Can Previous Associations of Single Nucleotide Polymorphisms in the TLR2, NOD1, CXCR5, and IL10 Genes in the Susceptibility to and Severity of Chlamydia trachomatis Infections Be Confirmed?
Abstract
:1. Introduction
2. Results
2.1. DNA Isolation and SNP Determination
2.2. Susceptibility to CT Infection
2.3. Severity of CT Infection
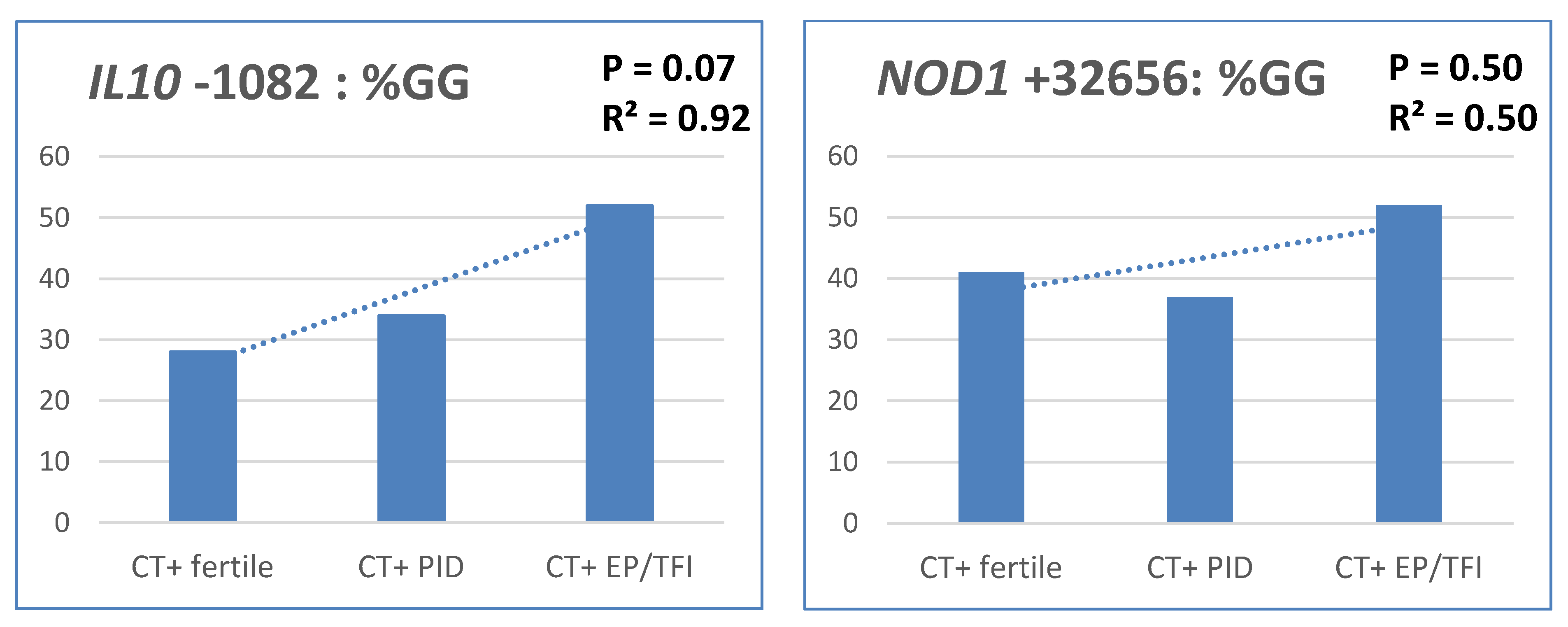
3. Discussion
4. Materials and Methods
4.1. Studied Cohorts
4.2. SNP Determination
4.3. Data Analyses
4.3.1. Susceptibility Analyses: Cohorts 1–3
4.3.2. Severity Analyses: Cohorts 1,2,4,5
4.3.3. Sensitivity Analysis
4.4. METC Approval
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Appendix A
| Susceptibility | NOD1 + 32656 | TLR2 + 2477 | CXCR5 + 10950 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Sensivisity Analysis | TT | TGG | GGGG | GG | GA | AA | TT | TC | CC | ||
| Cohort 1 (selection), 2 | 103 (53.6%) | 79 (41.1%) | 10 (5.2%) | Cohort 1 (negatives), 2 | 191 (91.1%) | 19 (9.0%) | 0 (0.0%) | 77 (36.7%) | 103 (49.0%) | 30 (14.3%) | |
| CT | Negative | 20 (51.3%) | 18 (46.2%) | 1 (2.6%) | Negative | 89 (90.8%) | 9 (9.2%) | 0 (0.0%) | 35 (35.5%) | 50 (51.0%) | 13 (13.3%) |
| Positive | 83 (54.2%) | 61 (39.9%) | 9 (5.6%) | Positive | 102 (91.1%) | 10 (8.9%) | 0 (0.0%) | 42 (37.5%) | 53 (47.3%) | 17 (15.2%) | |
| Severity | NOD1 + 32656 | TLR2 + 2477 | ||||||
|---|---|---|---|---|---|---|---|---|
| Analysis I: Cohorts 1 (selection), 2 | TT | TGG | GGGG | Analysis I: Cohort 2 | GG | GA | AA | |
| CT + total | 80 (54.1%) | 59 (39.9%) | 9 (6.1%) | CT + total | 98 (91.6%) | 9 (8.4%)) | 0 (0%) | |
| CT + AS | 55 (61.1%) | 29 (32.2%) | 6 (6.7%) | CT + AS | 60 (95.2%) | 3 (4.8%) | 0 (0%) | |
| CT + S | 25 (43.1%) | 30 (51.7%) | 3 (5.2%)) | CT + S | 38 (86.4%) | 6 (13.6%) | 0 (0%) | |
| p value | p for *GG vs. TT = 0.03 ** | p value | p for *A vs. GG = 0.08 | |||||
| Analysis II: Cohort 5 | TT | TGG | GGGG | |||||
| Total | 97 (59.9%) | 57 (35.2%) | 8 (4.9%) | |||||
| Controls | 69 (59.5%) | 40 (34.5%) | 7 (6.0%) | |||||
| Cases | 28 (60.9%) | 17 (37.0%) | 1 (2.2%) | |||||
| p value | for *GG vs. TT = 0.87 | |||||||
| Susceptibility | NOD1: *GG vs. TT | |
|---|---|---|
| Cohorts | OR crude | 0.9 (95%CI: 0.4−1.8) |
| 1 (selection), 2 | p value crude | 0.74 |
| OR MLR | 0.9 (95%CI: 0.4−1.9) | |
| p value MLR | 0.79 |
| Severity | NOD1: *GG vs. TT | TLR2: *A vs. GG | |||
|---|---|---|---|---|---|
| Cohorts | OR crude | 2.1 (95%CI: 1.1−4.1) | Cohort 2 | OR crude | 3.2 (95%CI: 0.7−13.4) |
| 1 (selection), 2 | p value crude | 0.03 ** | p value crude | 0.10 | |
| OR MLR | 2.0 (95%CI: 1.0−4.0) | OR MLR | 3.1 (95%CI: 0.7−13.1) | ||
| p value MPL | 0.04 ** | p value MPL | 0.13 | ||
| Cohort 5 | OR crude | 1.1 (95%CI: 0.5−2.1) | |||
| p value crude | 0.87 | ||||
| OR MLR | 1.0 (95%CI: 0.5−2.0) | ||||
| p value MLR | 0.91 | ||||

References
- Lal, J.A.; Malogajski, J.; Verweij, S.P.; De Boer, P.; Ambrosino, E.; Brand, A.; Ouburg, S.; Morre, S.A. Chlamydia trachomatis infections and subfertility: Opportunities to translate host pathogen genomic data into public health. Public Health Genom. 2013, 16, 50–61. [Google Scholar] [CrossRef]
- WHO. Sexually Transmitted Infections (STIs). Available online: http://www.who.int/mediacentre/factsheets/fs110/en/ (accessed on 5 January 2021).
- Staritsky, L.; van Aar, F.; Visser, M.; op de Coul, E.; Heijne, J.; Götz, H.; Nielen, M.; van Sighem, A.; van Benthem, B. Sexually transmitted infections in the Netherlands in 2019. In Seksueel Overdraagbare Aandoeningen in Nederland in 2019; Rijksinstituut voor Volksgezondheid en Milieu RIVM: Bilthoven, The Netherlands, 2020. [Google Scholar] [CrossRef]
- Black, C.M. Current methods of laboratory diagnosis of Chlamydia trachomatis infections. Clin. Microbiol. Rev. 1997, 10, 160–184. [Google Scholar] [CrossRef]
- Den Hartog, J.E.; Ouburg, S.; Land, J.A.; Lyons, J.M.; Ito, J.I.; Pena, A.S.; Morre, S.A. Do host genetic traits in the bacterial sensing system play a role in the development of Chlamydia trachomatis-associated tubal pathology in subfertile women? BMC Infect. Dis. 2006, 6, 122. [Google Scholar] [CrossRef] [Green Version]
- Spaargaren, J. A Multidisciplinary Approach to the Study of Chlamydia Trachomatis Infections: Female Urogenital and Male Anorectal Infections; VU Amsterdam: Amsterdam, The Netherlands, 2006; Available online: www.immunogenetics.nl (accessed on 12 May 2020).
- den Hartog, J.E.; Lyons, J.M.; Ouburg, S.; Fennema, J.S.; de Vries, H.J.; Bruggeman, C.A.; Ito, J.I.; Pena, A.S.; Land, J.A.; Morre, S.A. TLR4 in Chlamydia trachomatis infections: Knockout mice, STD patients and women with tubal factor subfertility. Drugs Today 2009, 45 (Suppl. B), 75–82. [Google Scholar]
- Hoenderboom, B.M.; van Benthem, B.H.B.; van Bergen, J.; Dukers-Muijrers, N.; Gotz, H.M.; Hoebe, C.; Hogewoning, A.A.; Land, J.A.; van der Sande, M.A.B.; Morre, S.A.; et al. Relation between Chlamydia trachomatis infection and pelvic inflammatory disease, ectopic pregnancy and tubal factor infertility in a Dutch cohort of women previously tested for chlamydia in a chlamydia screening trial. Sex. Transm. Infect. 2019, 95, 300–306. [Google Scholar] [CrossRef] [Green Version]
- Bailey, R.L.; Natividad-Sancho, A.; Fowler, A.; Peeling, R.W.; Mabey, D.C.; Whittle, H.C.; Jepson, A.P. Host genetic contribution to the cellular immune response to Chlamydia trachomatis: Heritability estimate from a Gambian twin study. Drugs Today 2009, 45 (Suppl. B), 45–50. [Google Scholar]
- Morre, S.A.; Karimi, O.; Ouburg, S. Chlamydia trachomatis: Identification of susceptibility markers for ocular and sexually transmitted infection by immunogenetics. FEMS Immunol. Med. Microbiol. 2009, 55, 140–153. [Google Scholar] [CrossRef] [PubMed]
- Mascellino, M.T.; Boccia, P.; Oliva, A. Immunopathogenesis in Chlamydia trachomatis Infected Women. ISRN Obstet. Gynecol. 2011, 2011, 436936. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Broeze, K.A.; Opmeer, B.C.; Coppus, S.F.; Van Geloven, N.; Den Hartog, J.E.; Land, J.A.; Van der Linden, P.J.; Ng, E.H.; Van der Steeg, J.W.; Steures, P.; et al. Integration of patient characteristics and the results of Chlamydia antibody testing and hysterosalpingography in the diagnosis of tubal pathology: An individual patient data meta-analysis. Hum. Reprod. 2012, 27, 2979–2990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Darville, T.; O’Neill, J.M.; Andrews, C.W., Jr.; Nagarajan, U.M.; Stahl, L.; Ojcius, D.M. Toll-like receptor-2, but not Toll-like receptor-4, is essential for development of oviduct pathology in chlamydial genital tract infection. J. Immunol. 2003, 171, 6187–6197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verweij, S.P.; Karimi, O.; Pleijster, J.; Lyons, J.M.; de Vries, H.J.; Land, J.A.; Morre, S.A.; Ouburg, S. TLR2, TLR4 and TLR9 genotypes and haplotypes in the susceptibility to and clinical course of Chlamydia trachomatis infections in Dutch women. Pathog. Dis. 2016, 74, ftv107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brankovic, I.; van Ess, E.F.; Noz, M.P.; Wiericx, W.A.; Spaargaren, J.; Morre, S.A.; Ouburg, S. NOD1 in contrast to NOD2 functional polymorphism influence Chlamydia trachomatis infection and the risk of tubal factor infertility. Pathog. Dis. 2015, 73, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karimi, O.; Jiang, J.; Ouburg, S.; Champion, C.; Khurana, A.; Liu, G.; Poya, H.; Freed, A.; Pleijster, J.; Rosengurt, N.; et al. CXCR5 Regulates Chlamydia Tubal Pahtology in Mice and Humans; 2011. Available online: https://research.vu.nl/files/42210838/chapter%2005.pdf (accessed on 12 May 2020).
- Ohman, H.; Tiitinen, A.; Halttunen, M.; Lehtinen, M.; Paavonen, J.; Surcel, H.M. Cytokine polymorphisms and severity of tubal damage in women with Chlamydia-associated infertility. J. Infect. Dis. 2009, 199, 1353–1359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ho, D.S.W.; Schierding, W.; Wake, M.; Saffery, R.; O’Sullivan, J. Machine Learning SNP Based Prediction for Precision Medicine. Front. Genet. 2019, 10, 267. [Google Scholar] [CrossRef] [Green Version]
- Zou, Y.; Lei, W.; He, Z.; Li, Z. The role of NOD1 and NOD2 in host defense against chlamydial infection. FEMS Microbiol. Lett. 2016, 363. [Google Scholar] [CrossRef] [Green Version]
- Kinnunen, A.H.; Surcel, H.M.; Lehtinen, M.; Karhukorpi, J.; Tiitinen, A.; Halttunen, M.; Bloigu, A.; Morrison, R.P.; Karttunen, R.; Paavonen, J. HLA DQ alleles and interleukin-10 polymorphism associated with Chlamydia trachomatis-related tubal factor infertility: A case-control study. Hum. Reprod. 2002, 17, 2073–2078. [Google Scholar] [CrossRef]
- Sabat, R.; Grutz, G.; Warszawska, K.; Kirsch, S.; Witte, E.; Wolk, K.; Geginat, J. Biology of interleukin-10. Cytokine Growth Factor Rev. 2010, 21, 331–344. [Google Scholar] [CrossRef] [Green Version]
- Scapini, P.; Lamagna, C.; Hu, Y.; Lee, K.; Tang, Q.; DeFranco, A.L.; Lowell, C.A. B cell-derived IL-10 suppresses inflammatory disease in Lyn-deficient mice. Proc. Natl. Acad. Sci. USA 2011, 108, E823–E832. [Google Scholar] [CrossRef] [Green Version]
- Moore, K.W.; de Waal Malefyt, R.; Coffman, R.L.; O’Garra, A. Interleukin-10 and the interleukin-10 receptor. Annu. Rev. Immunol. 2001, 19, 683–765. [Google Scholar] [CrossRef]
- Turner, D.M.; Williams, D.M.; Sankaran, D.; Lazarus, M.; Sinnott, P.J.; Hutchinson, I.V. An investigation of polymorphism in the interleukin-10 gene promoter. Eur. J. Immunogenet. 1997, 24, 1–8. [Google Scholar] [CrossRef]
- Ohman, H.; Tiitinen, A.; Halttunen, M.; Birkelund, S.; Christiansen, G.; Koskela, P.; Lehtinen, M.; Paavonen, J.; Surcel, H.M. IL-10 polymorphism and cell-mediated immune response to Chlamydia trachomatis. Genes Immun. 2006, 7, 243–249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gibson, A.W.; Edberg, J.C.; Wu, J.; Westendorp, R.G.; Huizinga, T.W.; Kimberly, R.P. Novel single nucleotide polymorphisms in the distal IL-10 promoter affect IL-10 production and enhance the risk of systemic lupus erythematosus. J. Immunol. 2001, 166, 3915–3922. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.; Tang, J.; Geisler, W.M.; Crowley-Nowick, P.A.; Wilson, C.M.; Kaslow, R.A. Human leukocyte antigen and cytokine gene variants as predictors of recurrent Chlamydia trachomatis infection in high-risk adolescents. J. Infect. Dis. 2005, 191, 1084–1092. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, E.; Mira, J.P.; Cornish, K.L.; Arbour, N.C.; Schwartz, D.A. A novel polymorphism in the toll-like receptor 2 gene and its potential association with staphylococcal infection. Infect. Immun. 2000, 68, 6398–6401. [Google Scholar] [CrossRef] [PubMed]
- Genome of the Netherlands, C. Whole-genome sequence variation, population structure and demographic history of the Dutch population. Nat. Genet. 2014, 46, 818–825. [Google Scholar] [CrossRef] [PubMed]
- Ouburg, S.; Spaargaren, J.; den Hartog, J.E.; Land, J.A.; Fennema, J.S.; Pleijster, J.; Pena, A.S.; Morre, S.A.; Consortium, I. The CD14 functional gene polymorphism -260 C>T is not involved in either the susceptibility to Chlamydia trachomatis infection or the development of tubal pathology. BMC Infect. Dis. 2005, 5, 114. [Google Scholar] [CrossRef] [Green Version]
- Verweij, S.P.; Bax, C.J.; Quint, K.D.; Quint, W.G.; van Leeuwen, A.P.; Peters, R.P.; Oostvogel, P.M.; Mutsaers, J.A.; Dorr, P.J.; Pleijster, J.; et al. Significantly higher serologic responses of Chlamydia trachomatis B group serovars versus C and I serogroups. Drugs Today 2009, 45 (Suppl. B), 135–140. [Google Scholar]
- Lanjouw, E.; Brankovic, I.; Pleijster, J.; Spaargaren, J.; Hoebe, C.J.; van Kranen, H.J.; Ouburg, S.; Morre, S.A. Specific polymorphisms in the vitamin D metabolism pathway are not associated with susceptibility to Chlamydia trachomatis infection in humans. Pathog. Dis. 2016, 74. [Google Scholar] [CrossRef] [Green Version]
- Jansen, M.E.; Brankovic, I.; Spaargaren, J.; Ouburg, S.; Morre, S.A. Potential protective effect of a G > A SNP in the 3’UTR of HLA-A for Chlamydia trachomatis symptomatology and severity of infection. Pathog. Dis. 2016, 74. [Google Scholar] [CrossRef] [Green Version]
- Hoenderboom, B.M.; van Oeffelen, A.A.; van Benthem, B.H.; van Bergen, J.E.; Dukers-Muijrers, N.H.; Gotz, H.M.; Hoebe, C.J.; Hogewoning, A.A.; van der Klis, F.R.; van Baarle, D.; et al. The Netherlands Chlamydia cohort study (NECCST) protocol to assess the risk of late complications following Chlamydia trachomatis infection in women. BMC Infect. Dis. 2017, 17, 264. [Google Scholar] [CrossRef]
- He, C.; Holme, J.; Anthony, J. SNP genotyping: The KASP assay. Methods Mol. Biol. 2014, 1145, 75–86. [Google Scholar] [CrossRef] [PubMed]

| Susceptibility | IL-10−1082 | NOD1 + 32656 | TLR2 + 2477 | CXCR5 + 10950 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AA | AG | GG | TT | TGG | GGGG | GG | GA | AA | TT | TC | CC | ||
| Cohorts 1,2 | 92 (30.3%) | 133 (43.8%) | 79 (26.0%) | 167 (54.9%) | 121 (39.8%) | 16 (5.3%) | 280 (92.1%) | 24 (7.9%) | 0 (0%) | 111 (36.5%) | 146 (48.0%) | 47 (15.5%) | |
| CT | Negative | 34 (34.7%) | 38 (38.8%) | 26 (26.5%) | 52 (53.1%) | 42 (42.9%) | 4 (4.1%) | 89 (90.8%) | 9 (9.2%) | 0 (0%) | 35 (35.7%) | 50 (51.0%) | 13 (13.3%) |
| Positive | 58 (28.2%) | 95 (46.1%) | 53 (25.7%) | 115 (55,8%) | 79 (38.3%) | 12 (5.8%) | 191 (92.7%) | 15 (7.3%)) | 0 (0%) | 76 (36.9%) | 96 (46.6%) | 34 (16.5%) | |
| Cohort 3 | 202 (28.6%) | 365 (51.6%) | 140 (19.8%) | 427 (60.4%) | 229 (32.4%) | 5 (7.2%) | 674 (95.3%) | 30 (4.2%) | 3 (0.4%) | 246 (34.8%) | 338 (47.8%) | 123 (17.4%) | |
| CT | Negative | 184 (29.4%) | 313 (50.0%) | 129 (20.6%) | 374 (59.7%) | 204 (32.6%) | 48 (7.7%) | 596 (95.2%) | 27 (4.3%) | 3 (0.5%) | 216 (34.5%) | 300 (47.9%) | 110 (17.6%) |
| Positve | 18 (22.2%) | 52 (64.2%) | 11 (13.6%) | 53 (65.4%) | 25 (30.9%) | 3 (3.7%) | 78 (96.3%) | 3 (3.7%) | 0 (0%) | 30 (37.0%) | 38 (46.9%) | 13 (16.0%) | |
| Severity | IL-10–1082 | NOD1 + 32656 | TLR2 + 2477 | CXCR5 + 10950 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Analysis I: Cohorts 1,2 | AA | AG | GG | TT | TGG | GGGG | GG | GA | AA | TT | TC | CC |
| CT + total | 56 (27.9%) | 94 (46.8%) | 51 (25.4%) | 112 (55.7%) | 77 (38,3%) | 12 (6.0%) | 187 (93.0%) | 14 (7.0%)) | 0 (0%) | 74 (36.8%) | 94 (46.8%) | 33 (16.4%) |
| CT + AS | 34 (29.6%) | 51 (44.3%) | 30 (26.1%) | 72 (62.6%) | 36 (31.3%) | 7 (6.1%) | 110 (95.7%) | 5 (4.3%) | 0 (0%) | 42 (36.5%) | 55 (47.8%) | 18 (15.7%) |
| CT + S | 22 (25.6%) | 43 (45.7%) | 21 (24.4%) | 40 (46.5%) | 41 (47.7%) | 5 (5.8%) | 77 (89.5%) | 9 (10.5%) | 0 (0%) | 32 (37.2%) | 39 (45.4%) | 15 (17.4%) |
| p value | p for GG vs. A* = 0.79 | p for *GG vs. TT = 0.02 ** | p for *A vs. GG = 0.10 | p for CC vs. T* = 0.74 | ||||||||
| Analysis II: Cohorts 4,5 | AA | AG | GG | TT | TGG | GGGG | GG | GA | AA | TT | TC | CC |
| Total | 45 (25.9%) | 73 (42.0%) | 56 (32.2%) | 102 (58.6%) | 64 (36.8%) | 8 (4.6%) | 165 (94.8%) | 7 (4.0%) | 2 (1.1%) | 63 (36.2%) | 80 (46.0%) | 31 (17.8%) |
| Controls | 31 (26.7%) | 53 (45.7%) | 32 (27.6%) | 69 (59.5%) | 40 (34.5%) | 7 (6.0%) | 109 (94.0%) | 5 (4.3%) | 2 (1.7%) | 44 (37.9%) | 55 (47.4%) | 17 (14.7%) |
| Cases | 14 (24.1%) | 20 (34.5%) | 24 (41.4%) | 33 (56.9%) | 24 (41.4%) | 1 (1.7%) | 56 (96,6%) | 2 (3,4%) | 0 (0.0%) | 19 (32.8%) | 25 (43.1%) | 14 (24.1%) |
| p value | p for GG vs. A* = 0.07 | p for *GG vs. TT = 0.74 | p for *A vs. GG = 0.47 | p for CC vs. T* = 0.12 | ||||||||
| Susceptibility | IL10: GG vs. A* | NOD1: *GG vs. TT | TLR2: *A vs. GG | CXCR5: *C vs. TT | |
|---|---|---|---|---|---|
| Cohorts 1,2 | OR crude | 1.0 (95%CI: 0.6–1.7) | 0.9 (95%CI: 0.6–1.4) | 0.8 (95%CI: 0.3–1.8) | 1.0 (95%CI: 0.6–1.6) |
| p value crude | 0.88 | 0.65 | 0.57 | 0.84 | |
| OR MLR | 1.0 (95%CI: 0.6–1.7) | 0.9 (95%CI: 0.6–1.5) | 0.8 (95%CI: 0.3–1.9 | 1.0 (95%CI: 0.6–1.6) | |
| p value MLR | 0.90 | 0.69 | 0.60 | 0.85 | |
| Cohort 3 | OR crude | 0.6 (95%CI: 0.3–1.2) | 0.8 (95%CI: 0.5–1.3) | 0.8 (95%CI: 0.3–2.9) | 0.9 (95%CI: 0.6–1.4) |
| p value crude | 0.14 | 0.33 | 0.79 | 0.65 | |
| OR MLR | 0.6 (95%CI: 0.3–1.2) | 0.8 (95%CI: 0.5–1.3) | 0.9 (95%CI: 0.3–2.9) | 0.9 (95%CI: 0.6–1.5) | |
| p value MLR | 0.15 | 0.32 | 0.82 | 0.66 | |
| Severity | IL10: GG vs. A* | NOD1: *GG vs. TT | TLR2: *A vs. GG | CXCR5: CC vs. *T | |
|---|---|---|---|---|---|
| Cohorts 1,2 | OR crude | 0.9 (95%CI: 0.5−1.7) | 1.9 (95%CI: 1.1−3.4) | 2.6 (95%CI: 0.8−8.0) | 1.1 (95%CI: 0.5−2.4) |
| p value crude | 0.79 | 0.02 ** | 0.10 | 0.73 | |
| OR MLR | 0.9 (95%CI: 0.5−1.7) | 1.9 (95%CI: 1.1−3.4) | 2.4 (95%CI: 0.8−7.5) | 1.1 (95%CI: 0.5−2.4) | |
| p value MLR | 0.68 | 0.03 ** | 0.14 | 0.79 | |
| Cohorts 4,5 | OR crude | 1.9 (95%CI: 1.0−3.6) | 1.1 (95%CI: 0.6−2.1)) | 0.6 (95%CI: 0.1−2.8) | 1.9 (95%CI: 0.8−4.1 |
| p value crude | 0.07 | 0.74 | 0.47 | 0.12 | |
| OR MLR | 1.9 (95%CI: 0.9−3.6) | 1.3 (95%CI: 0.6−2.4) | 0.5 (95%CI: 0.1−2.7) | 1.9 (95%CI: 0.8−4.2) | |
| p value MLR | 0.07 | 0.50 | 0.43 | 0.13 |
| Cohort | Cohort Description | n | CT Determination | Chlamydia Outcome | DNA Isolation | Severity Determination |
|---|---|---|---|---|---|---|
| Cohort 1: Patients from STI outpatient clinic Amsterdam | Women under the age of 33. Collected from July 2001 to December 2004 to investigate the role of a CD14 SNP in susceptibility to a CT infection [30]. | 192 | Cervical swabs were used for CT DNA detection by PCR (COBAS AMPLICOR; Hoffman–La Roche, Basel, Switzerland [6]. | Chlamydia negative n = 98 Chlamydia positive Symptomatic n = 42 Asymptomatic n = 52 | DNA was isolated from PBMC using isopropanol isolation [30]. | Women completed a questionnaire regarding their symptoms at that moment. |
| Cohort 2: Patients from STI outpatient clinic the Hague | Collected from January to October 2008 to investigate the differences in IgG response in reaction to an infection by CT serogroup B, Serogroup I or serogroup C [31] | 112 | Cervical, vaginal, and/or urethral swabs and urine specimens were used for CT detection via probe hybridization assays (pace2 assay, Genprobe) [31]. | Chlamydia positive Symptomatic n = 44 Asymptomatic n = 63 Unknown n = 5 | DNA was isolated from serum using Roche High Pure PCR Template Preparation kit. | Information about symptoms was collected at the STI clinic or at the Department of Obstetrics and Gynaecology. |
| Cohort 3: Patients from STI outpatient clinic South-Limburg | Women between 18 and 33 years old, originally used to investigate an association between susceptibility to a CT infection and specific mutations in the vitamin D metabolism [32] | 707 | CT status was assessed using Roche Cobas 4800 NAAT | Chlamydia negative n = 626 Chlamydia positive n = 81 | DNA was isolated from serum with a Hamilton Starlight isolation robot. | NA |
| Cohort 4: Gynaecology cohort from the University Medical Center Groningen | This women were part of a subfertility cohort aiming to investigate the influence of a HLA-A SNP to the severity of a CT infection. We used only the women with laparoscopically confirmed TFI [33]. | 12 | Serum was used for a CAT test (pELISA, Medac Diagnostika, Germany). [33] | DNA was isolated from serum using Roche High Pure PCR Template Preparation kit. | All had undergone laparoscopy | |
| Cohort 5: Subset of Netherlands Chlamydia Cohort Study (NECCST) [34] | Long-term prospective cohort aiming to determine CT complication risk and risk factors among women. Data collection (questionnaires, swabs and blood samples) from 2008–2016 [34]. | 178 | Chlamydia positivity was determined by either a self-reported chlamydia infection, a positive PCR-test outcome in the CSI study and/or the presence of CT IgG antibodies in serum | All positive Self-reported infection n = 164 and/or Positive PCR test n = 26 and/or Presence of CT IgG n = 53 | DNA was isolated from buccal swabs, vaginal swabs, or urine samples using Roche High Pure PCR Template Preparation kit [34]. | Women completed a questionnaire regarding long term complications. PID n = 42 And/or TFI n = 9 And/or Ectopic pregnancy n = 6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jukema, J.B.; Hoenderboom, B.M.; van Benthem, B.H.B.; van der Sande, M.A.B.; de Vries, H.J.C.; Hoebe, C.J.P.A.; Dukers-Muijrers, N.H.T.M.; Bax, C.J.; Morré, S.A.; Ouburg, S. Can Previous Associations of Single Nucleotide Polymorphisms in the TLR2, NOD1, CXCR5, and IL10 Genes in the Susceptibility to and Severity of Chlamydia trachomatis Infections Be Confirmed? Pathogens 2021, 10, 48. https://doi.org/10.3390/pathogens10010048
Jukema JB, Hoenderboom BM, van Benthem BHB, van der Sande MAB, de Vries HJC, Hoebe CJPA, Dukers-Muijrers NHTM, Bax CJ, Morré SA, Ouburg S. Can Previous Associations of Single Nucleotide Polymorphisms in the TLR2, NOD1, CXCR5, and IL10 Genes in the Susceptibility to and Severity of Chlamydia trachomatis Infections Be Confirmed? Pathogens. 2021; 10(1):48. https://doi.org/10.3390/pathogens10010048
Chicago/Turabian StyleJukema, Jelmer B., Bernice M. Hoenderboom, Birgit H. B. van Benthem, Marianne A. B. van der Sande, Henry J. C. de Vries, Christian J. P. A. Hoebe, Nicole H. T. M. Dukers-Muijrers, Caroline J. Bax, Servaas A. Morré, and Sander Ouburg. 2021. "Can Previous Associations of Single Nucleotide Polymorphisms in the TLR2, NOD1, CXCR5, and IL10 Genes in the Susceptibility to and Severity of Chlamydia trachomatis Infections Be Confirmed?" Pathogens 10, no. 1: 48. https://doi.org/10.3390/pathogens10010048






