Correlation between Pathogenic Determinants Associated with Clinically Isolated Non-Typhoidal Salmonella
Abstract
1. Introduction
2. Results
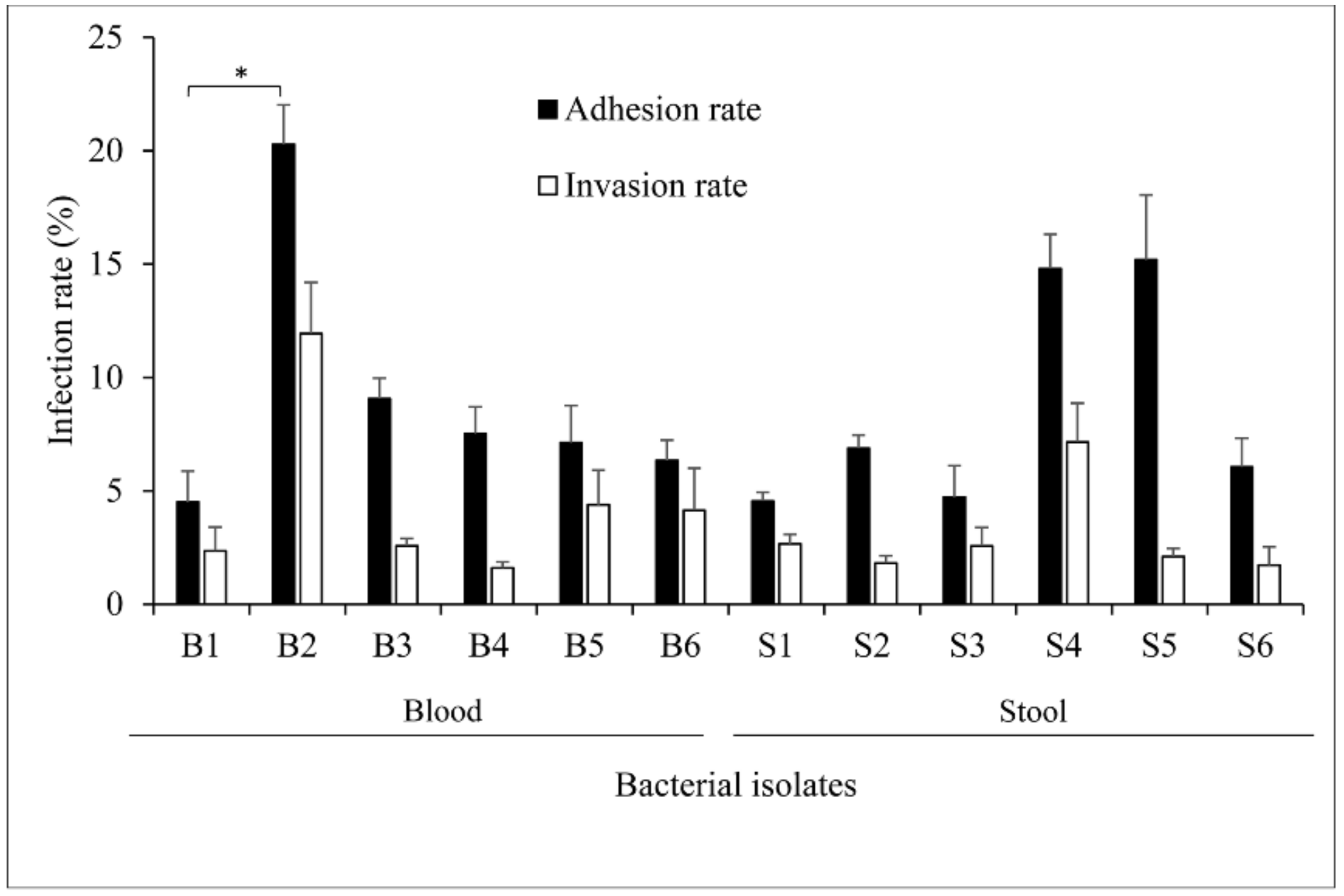
2.1. Overview of Salmonella Isolates Adherence and Invasiveness to Epithelial Cells
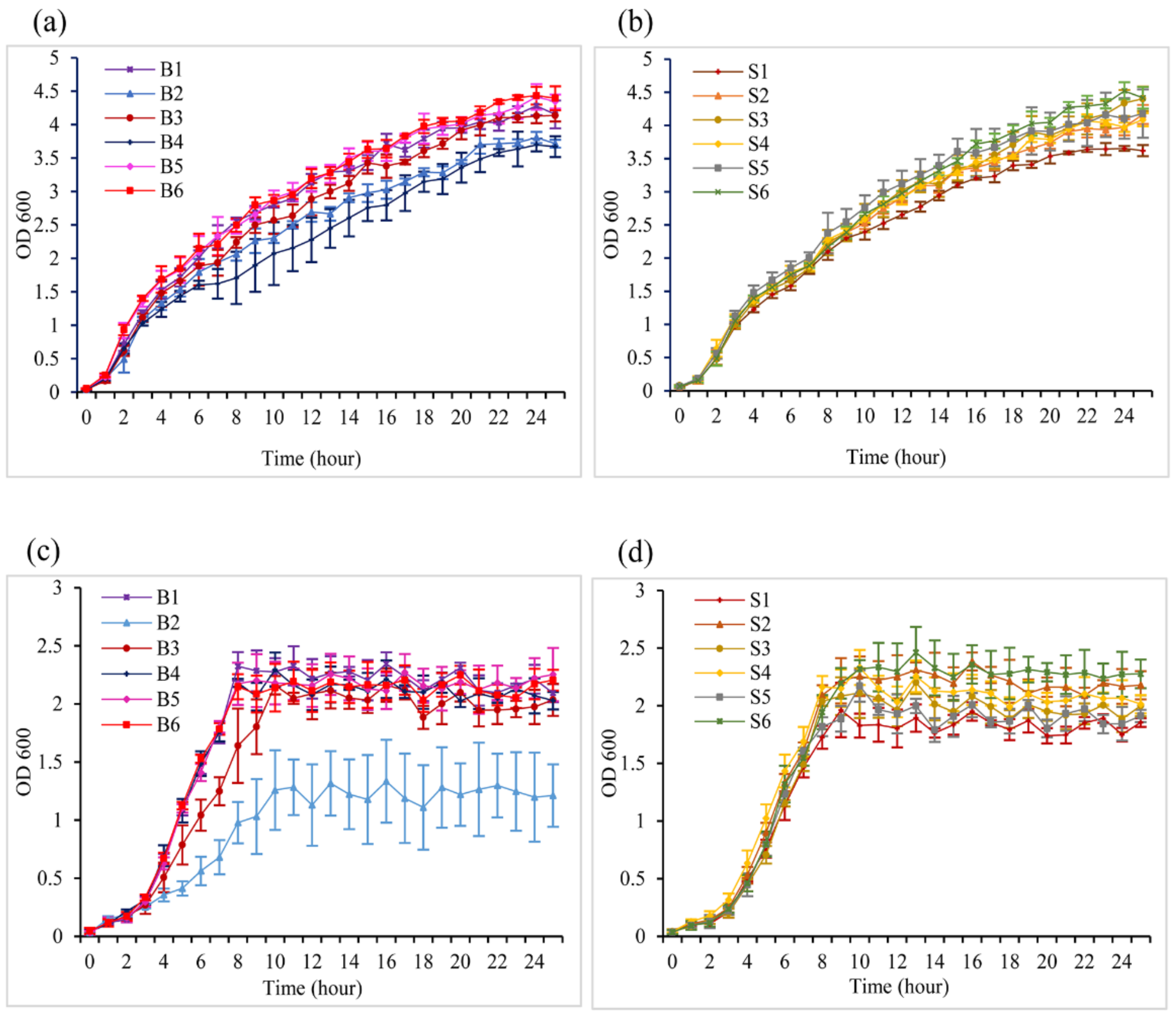
2.2. Environmental Conditions Affecting Bacterial Growth
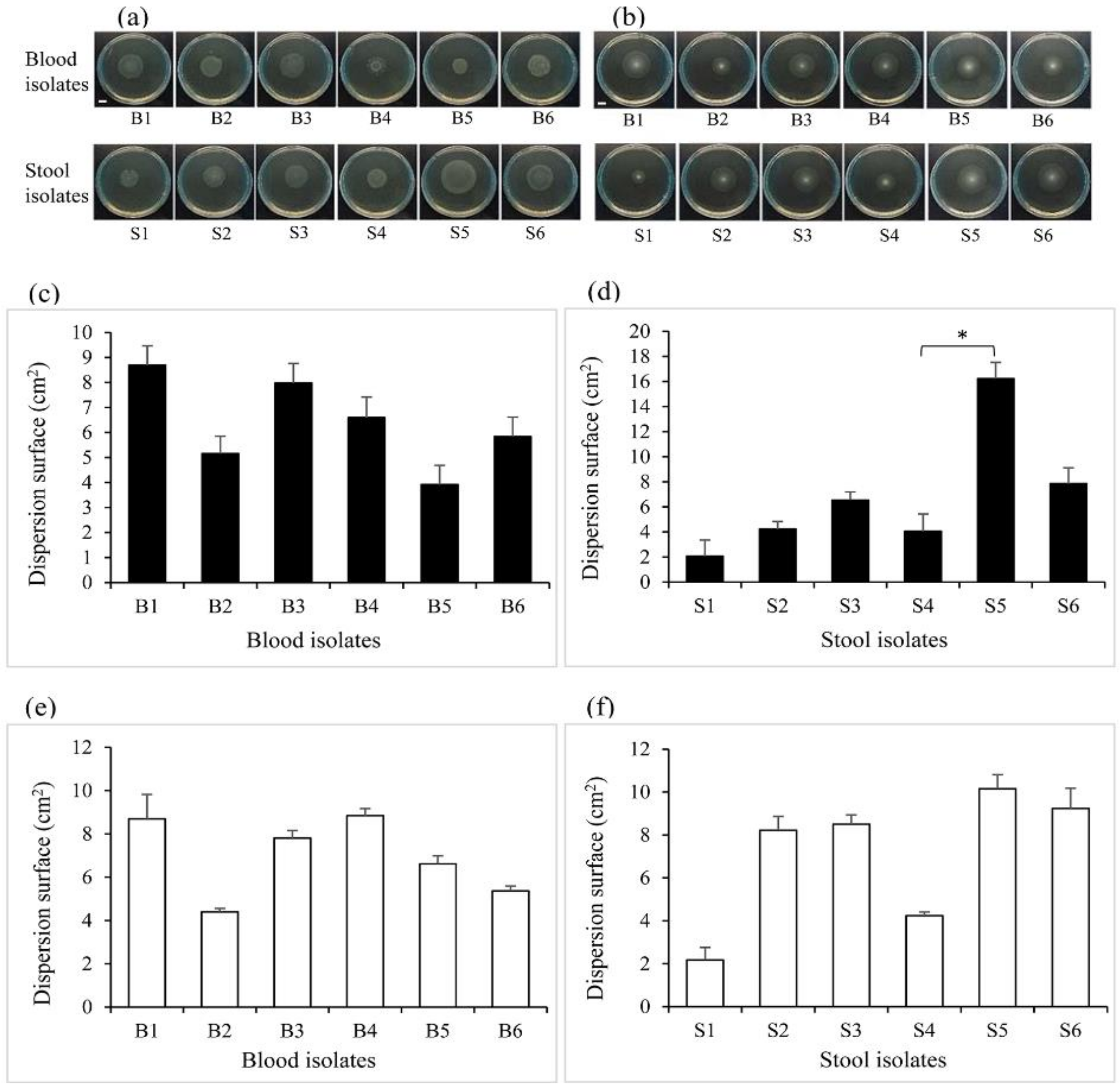
2.3. Motility of the Bacterial Isolates
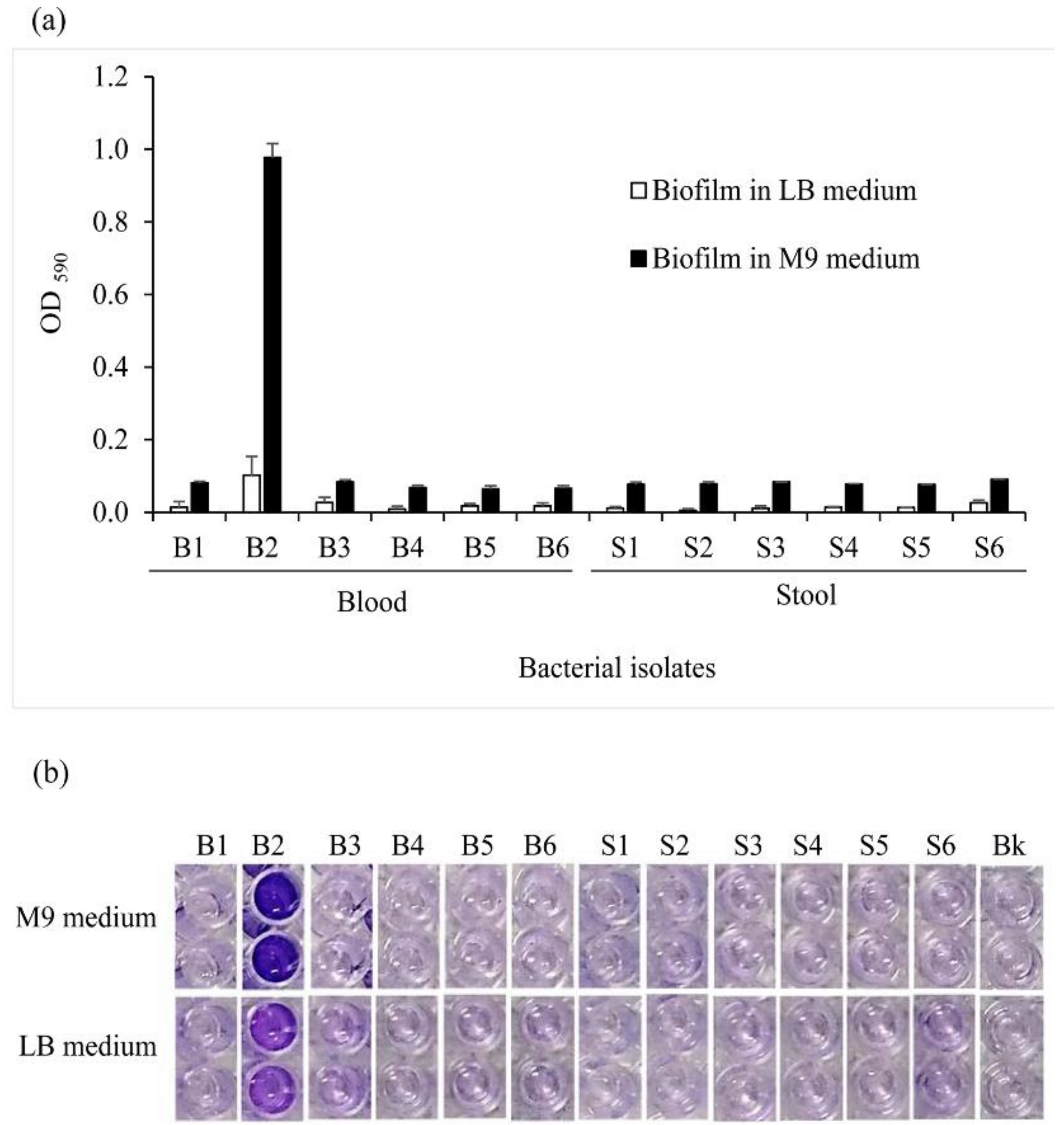
2.4. The Effects of the Intrinsic Nature of the Bacteria and Culture Medium on Biofilm Formation
2.5. Salmonella Isolates Genotyping by Multiplex PCR
2.6. Comparative Virulence Gene Expression and Serotyping
2.7. Salmonella Isolates In Vitro Susceptibility to Antibiotics
3. Discussion
4. Materials and Methods
4.1. Bacterial Samples and Cell Cultures
4.2. Bacterial Adhesion and Invasion Assay
4.3. Bacterial Growth Curve
4.4. Bacterial Motility
4.5. Salmonella Isolates Biofilm Formation
4.6. Virulence Genes Genotyping
4.7. Quantitative-PCR (qPCR) Analysis
4.8. Antibiotic Susceptibility Test and MIC
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Harth, R. In Africa, a Deadly Salmonella Strain Takes Hold. 2015. Available online: https://biodesign.asu.edu/news/africa-deadly-salmonella-strain-takes-hold (accessed on 22 May 2019).
- Yang, J.; Barrila, J.; Roland, K.L.; Kilbourne, J.; Ott, C.M.; Forsyth, R.J.; Nickerson, C.A. Characterization of the Invasive, Multidrug Resistant Non-typhoidal Salmonella Strain D23580 in a Murine Model of Infection. PLoS Negl. Trop. Dis. 2015, 9, e0003839. [Google Scholar] [CrossRef] [PubMed]
- Kagirita, A.A.; Baguma, A.; Owalla, T.J.; Bazira, J.; Majalija, S. Molecular Characterization of Salmonella from Human and Animal Origins in Uganda. Int. J. Bact. 2017, 4604789. [Google Scholar] [CrossRef]
- Kirk, M.D.; Pires, S.M.; Black, R.E.; Caipo, M.; Crump, J.A.; Devleesschauwer, B.; Döpfer, D.; Fazil, A.; Fischer-Walker, C.L.; Hald, T.; et al. World Health Organization Estimates of the Global and Regional Disease Burden of 22 Foodborne Bacterial, Protozoal, and Viral Diseases, 2010: A Data Synthesis. PLoS Med. 2015, 12, e1001921. [Google Scholar] [CrossRef]
- Besser, J.M. Salmonella epidemiology: A whirlwind of change. Food Microbiol. 2018, 71, 55–59. [Google Scholar] [CrossRef] [PubMed]
- Pires, S.M.; Vieira, A.R.; Hald, T.; Cole, D. Source Attribution of Human Salmonellosis: An Overview of Methods and Estimates. Foodborne Pathog. Dis. 2014, 11, 667–676. [Google Scholar] [CrossRef] [PubMed]
- Imanishi, M.; Anderson, T.C.; Routh, J.; Brown, C.; Conidi, G.; Glenn, L.; Reddy, V.; Waechter, H.; Malavet, M.; Nyaku, M.; et al. Salmonellosis and meat purchased at live-bird and animal-slaughter markets, United States, 2007–2012. Emerg. Infect. Dis. 2014, 20, 167–169. [Google Scholar] [CrossRef] [PubMed]
- De Knegt, L.V.; Pires, S.M.; Hald, T. Attributing foodborne salmonellosis in humans to animal reservoirs in the European Union using a multi-country stochastic model. Epidemiol. Infect. 2015, 143, 1175–1186. [Google Scholar] [CrossRef] [PubMed]
- Rezende, A.C.; Crucello, J.; Moreira, R.C.; Silva, B.S.; Sant’Ana, A.S. Incidence and growth of Salmonella enterica on the peel and pulp of avocado (Persea americana) and custard apple (Annona squamosa). Int. J. Food Microbiol. 2016, 235, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Chiou, C.S.; Liao, Y.S.; Liao, C.H.; Tsao, C.S.; Kuo, J.C. Salmonellosis Surveillance and Epidemiological Trend in Taiwan. Taiwan Epidemiol. Bullet. 2015, 31. [Google Scholar] [CrossRef]
- Wu, C.J.; Chen, P.L.; Tang, H.J.; Chen, H.M.; Tseng, F.C.; Shih, H.I.; Hung, Y.P.; Chung, C.H.; Ko, W.C. Incidence of Aeromonas bacteremia in Southern Taiwan: Vibrio and Salmonella bacteremia as comparators. J. Microbiol. Immunol. Infect. 2014, 47, 145–148. [Google Scholar] [CrossRef][Green Version]
- Kuo, H.C.; Lauderdale, T.L.; Lo, D.Y.; Chen, C.L.; Chen, P.C.; Liang, S.Y.; Kuo, J.C.; Liao, Y.S.; Liao, C.H.; Tsao, C.S.; et al. An Association of Genotypes and Antimicrobial Resistance Patterns among Salmonella Isolates from Pigs and Humans in Taiwan. PLoS ONE 2014, 9, e95772. [Google Scholar] [CrossRef] [PubMed]
- Nair, D.V.T.; Venkitanarayanan, K.; Johny, A.K. Antibiotic-Resistant Salmonella in the Food Supply and the Potential Role of Antibiotic Alternatives for Control. Foods 2018, 7, 167. [Google Scholar] [CrossRef]
- Hook, E.W. Salmonellosis: Certain factors influencing the interaction of Salmonella and the human host. Bull. N. Y. Acad. Med. 1961, 37, 499–512. [Google Scholar]
- Peterson, J.W. Bacterial Pathogenesis. In Medical Microbiology, 4th ed.; Baron, S., Ed.; University of Texas Medical Branch at Galveston: Galveston, TX, USA, 1996. [Google Scholar]
- Andino, A.; Hanning, I. Salmonella enterica: Survival, Colonization, and Virulence Differences among Serovars. Sci. World J. 2015, 2015, 16. [Google Scholar] [CrossRef] [PubMed]
- Tapia, M.D.; Tennant, S.M.; Bornstein, K.; Onwuchekwa, U.; Tamboura, B.; Maiga, A.; Sylla, M.; Sissoko, S.; Kourouma, N.; Toure, A.; et al. Invasive Nontyphoidal Salmonella Infections Among Children in Mali, 2002–2014: Microbiological and Epidemiologic Features Guide Vaccine Development. Clin. Infect. Dis. 2015, 61, S332–S338. [Google Scholar] [CrossRef] [PubMed]
- Hakim, S.; Davila, F.; Amin, M.; Hader, I.; Cappell, M.S. Infectious Aortitis: A Life-Threatening Endovascular Complication of Nontyphoidal Salmonella Bacteremia. Case. Rep. Med. 2018, 2018. [Google Scholar] [CrossRef] [PubMed]
- Hume, P.J.; Singh, V.; Davidson, A.C.; Koronakis, V. Swiss Army Pathogen: The Salmonella Entry Toolkit. Front. Cell Infect. Microbiol. 2017, 7, 348. [Google Scholar] [CrossRef]
- Gunn, J.S.; Marshall, J.M.; Baker, S.; Dongol, S.; Charles, R.C.; Ryan, E.T. Salmonella chronic carriage: Epidemiology, diagnosis and gallbladder persistence. Trends Microbiol. 2014, 22, 648–655. [Google Scholar] [CrossRef]
- Rhen, M.; Maskell, D.; Mastroeni, P.; Threlfall, J. Salmonella: Molecular Biology and Pathogenesis, 1st ed.; Horizon Bioscience: Wymondham, Norfolk, UK, 2007. [Google Scholar]
- Kearns, D.B. A field guide to bacterial swarming motility. Nat. Rev. Microbiol. 2010, 8, 634–644. [Google Scholar] [CrossRef]
- Crump, J.A.; Mintz, E.D. Global trends in typhoid and paratyphoid. Fever. Clin. Infect. Dis. 2010, 50, 241–246. [Google Scholar] [CrossRef]
- Suez, J.; Porwollik, S.; Dagan, A.; Marzel, A.; Schorr, Y.I.; Desai, P.T.; Agmon, V.; McClelland, M.; Rahav, G.; Gal-Mor, O. Virulence Gene Profiling and Pathogenicity Characterization of Non-Typhoidal Salmonella Accounted for Invasive Disease in Humans. PLoS ONE 2013, 8, e58449. [Google Scholar] [CrossRef] [PubMed]
- Varughese, E.A.; Bennett-Stamper, C.L.; Wymer, L.J.; Yadav, J.S. A new in vitro model using small intestinal epithelial cells to enhance infection of Cryptosporidium parvum. J. Microbiol. Methods 2014, 106, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Deditius, J.A.; Felgner, S.; Spöring, I.; Kühne, C.; Frahm, M.; Rohde, M.; Weiß, S.; Erhardt, M. Characterization of Novel Factors Involved in Swimming and Swarming Motility in Salmonella enterica Serovar Typhimurium. PLoS ONE 2015, 10, e0135351. [Google Scholar] [CrossRef] [PubMed]
- Harshey, R.M.; Partridge, J.D. Shelter in a swarm. J. Mol. Biol. 2015, 427, 3683–3694. [Google Scholar] [CrossRef]
- Chelvam, K.K.; Chai, L.C.; Thong, K.L. Variations in motility and biofilm formation of Salmonella enterica serovar Typhi. Gut Pathog. 2014, 6. [Google Scholar] [CrossRef]
- Park, S.Y.; Pontes, M.H.; Groisman, E.A. Flagella-independent surface motility in Salmonella enterica serovar Typhimurium. Proc. Natl. Acad. Sci. USA 2015, 112, 1850–1855. [Google Scholar] [CrossRef]
- Partridge, J.D.; Harshey, R.M. Swarming: Flexible roaming plans. J. Bacteriol. 2013, 195, 909–918. [Google Scholar] [CrossRef]
- Choi, E.; Han, Y.; Cho, Y.J.; Nam, D.; Lee, E.J. A trans-acting leader RNA from a Salmonella virulence gene. PNAS 2017, 114, 10232–10237. [Google Scholar] [CrossRef]
- Liu, S.L.; Ezaki, T.; Miura, H.; Matsui, K.; Yabuuchi, E. Intact Motility as a Salmonella typhi Invasion-Related Factor. Infect. Immun. 1988, 56, 1967–1973. [Google Scholar] [CrossRef]
- Chakroun, I.; Mahdhi, A.; Morcillo, P.; Cordero, H.; Cuesta, A.; Bakhrouf, A.; Mahdouani, K.; Esteban, M.Á. Motility, biofilm formation, apoptotic effect and virulence gene expression of atypical Salmonella Typhimurium outside and inside Caco-2 cells. Microb. Pathog. 2018, 114, 153–162. [Google Scholar] [CrossRef]
- MacKenzie, K.D.; Palmer, M.B.; Köster, W.L.; White, A.P. Examining the Link between Biofilm Formation and the Ability of Pathogenic Salmonella Strains to Colonize Multiple Host Species. Front. Vet. Sci. 2017, 4. [Google Scholar] [CrossRef] [PubMed]
- MacKenzie, K.D.; Wang, Y.; Musicha, P.; Hansen, E.G.; Palmer, M.B.; Herman, D.J.; Feasey, N.A.; White, A.P. Parallel evolution leading to impaired biofilm formation in invasive Salmonella strains. PLoS Genet. 2019, 15, e1008233. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, G.; Aheto, K.; Shirtliff, M.E.; Tennant, S.M. Poor biofilm-forming ability and long-term survival of invasive Salmonella Typhimurium ST313. Pathog. Dis. 2016, 74. [Google Scholar] [CrossRef]
- Paudyal, N.; Pan, H.; Elbediwi, M.; Zhou, X.; Peng, X.; Li, X.; Fang, W.; Yue, M. Characterization of Salmonella Dublin isolated from bovine and human hosts. BMC Microbiol. 2019, 19, 226. [Google Scholar] [CrossRef]
- Speranza, B.; Corbo, M.R.; Sinigaglia, M. Effects of nutritional and environmental conditions on Salmonella sp. biofilm formation. J. Food Sci. 2010, 76. [Google Scholar] [CrossRef]
- González, J.F.; Alberts, H.; Lee, J.; Doolittle, L.; Gunn, J.S. Biofilm Formation Protects Salmonella from the Antibiotic Ciprofloxacin In Vitro and In Vivo in the Mouse Model of chronic Carriage. Sci Rep. 2018, 8. [Google Scholar] [CrossRef]
- Boddicker, J.D.; Knosp, B.M.; Jones, B.D. Transcription of the Salmonella Invasion Gene Activator, hilA, Requires HilD Activation in the Absence of Negative Regulators. J. Bacteriol. 2003, 185, 525–533. [Google Scholar] [CrossRef]
- Kisiela, D.; Laskowska, A.; Sapeta, A.; Kuczkowski, M.; Wieliczko, A.; Ugorski, M. Functional characterization of the FimH adhesin from Salmonella enterica serovar Enteritidis. Microbiology 2006, 152, 1337–1346. [Google Scholar] [CrossRef]
- Guiney, D.G.; Fierer, J. The Role of the spv Genes in Salmonella Pathogenesis. Front. Microbiol. 2011, 2, 129. [Google Scholar] [CrossRef]
- Su, L.H.; Chiu, C.H.; Wu, T.L.; Chu, C.; Chia, J.H.; Kuo, A.J.; Lee, C.C.; Sun, C.F.; Ou, J.T. Molecular Epidemiology of Salmonella entericaSerovar Enteritidis Isolated in Taiwan. Microbiol. Immunol. 2002, 46, 833–840. [Google Scholar] [CrossRef]
- Tseng, C.F.; Chiu, N.C.; Huang, C.Y.; Huang, D.T.N.; Chang, L.; Kung, Y.H.; Huang, F.Y.; Chi, H. The epidemiology of non-typhoidal Salmonella gastroenteritis and Campylobacter gastroenteritis in pediatric inpatients in northern Taiwan. J. Microbiol. Immunol. Infect. 2019, 52, 449–455. [Google Scholar] [CrossRef] [PubMed]
- Eng, S.K.; Pusparajah, P.; Ab Mutalib, N.S.; Ser, H.L.; Chan, K.G.; Lee, L.H. Salmonella: A review on pathogenesis, epidemiology and antibiotic resistance. Front. Life Sci. 2015, 8, 284–293. [Google Scholar] [CrossRef]
- Christenson, J.C. Salmonella infections. Pediatr. Rev. 2013, 34, 375–383. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Li, W.C.; Huang, F.Y.; Liu, C.P.; Weng, L.C.; Wang, N.Y.; Chiu, N.C.; Chiang, C.S. Ceftriaxone Resistance of Nontyphoidal Salmonella enterica Isolates in Northern Taiwan Attributable to Production of CTX-M-14 and CMY-2 β-Lactamases. J. Clin. Microbiol. 2005, 43, 3237–3243. [Google Scholar] [CrossRef]
- Lo, H.Y.; Lai, F.P.; Yang, Y.J. Changes in epidemiology and antimicrobial susceptibility of nontyphoid Salmonella in children in southern Taiwan, 1997–2016. J. Microbiol. Immunol. Infect. 2018, 54, 585–591. [Google Scholar] [CrossRef]
- Su, L.H.; Teng, W.S.; Chen, C.L.; Lee, H.Y.; Li, H.C.; Wu, T.L.; Chiu, C.H. Increasing Ceftriaxone Resistance in Salmonellae, Taiwan. Emerg. Infect. Dis. 2011, 17, 1086–1090. [Google Scholar] [CrossRef]
- Rossi, O.; Dybowski, R.; Maskell, D.J.; Grant, A.J.; Restif, O.; Mastroeni, P. Within-host spatiotemporal dynamics of systemic Salmonella infection during and after antimicrobial treatment. J. Antimicrob. Chemother. 2017, 72, 3390–3397. [Google Scholar] [CrossRef]
- Khanal, P.R.; Satyal, D.; Bhetwal, A.; Maharjan, A.; Shakya, S.; Tandukar, S.; Parajuli, N.P. Renaissance of Conventional First-Line Antibiotics in Salmonella enterica Clinical Isolates: Assessment of MICs for Therapeutic Antimicrobials in Enteric Fever Cases from Nepal. Biomed. Res. Int. 2017, 2017. [Google Scholar] [CrossRef]
- Ed-Dra, A.; Karraouan, B.; Allaoui, A.E.; Khayatti, M.; Ossmani, H.E.; Filali, F.R.; ElMdaghri, N.; Bouchrif, B. Isolation, antimicrobial resistance and genetic diversity of Salmonella Infantis isolates from foods and human samples in Morocco. J. Glob. Antimicrob. Resist. 2018, 14, 297–301. [Google Scholar] [CrossRef]
- Sjölund-Karlsson, M.; Joyce, K.; Blickenstaff, K.; Ball, T.; Haro, J.; Medalla, F.M.; Fedorka-Cray, P.; Zhao, S.; Crump, J.A.; Whichard, J.M. Antimicrobial Susceptibility to Azithromycin among Salmonella enterica Isolates from the United States. Antimicrob. Agents Chemother. 2011, 55, 3985–3989. [Google Scholar] [CrossRef]
- Chiu, C.H.; Su, L.H.; Hung, C.C.; Chen, K.L.; Chu, C. Prevalence and Antimicrobial Susceptibility of Serogroup D Nontyphoidal Salmonella in a University Hospital in Taiwan. J. Clin. Microbiol. 2004, 42, 415–417. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Murray, P.R. (Ed.) Enterobacteriaceae: Introduction and identification. In Manual of Clinical Microbiology, 6th ed.; ASM Press: Washington, DC, USA, 1995; pp. 438–449. [Google Scholar]
- Gagnon, M.; Zihler, B.A.; Chervet, N.; Chassard, C.; Lacroix, C. Comparison of the Caco-2, HT-29 and the mucus-secreting HT29-MTX intestinal cell models to investigate Salmonella adhesion and invasion. J. Microbiol. Methods 2013, 94, 274–279. [Google Scholar] [CrossRef] [PubMed]
- Peng, D. Biofilm Formation of Salmonella. In Microbial Biofilms—Importance and Applications; Dhanasekaran, D., Thajuddin, N., Eds.; IntechOpen: London, UK, 2016. [Google Scholar] [CrossRef]
- Díez-García, M.; Capita, R.; Alonso-Calleja, C. Influence of serotype on the growth kinetics and the ability to form biofilms of Salmonella isolates from poultry. Food Microbiol. 2012, 31, 173–180. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Magalhães, A.P.; França, A.; Pereira, M.O.; Cerca, N. RNA-based qPCR as a tool to quantify and to characterize dual-species biofilms. Sci. Rep. 2019, 9. [Google Scholar] [CrossRef]
- Hudzicki, J. Kirby-Bauer Disk Diffusion Susceptibility Test Protocol in A.S.M. 2009. Available online: https://asm.org/getattachment/2594ce26-bd44-47f6-8287-0657aa9185ad/Kirby-Bauer-Disk-Diffusion-Susceptibility-Test-Protocol-pdf.pdf (accessed on 13 September 2018).

| Bacterial Isolates | FliC | SpvC | InvA | FimH | SopB | HilA |
|---|---|---|---|---|---|---|
| B1 | - | + | + | + | + | + |
| B2 | + | - | + | + | + | + |
| B3 | - | + | + | + | + | + |
| B4 | - | + | + | + | + | + |
| B5 | + | - | + | + | + | + |
| B6 | + | - | + | + | + | + |
| S1 | + | + | + | + | + | + |
| S2 | - | + | + | + | + | + |
| S3 | - | + | + | + | + | + |
| S4 | + | - | + | + | + | + |
| S5 | - | - | + | + | - | + |
| S6 | - | + | + | + | + | + |
| Antimicrobials | Susceptibility Characteristics (n = 50) | ||
|---|---|---|---|
| Sensitive N (%) | Intermediate N (%) | Resistant N (%) | |
| Chloramphenicol | 32 (64%) | 0 (0%) | 18 (36%) |
| Ciprofloxacin | 2 (4%) | 38 (76%) | 10 (20%) |
| Ampicillin | 24 (48%) | 1 (2%) | 25 (50%) |
| Gentamicin | 22 (44%) | 24 (48%) | 4 (8%) |
| Ceftriaxone | 45 (90%) | 4 (8%) | 1 (2%) |
| Trimethoprim-Sulfamethoxazole | 36 (72%) | 0 (0%) | 14 (28%) |
| Bacterial Identification Name | Bacterial Collection Number | Isolation Source | Serotyping Group |
|---|---|---|---|
| B1 | 1517 | Blood | D1 |
| B2 | 1596 | Blood | B |
| B3 | 1462 | Blood | D1 |
| B4 | 1793 | Blood | D1 |
| B5 | 1829 | Blood | B |
| B6 | 1833 | Blood | B |
| S1 | 1792 | Stool | B |
| S2 | 1785 | Stool | D1 |
| S3 | 1794 | Stool | D1 |
| S4 | 1795 | Stool | B |
| S5 | 1798 | Stool | B |
| S6 | 1762 | Stool | D1 |
| Target Gene | Oligo Sequence | Amplicon Size (bp) | NCBI Reference Sequence |
|---|---|---|---|
| FliC | F: AACGCAGTAAAGAGAGGACG | 162 | NC 003197.2 |
| R: ACACCGTAAACAACCTGACT | |||
| SpvC | F: TCACGTAAAGCCTGTCTCTG | 214 | NC 003277.2 |
| R: CAGACCAGGAAAATTCGCAG | |||
| FimH | F: CGGAAAAATCAGGTTGGGTC | 360 | NC 003197.2 |
| R: CGGTAGAGGTCGTCACATAG | |||
| SopB | F: CTTTTGCAGGTAAGCCATCC | 235 | NC 003197.2 |
| R: CACTCGCTGCATAACCTCTA | |||
| InvA | F: TCCACGAATATGCTCCACAA | 381 | NC 003197.2 |
| R: CCAACAATCCATCAGCAAGG | |||
| HilA | F: GCTTTAGGATTACTGGGGCT | 407 | NC003197.2 |
| R: AGTCCTGTTATTTCCTGCGT |
| Target Gene | Oligo Sequence |
|---|---|
| FliC | F: TCTGTACCGCACCCAGGTCA |
| R: CGGCTGCTACAACCACCGAA | |
| SpvC | F: CACTCTGGCGCATCCCTGAA |
| R: GCTGCTTATGATGGGGCGGA | |
| FimH | F: TGACGATCCCTCGCCAGACA |
| R: ACGACCTGTCCGGCATTCAC | |
| SopB | F: GTATTGCGCCAGCCATTCGC |
| R: GCCGAGGCGCTACATCAGTT | |
| InvA | F: CGACTTCCGCGACACGTTCT |
| R: TAGCCTGGCGGTGGGTTTTG | |
| HilA | F: TCGTAGTGGTGTCTCCGCCA |
| R: CTCAGGCCAAAGGGCGCATA | |
| RpoD | F: GTTGACCCGGGAAGGCGAAA |
| R: GGGTATTCGGCAACGGAGCA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ouali, B.E.F.; Chiou, T.-H.; Chen, J.-W.; Lin, I.-C.; Liu, C.-C.; Chiang, Y.-C.; Ho, T.-S.; Wang, H.-V. Correlation between Pathogenic Determinants Associated with Clinically Isolated Non-Typhoidal Salmonella. Pathogens 2021, 10, 74. https://doi.org/10.3390/pathogens10010074
Ouali BEF, Chiou T-H, Chen J-W, Lin I-C, Liu C-C, Chiang Y-C, Ho T-S, Wang H-V. Correlation between Pathogenic Determinants Associated with Clinically Isolated Non-Typhoidal Salmonella. Pathogens. 2021; 10(1):74. https://doi.org/10.3390/pathogens10010074
Chicago/Turabian StyleOuali, Boimpoundi Eunice Flavie, Tsyr-Huei Chiou, Jenn-Wei Chen, I-Chu Lin, Ching-Chuan Liu, Yu-Chung Chiang, Tzong-Shiann Ho, and Hao-Ven Wang. 2021. "Correlation between Pathogenic Determinants Associated with Clinically Isolated Non-Typhoidal Salmonella" Pathogens 10, no. 1: 74. https://doi.org/10.3390/pathogens10010074
APA StyleOuali, B. E. F., Chiou, T.-H., Chen, J.-W., Lin, I.-C., Liu, C.-C., Chiang, Y.-C., Ho, T.-S., & Wang, H.-V. (2021). Correlation between Pathogenic Determinants Associated with Clinically Isolated Non-Typhoidal Salmonella. Pathogens, 10(1), 74. https://doi.org/10.3390/pathogens10010074






