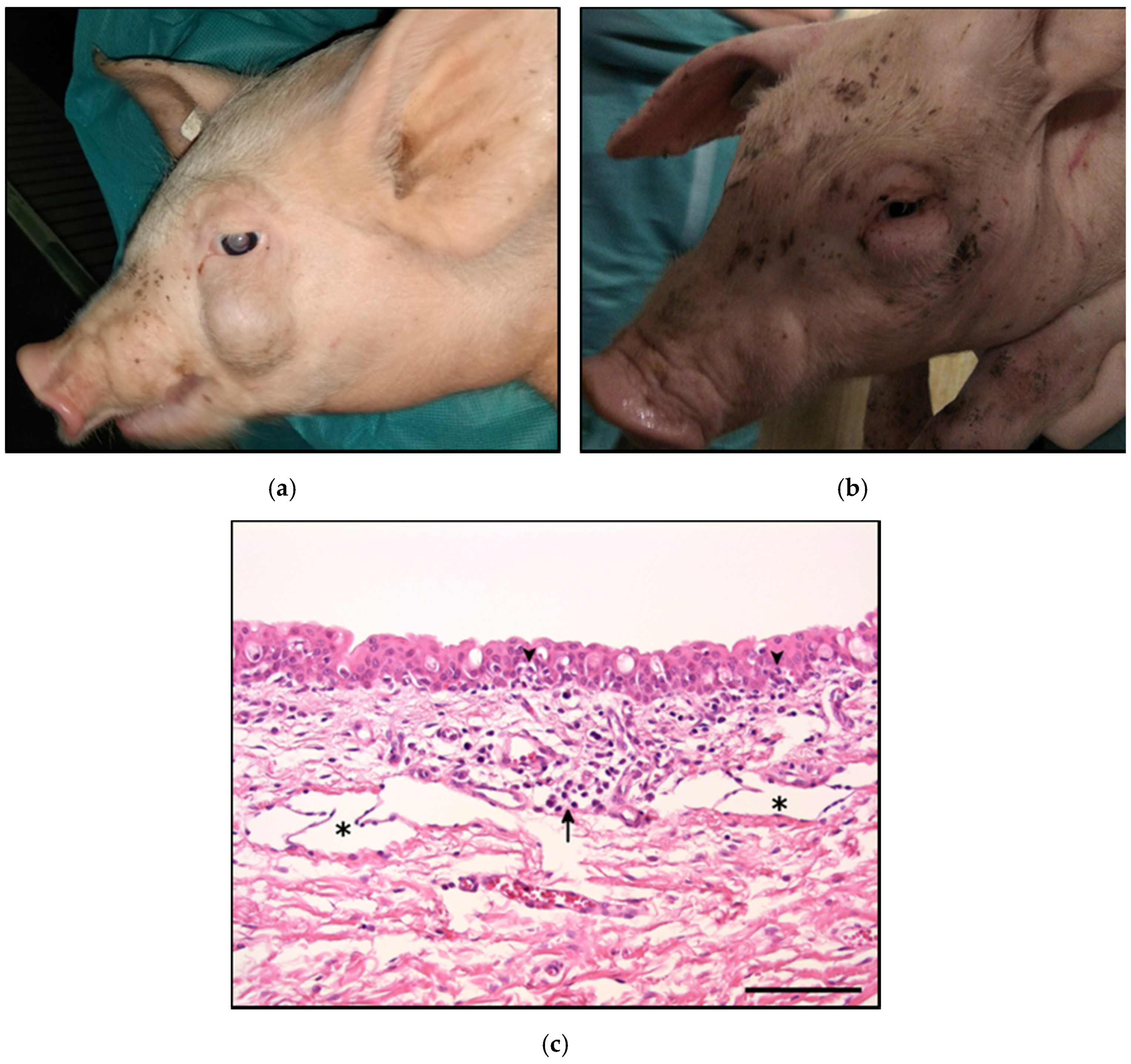
2.1. Macroscopic and Histopathological Findings
2.2. Characterization of Mycoplasma sp. Isolates Cultivated from Eye Swabs
3. Discussion
4. Materials and Methods
4.1. Clinical Examination on Farms and Sampling
4.2. Ethical Statement
4.3. Characterization of Farms
4.4. Cultivation and MALDI-TOF Mass Spectrometry
4.5. Determination of MIC Values
4.6. PCR Analysis of Swab Samples and Mycoplasma Cultures
4.7. Phylogenetic Analysis of Selected Mycoplasma Isolates
4.8. Genome Sequencing of Strain 1654_15
4.9. Genome Comparison Analysis
4.10. Accession Number
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kobisch, M.; Friis, N.F. Swine mycoplasmoses. Rev. Sci. Tech. 1996, 15, 1569–1605. [Google Scholar] [CrossRef] [PubMed]
- Bové, J.M. Molecular features of mollicutes. Clin. Infect. Dis. 1993, 17 (Suppl. 1), S10–S31. [Google Scholar] [CrossRef] [PubMed]
- Razin, S.; Yogev, D.; Naot, Y. Molecular biology and pathogenicity of mycoplasmas. Microbiol. Mol. Biol. Rev. 1998, 62, 1094–1156. [Google Scholar] [CrossRef] [PubMed]
- Friis, N.F.; Feenstra, A.A. Mycoplasma hyorhinis in the etiology of serositis among piglets. Acta Vet. Scand. 1994, 35, 93–98. [Google Scholar]
- Clavijo, M.J.; Murray, D.; Oliveira, S.; Rovira, A. Infection dynamics of Mycoplasma hyorhinis in three commercial pig populations. Vet. Rec. 2017, 181, 68. [Google Scholar] [CrossRef] [PubMed]
- Kinne, J.; Johannsen, U.; Neumann, R.; Mehlhorn, G.; Pfützner, H. The pathology and pathogenesis of experimental Mycoplasma hyorhinis infection of piglets with and without thermomotoric stress. 1. Pathologico-anatomic, histologic and immunomorphologic study results. Zentralbl. Veterinarmed. A 1991, 38, 306–320. [Google Scholar] [CrossRef]
- Johannsen, U.; Menger, S.; Kinne, J.; Neumann, R.; Mehlhorn, G.; Pfützner, H. The pathology and pathogenesis of experimental Mycoplasma hyorhinis infection of piglets with and without thermomotor stress. 2. Electron microscopic study results. Zentralbl. Veterinarmed. A 1991, 38, 321–336. [Google Scholar] [CrossRef]
- Lin, J.H.; Chen, S.P.; Yeh, K.S.; Weng, C.N. Mycoplasma hyorhinis in Taiwan: Diagnosis and isolation of swine pneumonia pathogen. Vet. Microbiol. 2006, 115, 111–116. [Google Scholar] [CrossRef]
- Muhlradt, P.F.; Kiess, M.; Meyer, H.; Sussmuth, R.; Jung, G. Structure and specific activity of macrophage-stimulating lipopeptides from Mycoplasma hyorhinis. Infect. Immun. 1998, 66, 4804–4810. [Google Scholar] [CrossRef]
- Gois, M.; Kuksa, F. Intranasal infection of gnotobiotic piglets with Mycoplasma hyorhinis: Differences in virulence of the strains and influence of age on the development of infection. Zentralbl. Veterinarmed. B 1974, 21, 352–361. [Google Scholar] [CrossRef]
- Lee, J.A.; Hwang, M.A.; Han, J.H.; Cho, E.H.; Lee, J.B.; Park, S.Y.; Song, C.S.; Choi, I.S.; Lee, S.W. Reduction of mycoplasmal lesions and clinical signs by vaccination against Mycoplasma hyorhinis. Vet. Immunol. Immunopathol. 2018, 196, 14–17. [Google Scholar] [CrossRef] [PubMed]
- Friis, N.F.; Kokotovic, B.; Svensmark, B. Mycoplasma hyorhinis isolation from cases of otitis media in piglets. Acta Vet. Scand. 2002, 43, 191–193. [Google Scholar] [PubMed]
- Morita, T.; Ohiwa, S.; Shimada, A.; Kazama, S.; Yagihashi, T.; Umemura, T. Intranasally inoculated Mycoplasma hyorhinis causes eustachitis in pigs. Vet. Pathol. 1999, 36, 174–178. [Google Scholar] [CrossRef] [PubMed]
- Morita, T.; Fukuda, H.; Awakura, T.; Shimada, A.; Umemura, T.; Kazama, S.; Yagihashi, T. Demonstration of Mycoplasma hyorhinis as a possible primary pathogen for porcine otitis media. Vet. Pathol. 1995, 32, 107–111. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.H.; Joo, H.S.; Lee, W.H.; Seok, H.B.; Calsamig, M.; Pijoan, C.; Molitor, T.W. Identification and characterization of cytopathogenic Mycoplasma hyorhinis from swine farms with a history of abortions. J. Vet. Med. Sci. 2003, 65, 501–509. [Google Scholar] [CrossRef]
- Resende, T.P.; Pieters, M.; Vannucci, F.A. Swine conjunctivitis outbreaks associated with Mycoplasma hyorhinis. J. Vet. Diagn. Investig. 2019, 31, 766–769. [Google Scholar] [CrossRef]
- Madson, D.M.A.; Arruda, P.H.E.; Arruda, B.L. Nervous and Locomotor System. In Diseases of Swine, 11th ed.; Zimmerman, J.J., Karriker, L.A., Ramirez, A., Schwartz, K.J., Stevenson, G.W., Zhang, J., Eds.; John Wiley & Sons Inc.: Ames, IA, USA, 2019; pp. 339–372. [Google Scholar]
- Alberti, A.; Addis, M.F.; Chessa, B.; Cubeddu, T.; Profiti, M.; Rosati, S.; Ruiu, A.; Pittau, M. Molecular and antigenic characterization of a Mycoplasma bovis strain causing an outbreak of infectious keratoconjunctivitis. J. Vet. Diagn. Investig. 2006, 18, 41–51. [Google Scholar] [CrossRef]
- Angelos, J.A. Infectious bovine keratoconjunctivitis (pinkeye). Vet. Clin. N. Am. Food Anim. Pract. 2015, 31, 61–79. [Google Scholar] [CrossRef]
- Haesebrouck, F.; Devriese, L.A.; van Rijssen, B.; Cox, E. Incidence and significance of isolation of Mycoplasma felis from conjunctival swabs of cats. Vet. Microbiol. 1991, 26, 95–101. [Google Scholar] [CrossRef]
- Giacometti, M.; Janovsky, M.; Belloy, L.; Frey, J. Infectious keratoconjunctivitis of ibex, chamois and other Caprinae. Rev. Sci. Tech. 2002, 21, 335–345. [Google Scholar] [CrossRef]
- Dhondt, A.A.; Altizer, S.; Cooch, E.G.; Davis, A.K.; Dobson, A.; Driscoll, M.J.; Hartup, B.K.; Hawley, D.M.; Hochachka, W.M.; Hosseini, P.R.; et al. Dynamics of a novel pathogen in an avian host: Mycoplasmal conjunctivitis in house finches. Acta Trop. 2005, 94, 77–93. [Google Scholar] [CrossRef] [PubMed]
- Rogers, D.G.; Frey, M.L.; Hogg, A. Conjunctivitis associated with a Mycoplasma-like organism in swine. J. Am. Vet. Med. Assoc. 1991, 198, 450–452. [Google Scholar] [PubMed]
- Friis, N.F. A serologic variant of Mycoplasma hyorhinis recovered from the conjunctiva of swine. Acta Vet. Scand. 1976, 17, 343–353. [Google Scholar] [PubMed]
- Spergser, J.; Hess, C.; Loncaric, I.; Ramírez, A.S. Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry Is a Superior Diagnostic Tool for the Identification and Differentiation of Mycoplasmas Isolated from Animals. J. Clin. Microbiol. 2019, 57, e00316-19. [Google Scholar] [CrossRef] [PubMed]
- Beko, K.; Felde, O.; Sulyok, K.M.; Kreizinger, Z.; Hrivnak, V.; Kiss, K.; Biksi, I.; Jerzsele, A.; Gyuranecz, M. Antibiotic susceptibility profiles of Mycoplasma hyorhinis strains isolated from swine in Hungary. Vet. Microbiol. 2019, 228, 196–201. [Google Scholar] [CrossRef] [PubMed]
- Gautier-Bouchardon, A.V. Antimicrobial Resistance in Mycoplasma spp. Microbiol. Spectr. 2018, 6, 425–446. [Google Scholar] [CrossRef]
- Volokhov, D.V.; Simonyan, V.; Davidson, M.K.; Chizhikov, V.E. RNA polymerase beta subunit (rpoB) gene and the 16S-23S rRNA intergenic transcribed spacer region (ITS) as complementary molecular markers in addition to the 16S rRNA gene for phylogenetic analysis and identification of the species of the family Mycoplasmataceae. Mol. Phylogenet. Evol. 2012, 62, 515–528. [Google Scholar] [CrossRef]
- Adekambi, T.; Drancourt, M.; Raoult, D. The rpoB gene as a tool for clinical microbiologists. Trends Microbiol. 2009, 17, 37–45. [Google Scholar] [CrossRef]
- Volokhov, D.V.; Grózner, D.; Gyuranecz, M.; Ferguson-Noel, N.; Gao, Y.; Bradbury, J.M.; Whittaker, P.; Chizhikov, V.E.; Szathmary, S.; Stipkovits, L. Mycoplasma anserisalpingitidis sp. nov., isolated from European domestic geese (Anser anser domesticus) with reproductive pathology. Int. J. Syst. Evol. Microbiol. 2020, 70, 2369–2381. [Google Scholar] [CrossRef]
- Richter, M.; Rosselló-Móra, R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA 2009, 106, 19126–19131. [Google Scholar] [CrossRef]
- Konstantinidis, K.T.; Tiedje, J.M. Towards a genome-based taxonomy for prokaryotes. J. Bacteriol. 2005, 187, 6258–6264. [Google Scholar] [CrossRef] [PubMed]
- Galvao Ferrarini, M.; Mucha, S.G.; Parrot, D.; Meiffrein, G.; Ruggiero Bachega, J.F.; Comte, G.; Zaha, A.; Sagot, M.F. Hydrogen peroxide production and myo-inositol metabolism as important traits for virulence of Mycoplasma hyopneumoniae. Mol. Microbiol. 2018, 108, 683–696. [Google Scholar] [CrossRef] [PubMed]
- Blotz, C.; Stulke, J. Glycerol metabolism and its implication in virulence in Mycoplasma. FEMS Microbiol. Rev. 2017, 41, 640–652. [Google Scholar] [CrossRef] [PubMed]
- Trueeb, B.S.; Gerber, S.; Maes, D.; Gharib, W.H.; Kuhnert, P. Tn-sequencing of Mycoplasma hyopneumoniae and Mycoplasma hyorhinis mutant libraries reveals non-essential genes of porcine mycoplasmas differing in pathogenicity. Vet. Res. 2019, 50, 55. [Google Scholar] [CrossRef] [PubMed]
- Arfi, Y.; Minder, L.; Di Primo, C.; Le Roy, A.; Ebel, C.; Coquet, L.; Claverol, S.; Vashee, S.; Jores, J.; Blanchard, A.; et al. MIB-MIP is a mycoplasma system that captures and cleaves immunoglobulin G. Proc. Natl. Acad. Sci. USA 2016, 113, 5406–5411. [Google Scholar] [CrossRef] [PubMed]
- Goyal, S.M. Porcine reproductive and respiratory syndrome. J. Vet. Diagn. Investig. 1993, 5, 656–664. [Google Scholar] [CrossRef] [PubMed]
- Kawashima, K.; Yamada, S.; Kobayashi, H.; Narita, M. Detection of porcine reproductive and respiratory syndrome virus and Mycoplasma hyorhinis antigens in pulmonary lesions of pigs suffering from respiratory distress. J. Comp. Pathol. 1996, 114, 315–323. [Google Scholar] [CrossRef]
- Fahrion, A.S.; Grosse Beilage, E.; Nathues, H.; Dürr, S.; Doherr, M.G. Evaluating perspectives for PRRS virus elimination from pig dense areas with a risk factor based herd index. Prev. Vet. Med. 2014, 114, 247–258. [Google Scholar] [CrossRef]
- Grosse Beilage, E.G.; Bätza, H.J. PRRSV-eradication: An option for pigherds in Germany? Berl. Munch. Tierarztl. Wochenschr. 2007, 120, 470–479. [Google Scholar]
- Luo, C.; Shen, G.; Liu, N.; Gong, F.; Wei, X.; Yao, S.; Liu, D.; Teng, X.; Ye, N.; Zhang, N.; et al. Ammonia drives dendritic cells into dysfunction. J. Immunol. 2014, 193, 1080–1089. [Google Scholar] [CrossRef]
- Zielińska, M.; Milewski, K.; Skowrońska, M.; Gajos, A.; Ziemińska, E.; Beręsewicz, A.; Albrecht, J. Induction of inducible nitric oxide synthase expression in ammonia-exposed cultured astrocytes is coupled to increased arginine transport by upregulated y+LAT2 transporter. J. Neurochem. 2015, 135, 1272–1281. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.B.; Burgess, L.R.; Webster, A.J.F.; Wathes, C.M. Behavioural responses of pigs to atmospheric ammonia in a chronic choice test. Anim. Sci. 1996, 63, 437–445. [Google Scholar] [CrossRef]
- Drummond, J.G.; Curtis, S.E.; Simon, J.; Norton, H.W. Effects of Aerial Ammonia on Growth and Health of Young Pigs1. J. Anim. Sci. 1980, 50, 1085–1091. [Google Scholar] [CrossRef]
- Michiels, A.; Piepers, S.; Ulens, T.; Van Ransbeeck, N.; Del Pozo Sacristán, R.; Sierens, A.; Haesebrouck, F.; Demeyer, P.; Maes, D. Impact of particulate matter and ammonia on average daily weight gain, mortality and lung lesions in pigs. Prev. Vet. Med. 2015, 121, 99–107. [Google Scholar] [CrossRef]
- Takai, H.; Nekomoto, K.; Dahl, P.J.; Okamoto, E.; Morita, S.; Hoshiba, S. Ammonia contents and desorption from dusts collected in livestock buildings. Agric. Eng. Int. CIGR J. 2002, 4, 1–11. [Google Scholar]
- Nathues, H.; Spergser, J.; Rosengarten, R.; Kreienbrock, L.; Grosse Beilage, E. Value of the clinical examination in diagnosing enzootic pneumonia in fattening pigs. Vet. J. 2012, 193, 443–447. [Google Scholar] [CrossRef]
- Stein, H.; Schulz, J.; Kemper, N.; Tichy, A.; Krauss, I.; Knecht, C.; Hennig-Pauka, I. Fogging low concentrated organic acid in a fattening pig unit—Effect on animal health and microclimate. Ann. Agric. Environ. Med. 2016, 23, 581–586. [Google Scholar] [CrossRef]
- Christianson, W.T.; Joo, H.S. Porcine reproductive and respiratory syndrome: A review. J. Swine Health Prod. 1994, 2, 10–28. [Google Scholar]
- Caron, J.; Ouardani, M.; Dea, S. Diagnosis and differentiation of Mycoplasma hyopneumoniae and Mycoplasma hyorhinis infections in pigs by PCR amplification of the p36 and p46 genes. J. Clin. Microbiol. 2000, 38, 1390–1396. [Google Scholar] [CrossRef]
- Tocqueville, V.; Ferré, S.; Nguyen, N.H.; Kempf, I.; Marois-Créhan, C. Multilocus sequence typing of Mycoplasma hyorhinis strains identified by a real-time TaqMan PCR assay. J. Clin. Microbiol. 2014, 52, 1664–1671. [Google Scholar] [CrossRef]
- Fourour, S.; Fablet, C.; Tocqueville, V.; Dorenlor, V.; Eono, F.; Eveno, E.; Kempf, I.; Marois-Créhan, C. A new multiplex real-time TaqMan®PCR for quantification of Mycoplasma hyopneumoniae, M. hyorhinis and M. flocculare: Exploratory epidemiological investigations to research mycoplasmal association in enzootic pneumonia-like lesions in slaughtered pigs. J. Appl. Microbiol. 2018, 125, 345–355. [Google Scholar] [CrossRef] [PubMed]
- Rosengarten, R.; Wise, K.S. The Vlp system of Mycoplasma hyorhinis: Combinatorial expression of distinct size variant lipoproteins generating high-frequency surface antigenic variation. J. Bacteriol. 1991, 173, 4782–4793. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Q.; Wang, J.; Ji, Y.; Ni, B.; Zhang, B.; Ma, Q.; Wei, Y.; Xiao, S.; Feng, Z.; Liu, M.; et al. The functions of the variable lipoprotein family of Mycoplasma hyorhinis in adherence to host cells. Vet. Microbiol. 2016, 186, 82–89. [Google Scholar] [CrossRef] [PubMed]
- Citti, C.; Kim, M.F.; Wise, K.S. Elongated versions of Vlp surface lipoproteins protect Mycoplasma hyorhinis escape variants from growth-inhibiting host antibodies. Infect. Immun. 1997, 65, 1773–1785. [Google Scholar] [CrossRef] [PubMed]
- Spergser, J.; Loncaric, I.; Tichy, A.; Fritz, J.; Scope, A. The cultivable autochthonous microbiota of the critically endangered Northern bald ibis (Geronticus eremita). PLoS ONE 2018, 13, e0195255. [Google Scholar] [CrossRef] [PubMed]
- Ramírez, A.S.; Gonzales, M.; Déniz, S.; Fernández, A.; Poveda, J.B. Evaluation of a modified SP-4 medium in the replication of Mycoplasma spp. In Mycoplasmas of Ruminants: Pathogenicity, Diagnostics, Epidemiology and Molecular Genetics; Frey, J., Sarris, K., Eds.; European Cooperation on Scientific and Technical Research: Luxembourg, 1997; Volume 2, pp. 36–39. [Google Scholar]
- Hannan, P.C. Guidelines and recommendations for antimicrobial minimum inhibitory concentration (MIC) testing against veterinary mycoplasma species. International Research Programme on Comparative Mycoplasmology. Vet. Res. 2000, 31, 373–395. [Google Scholar] [CrossRef] [PubMed]
- Hartley, J.C.; Kaye, S.; Stevenson, S.; Bennett, J.; Ridgway, G. PCR detection and molecular identification of Chlamydiaceae species. J. Clin. Microbiol. 2001, 39, 3072–3079. [Google Scholar] [CrossRef]
- Lane, D.J. 16S/23S rRNA sequencing. In Nucleic Acid Techniques in Bacterial Systematics; Stackebrandt, E., Goodfellow, M., Eds.; Wiley: Chichester, UK, 1991; pp. 115–175. [Google Scholar]
- Yoon, S.H.; Ha, S.M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef] [PubMed]
- Wattam, A.R.; Abraham, D.; Dalay, O.; Disz, T.L.; Driscoll, T.; Gabbard, J.L.; Gillespie, J.J.; Gough, R.; Hix, D.; Kenyon, R.; et al. PATRIC, the bacterial bioinformatics database and analysis resource. Nucleic Acids Res. 2014, 42, D581–D591. [Google Scholar] [CrossRef] [PubMed]
- Richter, M.; Rosselló-Móra, R.; Oliver Glöckner, F.; Peplies, J. JSpeciesWS: A web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 2016, 32, 929–931. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef]
- Wattam, A.R.; Gabbard, J.L.; Shukla, M.; Sobral, B.W. Comparative genomic analysis at the PATRIC, a bioinformatic resource center. Methods Mol. Biol. 2014, 1197, 287–308. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Stamatakis, A.; Hoover, P.; Rougemont, J. A rapid bootstrap algorithm for the RAxML Web servers. Syst. Biol. 2008, 57, 758–771. [Google Scholar] [CrossRef]
- Davis, J.J.; Gerdes, S.; Olsen, G.J.; Olson, R.; Pusch, G.D.; Shukla, M.; Vonstein, V.; Wattam, A.R.; Yoo, H. PATtyFams: Protein Families for the Microbial Genomes in the PATRIC Database. Front. Microbiol. 2016, 7, 118. [Google Scholar] [CrossRef]
- Wattam, A.R.; Davis, J.J.; Assaf, R.; Boisvert, S.; Brettin, T.; Bun, C.; Conrad, N.; Dietrich, E.M.; Disz, T.; Gabbard, J.L.; et al. Improvements to PATRIC, the all-bacterial Bioinformatics Database and Analysis Resource Center. Nucleic Acids Res. 2017, 45, D535–D542. [Google Scholar] [CrossRef]
- Brettin, T.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Olsen, G.J.; Olson, R.; Overbeek, R.; Parrello, B.; Pusch, G.D.; et al. RASTtk: A modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 2015, 5, 8365. [Google Scholar] [CrossRef] [PubMed]
- Darling, A.C.; Mau, B.; Blattner, F.R.; Perna, N.T. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004, 14, 1394–1403. [Google Scholar] [CrossRef] [PubMed]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, M.J.; Petty, N.K.; Beatson, S.A. Easyfig: A genome comparison visualizer. Bioinformatics 2011, 27, 1009–1010. [Google Scholar] [CrossRef] [PubMed]










| Sample ID | Farm | Geography | Collection Date (Month/Year) | Mycoplasmas (Culture) | Mycoplasma sp. 1654_15-Specific PCR (p37) | M. hyorhinis-Specific PCR (p37) | Chlamydia spp.-Specific PCR | Bacteria (Culture) |
|---|---|---|---|---|---|---|---|---|
| 1654_1 | A | Lower Saxony | 4/2018 | Mycoplasma sp. (ID 1654_1) | positive | negative | negative | negative |
| 1654_2 | A | Lower Saxony | 4/2018 | Positive b | positive | negative | negative | Escherichia coli |
| 1654_3 | A | Lower Saxony | 4/2018 | Positive b | positive | negative | negative | St. chromogenes |
| 1654_4 | A | Lower Saxony | 4/2018 | Mycoplasma sp. (ID 1654_4) | positive | negative | positive | negative |
| 1654_5 | A | Lower Saxony | 4/2018 | Positive b | positive | negative | negative | E. coli |
| 1654_6 | A | Lower Saxony | 4/2018 | Mycoplasma sp. (ID 1654_6) a | positive | negative | negative | negative |
| 1654_7 | A | Lower Saxony | 4/2018 | Mycoplasma sp. (ID 1654_7) | positive | positive | negative | negative |
| 1654_8 | A | Lower Saxony | 4/2018 | Mycoplasma sp. (ID 1654_8) | positive | negative | positive | negative |
| 1654_9 | A | Lower Saxony | 4/2018 | Mycoplasma sp. (ID 1654_9) | positive | negative | negative | negative |
| 1654_10 | A | Lower Saxony | 4/2018 | negative | positive | negative | negative | negative |
| 1654_11 | A | Lower Saxony | 4/2018 | positiveb | positive | negative | negative | St. chromogenes |
| 1654_12 | A | Lower Saxony | 4/2018 | Mycoplasma sp. (ID 1654_12) | positive | positive | positive | negative |
| 1654_13 | A | Lower Saxony | 4/2018 | Mycoplasma sp. (ID 1654_13) a | positive | negative | negative | negative |
| 1654_14 | A | Lower Saxony | 4/2018 | Positive b | positive | negative | negative | E. coli |
| 1654_15 | A | Lower Saxony | 4/2018 | Mycoplasma sp. (ID 1654_15) * a | positive | negative | negative | negative |
| 2184_1 | B | North Rhine-Westphalia | 11/2018 | Mycoplasma sp. (ID 2184_1) a | positive | negative | negative | negative |
| 2184_2 | B | North Rhine-Westphalia | 11/2018 | Mycoplasma sp. (ID 2184_2) | positive | negative | negative | negative |
| 2184_3 | B | North Rhine-Westphalia | 11/2018 | Mycoplasma sp. (ID 2184_3) a | positive | negative | negative | negative |
| 2184_4 | B | North Rhine-Westphalia | 11/2018 | Mycoplasma sp. (ID 2184_4) | positive | negative | negative | negative |
| 2184_5 | B | North Rhine-Westphalia | 11/2018 | Mycoplasma sp. (ID 2184_5) | positive | negative | positive | negative |
| 2184_6 | B | North Rhine-Westphalia | 11/2018 | Mycoplasma sp. (ID 2184_6) a | positive | positive | positive | negative |
| 2184_7 | B | North Rhine-Westphalia | 11/2018 | Positive b | positive | negative | negative | E. coli |
| 2184_8 | B | North Rhine-Westphalia | 11/2018 | negative | positive | negative | negative | negative |
| 338_1 | C | North Rhine-Westphalia | 1/2020 | Mycoplasma sp. (ID 338_1) | positive | positive | positive | negative |
| 338_2 | C | North Rhine-Westphalia | 1/2020 | Mycoplasma sp. (ID 338_2) | positive | negative | negative | negative |
| 338_3 | C | North Rhine-Westphalia | 1/2020 | Mycoplasma sp. (ID 338_3) | positive | negative | negative | negative |
| 338_4 | C | North Rhine-Westphalia | 1/2020 | Mycoplasma sp. (ID 338_4) a | positive | negative | positive | negative |
| 338_5 | C | North Rhine-Westphalia | 1/2020 | Mycoplasma sp. (ID 338_5) a | positive | negative | negative | negative |
| 338_6 | C | North Rhine-Westphalia | 1/2020 | Mycoplasma sp. (ID 338_6) | positive | negative | negative | St. hyicus |
| 338_7 | C | North Rhine-Westphalia | 1/2020 | Mycoplasma sp. (ID 338_7) | positive | negative | negative | St. hyicus |
| 338_8 | C | North Rhine-Westphalia | 1/2020 | Mycoplasma sp. (ID 338_8) a | positive | negative | negative | negative |
| 338_9 | C | North Rhine-Westphalia | 1/2020 | Mycoplasma sp. (ID 338_9) | positive | negative | negative | negative |
| 338_10 | C | North Rhine-Westphalia | 1/2020 | Mycoplasma sp. (ID 338_10) | positive | positive | negative | negative |
| 338_11 | C | North Rhine-Westphalia | 1/2020 | Mycoplasma sp. (ID 338_11) | positive | negative | positive | negative |
| 338_12 | C | North Rhine-Westphalia | 1/2020 | Mycoplasma sp. (ID 338_12) | positive | negative | negative | negative |
| Mycoplasma Isolate/Strain | TYLT | TIL | TUL | TIA | OXY | GEN | SPE | LINC | FFN | ENR |
|---|---|---|---|---|---|---|---|---|---|---|
| 1654_6 | 8 | >64 | >64 | ≤0.25 | 0.5 | 1 | 4 | 2 | 2 | 4 |
| 1654_13 | 8 | >64 | >64 | ≤0.25 | 0.5 | 1 | 4 | 2 | 2 | 4 |
| 1654_15 | 8 | >64 | >64 | ≤0.25 | 0.5 | 1 | 4 | 2 | 2 | 4 |
| 2184_1 | 16 | >64 | >64 | ≤0.25 | 0.5 | 2 | 4 | 4 | 2 | 8 |
| 2184_3 | 16 | >64 | >64 | ≤0.25 | 0.5 | 2 | 4 | 4 | 2 | 8 |
| 2184_6 | 16 | >64 | >64 | ≤0.25 | 0.5 | 2 | 4 | 4 | 2 | 8 |
| 338_4 | 16 | >64 | >64 | ≤0.25 | 0.5 | 1 | 4 | 2 | 2 | 4 |
| 338_5 | 16 | >64 | >64 | ≤0.25 | 0.5 | 1 | 4 | 2 | 2 | 4 |
| 338_8 | 16 | >64 | >64 | ≤0.25 | 0.5 | 1 | 4 | 2 | 2 | 4 |
| BTS-7T | ≤0.25 | 1 | 2 | ≤0.25 | ≤0.25 | 1 | 4 | ≤0.25 | 1 | 0.50 |
| Primer | Sequence | Product Size |
|---|---|---|
| p37-F | 5′-TTTCACCGGCAGACTGAGAC-3′ | 322 bp |
| p37-R | 5′-GCTGGAGTCACAACATCTGGA-3′ | |
| iolA-F | 5′-GCTGCTGTTTCAATGGGAGC-3′ | 225 bp |
| iolA-R | 5′-AGCAGGATTACCAAGCGGAA-3′ | |
| iolB-F | 5′-ACAAGTGCTCTTCTGCTTCGA-3′ | 239 bp |
| iolB-R | 5′-ACATCACCATCCACAGCCTG-3′ | |
| iolC-F | 5′-CACCGCCACCGTATCCTTTT-3′ | 696 bp |
| iolC-R | 5′-ACATCGGAGGATCAACTGCA-3′ | |
| iolD-F | 5′-AGCGCAGAACTAGCTTGTGA-3′ | 464 bp |
| iolD-R | 5′-GCAGCGAATATGCTAACCGC-3′ | |
| iolE-F | 5′-ATGTCTTCACCTGTGTGGG-3′ | 478 bp |
| iolE-R | 5′-CACAAGCGGGCTATCAAGGA-3′ | |
| iolG-F | 5′-AGTCCAACAGCACATCATCCA-3′ | 333 bp |
| iolG-R | 5′-TAAATCGTGCGCTCCGACAT-3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hennig-Pauka, I.; Sudendey, C.; Kleinschmidt, S.; Ruppitsch, W.; Loncaric, I.; Spergser, J. Swine Conjunctivitis Associated with a Novel Mycoplasma Species Closely Related to Mycoplasma hyorhinis. Pathogens 2021, 10, 13. https://doi.org/10.3390/pathogens10010013
Hennig-Pauka I, Sudendey C, Kleinschmidt S, Ruppitsch W, Loncaric I, Spergser J. Swine Conjunctivitis Associated with a Novel Mycoplasma Species Closely Related to Mycoplasma hyorhinis. Pathogens. 2021; 10(1):13. https://doi.org/10.3390/pathogens10010013
Chicago/Turabian StyleHennig-Pauka, Isabel, Christoph Sudendey, Sven Kleinschmidt, Werner Ruppitsch, Igor Loncaric, and Joachim Spergser. 2021. "Swine Conjunctivitis Associated with a Novel Mycoplasma Species Closely Related to Mycoplasma hyorhinis" Pathogens 10, no. 1: 13. https://doi.org/10.3390/pathogens10010013
APA StyleHennig-Pauka, I., Sudendey, C., Kleinschmidt, S., Ruppitsch, W., Loncaric, I., & Spergser, J. (2021). Swine Conjunctivitis Associated with a Novel Mycoplasma Species Closely Related to Mycoplasma hyorhinis. Pathogens, 10(1), 13. https://doi.org/10.3390/pathogens10010013





