An In Vitro Cell Culture Model for Pyoverdine-Mediated Virulence
Abstract
1. Introduction
2. Results
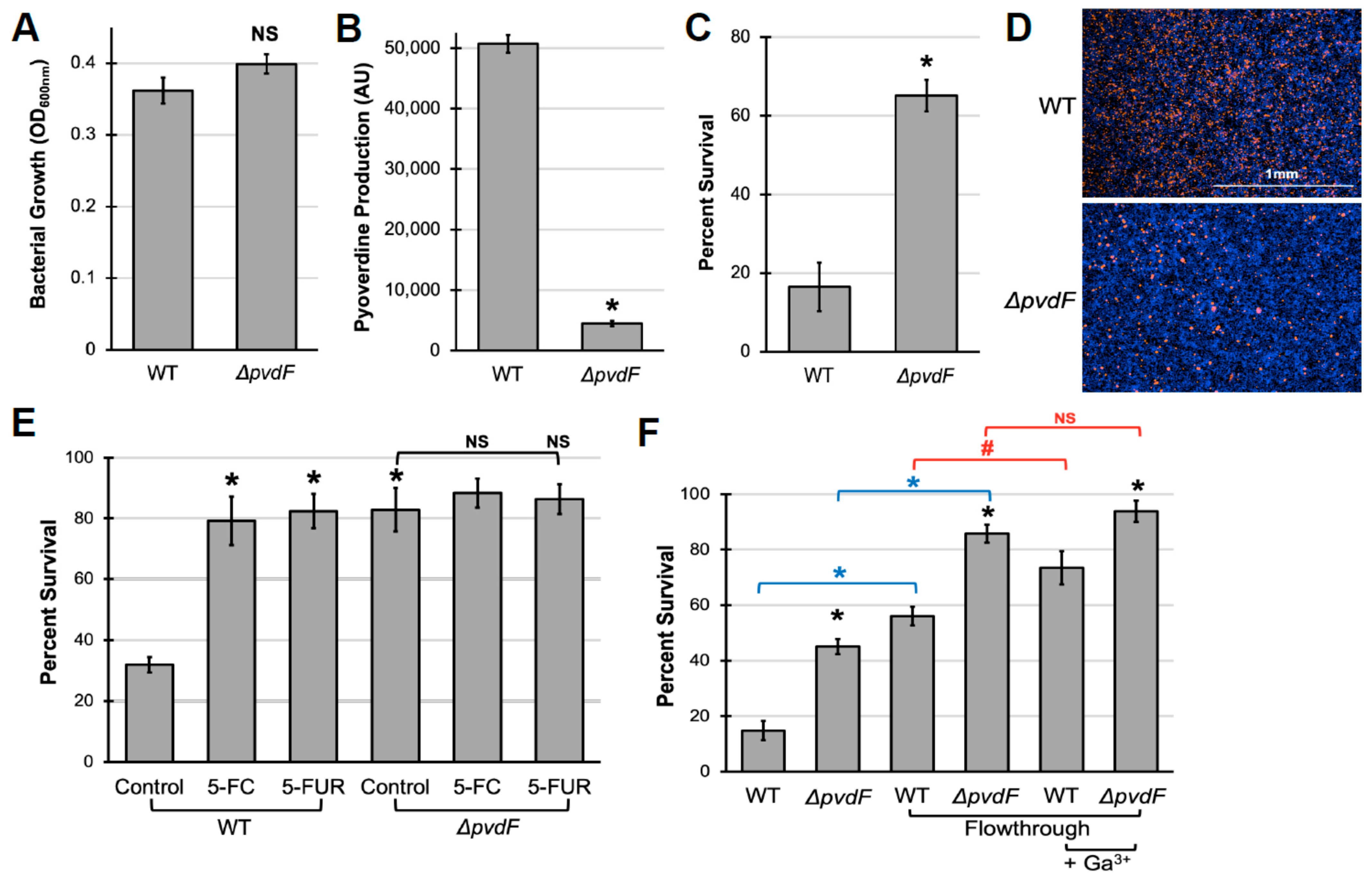
2.1. Pyoverdine Production Is Important for P. aeruginosa Virulence against Murine Macrophages
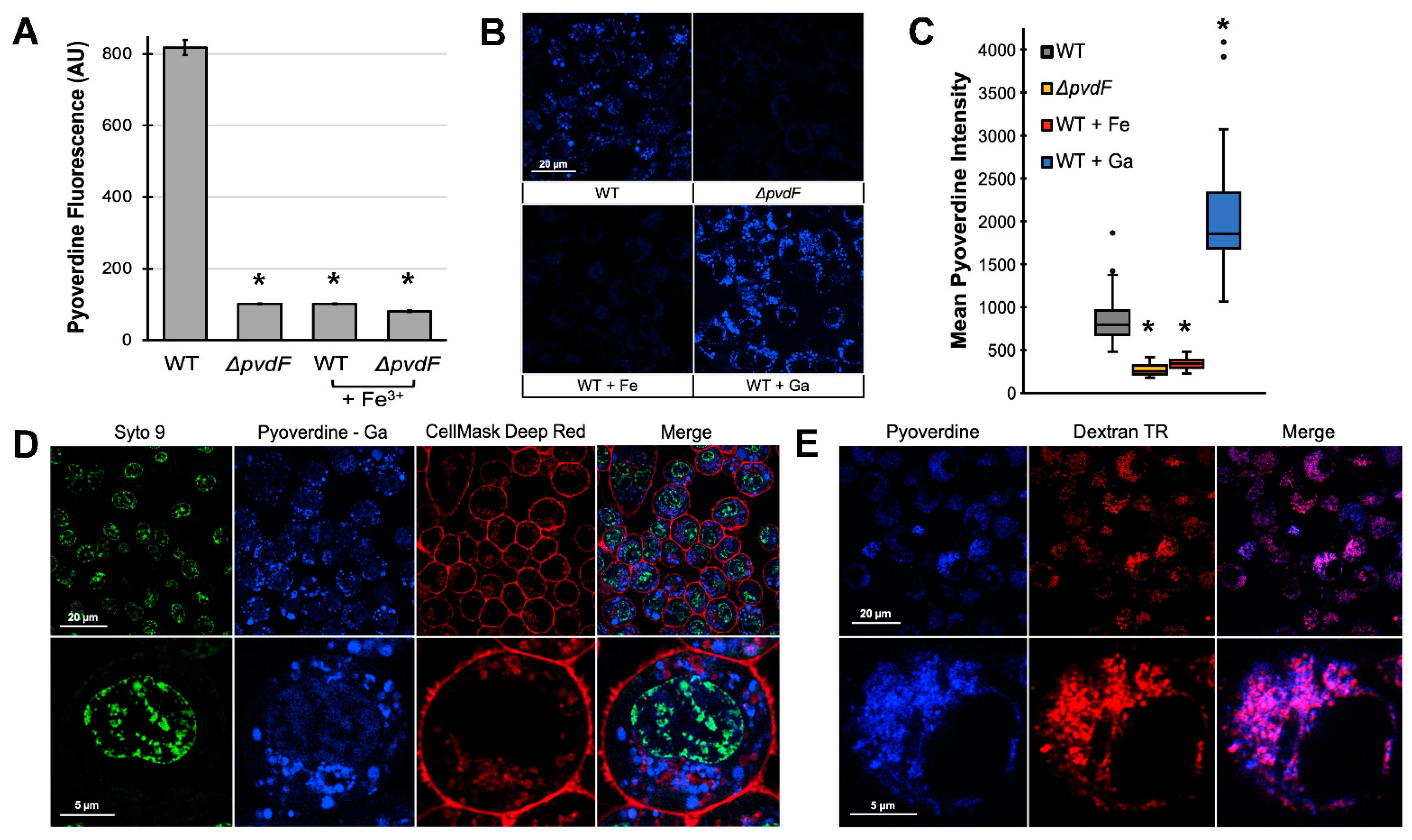
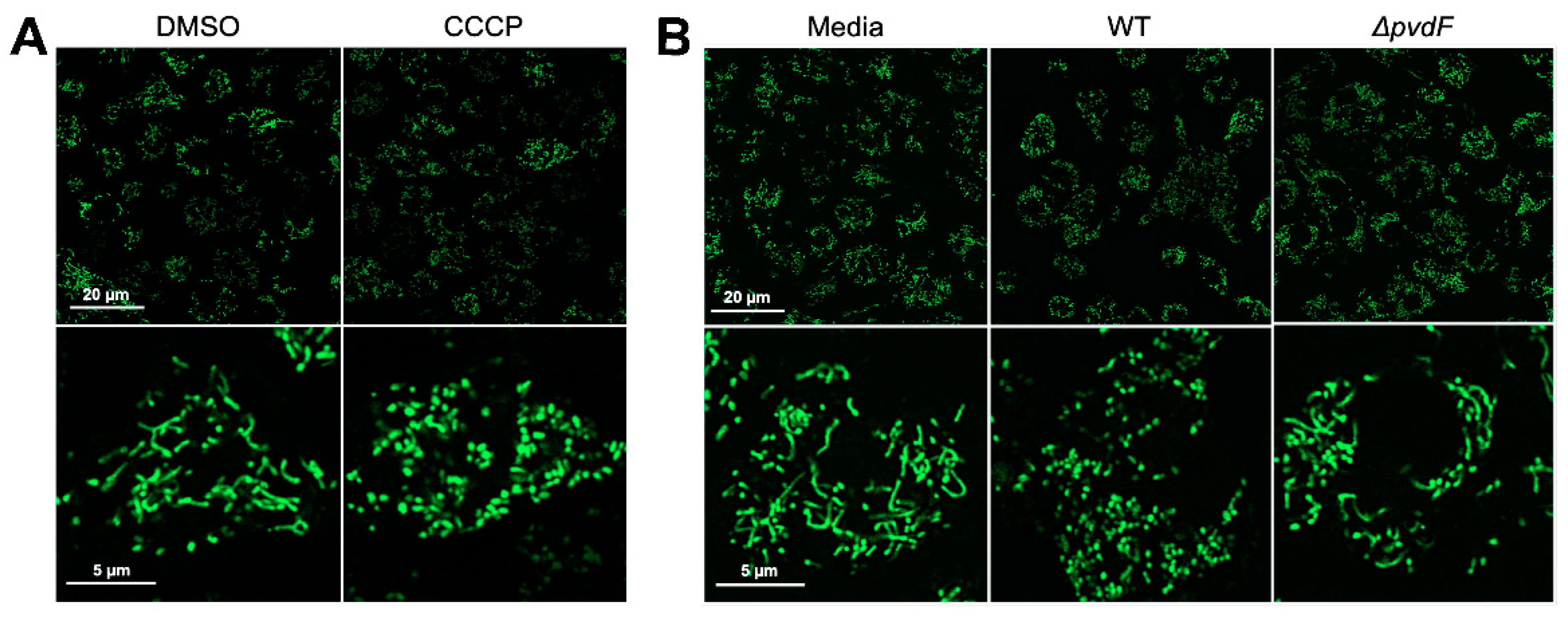
2.2. Pyoverdine Translocates into Macrophages and Disrupts Mitochondrial Function
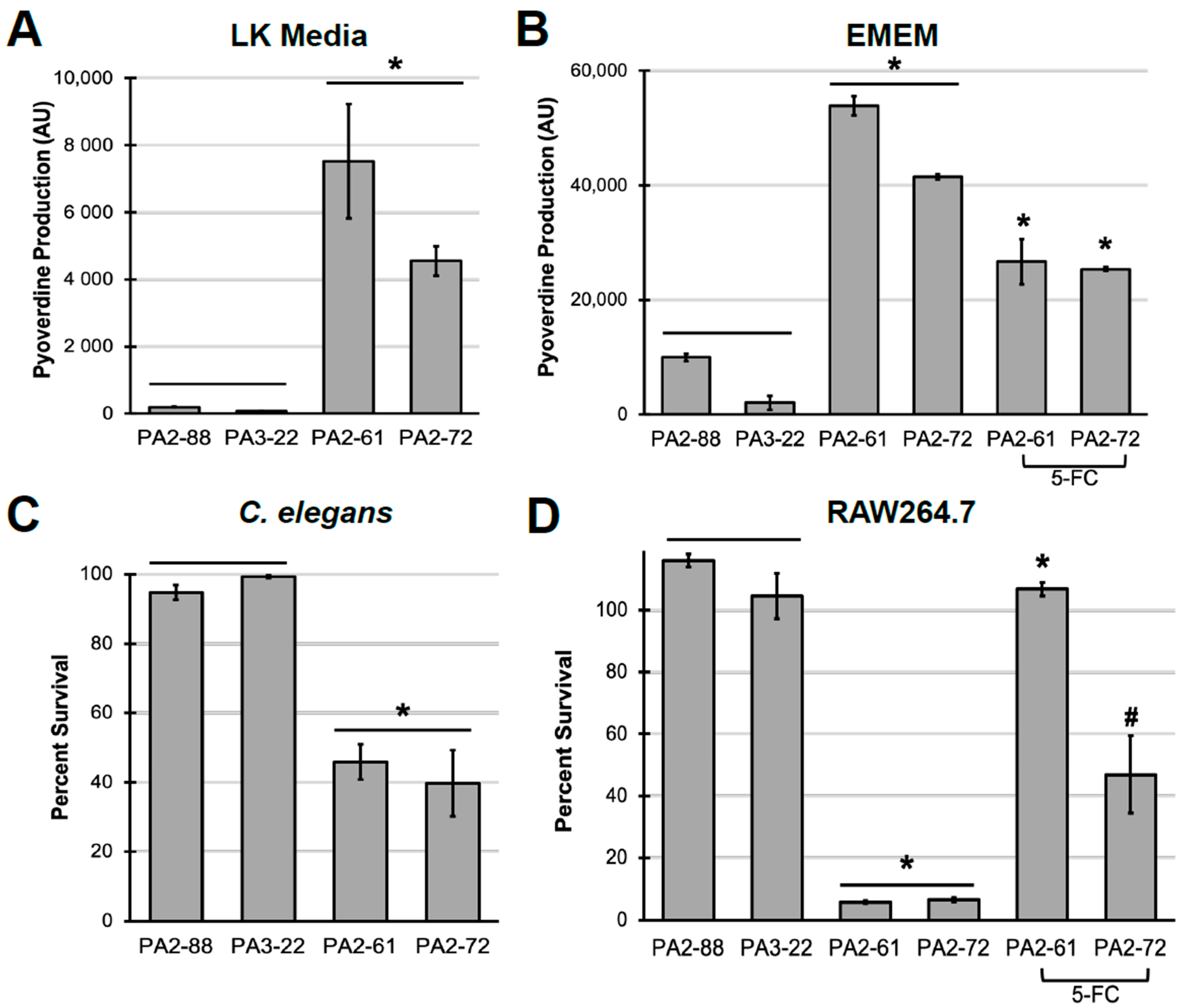
2.3. Pyoverdine Production Correlates to Virulence in P. aeruginosa Clinical Isolates
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Growth Conditions
4.2. Macrophage Cell Lines and Growth Conditions
4.3. Cell Viability Assay
4.4. Measuring Pyoverdine Content
4.5. Confocal Microscopy
4.6. C. elegans Liquid Killing
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lyczak, J.B.; Cannon, C.L.; Pier, G.B. Lung infections associated with cystic fibrosis. Clin. Microbiol. Rev. 2002, 15, 194–222. [Google Scholar] [CrossRef] [PubMed]
- Kollef, M.H.; Chastre, J.; Fagon, J.Y.; Francois, B.; Niederman, M.S.; Rello, J.; Torres, A.; Vincent, J.L.; Wunderink, R.G.; Go, K.W.; et al. Global prospective epidemiologic and surveillance study of ventilator-associated pneumonia due to Pseudomonas aeruginosa. Crit. Care Med. 2014, 42, 2178–2187. [Google Scholar] [CrossRef] [PubMed]
- Saint-Criq, V.; Villeret, B.; Bastaert, F.; Kheir, S.; Hatton, A.; Cazes, A.; Xing, Z.; Sermet-Gaudelus, I.; Garcia-Verdugo, I.; Edelman, A.; et al. LasB protease impairs innate immunity in mice and humans by targeting a lung epithelial cystic fibrosis transmembrane regulator-IL-6-antimicrobial-repair pathway. Thorax 2018, 73, 49–61. [Google Scholar] [CrossRef] [PubMed]
- Casilag, F.; Lorenz, A.; Krueger, J.; Klawonn, F.; Weiss, S.; Häussler, S. The LasB Elastase of Pseudomonas aeruginosa Acts in Concert with Alkaline Protease AprA To Prevent Flagellin-Mediated Immune Recognition. Infect. Immun. 2016, 84, 162–171. [Google Scholar] [CrossRef] [PubMed]
- Minandri, F.; Imperi, F.; Frangipani, E.; Bonchi, C.; Visaggio, D.; Facchini, M.; Pasquali, P.; Bragonzi, A.; Visca, P. Role of Iron Uptake Systems in Pseudomonas aeruginosa Virulence and Airway Infection. Infect. Immun. 2016, 84, 2324–2335. [Google Scholar] [CrossRef]
- Hauser, A.R. The type III secretion system of Pseudomonas aeruginosa: Infection by injection. Nat. Rev. Microbiol. 2009, 7, 654–665. [Google Scholar] [CrossRef]
- Moradali, M.F.; Ghods, S.; Rehm, B.H. Pseudomonas aeruginosa Lifestyle: A Paradigm for Adaptation, Survival, and Persistence. Front. Cell Infect. Microbiol. 2017, 7, 39. [Google Scholar] [CrossRef]
- Xiao, R.; Kisaalita, W.S. Iron acquisition from transferrin and lactoferrin by Pseudomonas aeruginosa pyoverdin. Microbiology 1997, 143 Pt 7, 2509–2515. [Google Scholar] [CrossRef]
- Skaar, E.P. The battle for iron between bacterial pathogens and their vertebrate hosts. PLoS Pathog. 2010, 6, e1000949. [Google Scholar] [CrossRef]
- Takase, H.; Nitanai, H.; Hoshino, K.; Otani, T. Impact of siderophore production on Pseudomonas aeruginosa infections in immunosuppressed mice. Infect. Immun. 2000, 68, 1834–1839. [Google Scholar] [CrossRef]
- Ankenbauer, R.; Sriyosachati, S.; Cox, C.D. Effects of siderophores on the growth of Pseudomonas aeruginosa in human serum and transferrin. Infect. Immun. 1985, 49, 132–140. [Google Scholar] [CrossRef] [PubMed]
- Banin, E.; Vasil, M.L.; Greenberg, E.P. Iron and Pseudomonas aeruginosa biofilm formation. Proc. Natl. Acad. Sci. USA 2005, 102, 11076–11081. [Google Scholar] [CrossRef] [PubMed]
- Kang, D.; Kirienko, N.V. Interdependence between iron acquisition and biofilm formation in Pseudomonas aeruginosa. J. Microbiol. 2018, 56, 449–457. [Google Scholar] [CrossRef] [PubMed]
- Lamont, I.L.; Beare, P.A.; Ochsner, U.; Vasil, A.I.; Vasil, M.L. Siderophore-mediated signaling regulates virulence factor production in Pseudomonasaeruginosa. Proc. Natl. Acad. Sci. USA 2002, 99, 7072–7077. [Google Scholar] [CrossRef] [PubMed]
- Wilderman, P.J.; Vasil, A.I.; Johnson, Z.; Wilson, M.J.; Cunliffe, H.E.; Lamont, I.L.; Vasil, M.L. Characterization of an endoprotease (PrpL) encoded by a PvdS-regulated gene in Pseudomonas aeruginosa. Infect. Immun. 2001, 69, 5385–5394. [Google Scholar] [CrossRef] [PubMed]
- Ochsner, U.A.; Johnson, Z.; Lamont, I.L.; Cunliffe, H.E.; Vasil, M.L. Exotoxin A production in Pseudomonas aeruginosa requires the iron-regulated pvdS gene encoding an alternative sigma factor. Mol. Microbiol. 1996, 21, 1019–1028. [Google Scholar] [CrossRef]
- Jenkins, C.E.; Swiatoniowski, A.; Issekutz, A.C.; Lin, T.J. Pseudomonas aeruginosa exotoxin A induces human mast cell apoptosis by a caspase-8 and -3-dependent mechanism. J. Biol. Chem. 2004, 279, 37201–37207. [Google Scholar] [CrossRef]
- Michalska, M.; Wolf, P. Pseudomonas Exotoxin A: Optimized by evolution for effective killing. Front. Microbiol. 2015, 6, 963. [Google Scholar] [CrossRef]
- McEwan, D.L.; Kirienko, N.V.; Ausubel, F.M. Host translational inhibition by Pseudomonas aeruginosa Exotoxin A Triggers an immune response in Caenorhabditis elegans. Cell Host Microbe 2012, 11, 364–374. [Google Scholar] [CrossRef]
- Bradshaw, J.L.; Caballero, A.R.; Bierdeman, M.A.; Adams, K.V.; Pipkins, H.R.; Tang, A.; O’Callaghan, R.J.; McDaniel, L.S. Pseudomonas aeruginosa Protease IV Exacerbates Pneumococcal Pneumonia and Systemic Disease. mSphere 2018, 3. [Google Scholar] [CrossRef]
- Malloy, J.L.; Veldhuizen, R.A.; Thibodeaux, B.A.; O’Callaghan, R.J.; Wright, J.R. Pseudomonas aeruginosa protease IV degrades surfactant proteins and inhibits surfactant host defense and biophysical functions. Am. J. Physiol. Lung Cell Mol. Physiol. 2005, 288, L409–L418. [Google Scholar] [CrossRef] [PubMed]
- Guillon, A.; Brea, D.; Morello, E.; Tang, A.; Jouan, Y.; Ramphal, R.; Korkmaz, B.; Perez-Cruz, M.; Trottein, F.; O’Callaghan, R.J.; et al. Pseudomonas aeruginosa proteolytically alters the interleukin 22-dependent lung mucosal defense. Virulence 2017, 8, 810–820. [Google Scholar] [CrossRef] [PubMed]
- Kirienko, N.V.; Kirienko, D.R.; Larkins-Ford, J.; Wahlby, C.; Ruvkun, G.; Ausubel, F.M. Pseudomonas aeruginosa disrupts Caenorhabditis elegans iron homeostasis, causing a hypoxic response and death. Cell Host Microbe 2013, 13, 406–416. [Google Scholar] [CrossRef] [PubMed]
- Kang, D.; Kirienko, D.R.; Webster, P.; Fisher, A.L.; Kirienko, N.V. Pyoverdine, a siderophore from Pseudomonas aeruginosa, translocates into C. elegans, removes iron, and activates a distinct host response. Virulence 2018, 9, 804–817. [Google Scholar] [CrossRef] [PubMed]
- Kirienko, N.V.; Ausubel, F.M.; Ruvkun, G. Mitophagy confers resistance to siderophore-mediated killing by Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2015, 112, 1821–1826. [Google Scholar] [CrossRef] [PubMed]
- Tjahjono, E.; Kirienko, N.V. A conserved mitochondrial surveillance pathway is required for defense against Pseudomonas aeruginosa. PLoS Genet. 2017, 13, e1006876. [Google Scholar] [CrossRef]
- Tjahjono, E.; McAnena, A.P.; Kirienko, N.V. The evolutionarily conserved ESRE stress response network is activated by ROS and mitochondrial damage. BMC Biol. 2020, 18, 74. [Google Scholar] [CrossRef]
- Weigert, M.; Ross-Gillespie, A.; Leinweber, A.; Pessi, G.; Brown, S.P.; Kummerli, R. Manipulating virulence factor availability can have complex consequences for infections. Evol. Appl. 2017, 10, 91–101. [Google Scholar] [CrossRef]
- Meyer, J.M.; Neely, A.; Stintzi, A.; Georges, C.; Holder, I.A. Pyoverdin is essential for virulence of Pseudomonas aeruginosa. Infect. Immun. 1996, 64, 518–523. [Google Scholar] [CrossRef]
- Kirienko, D.R.; Revtovich, A.V.; Kirienko, N.V. A High-Content, Phenotypic Screen Identifies Fluorouridine as an Inhibitor of Pyoverdine Biosynthesis and Pseudomonas aeruginosa Virulence. mSphere 2016, 1. [Google Scholar] [CrossRef]
- Kang, D.; Revtovich, A.V.; Chen, Q.; Shah, K.N.; Cannon, C.L.; Kirienko, N.V. Pyoverdine-Dependent Virulence of Pseudomonas aeruginosa Isolates from Cystic Fibrosis Patients. Front. Microbiol. 2019, 10, 2048. [Google Scholar] [CrossRef] [PubMed]
- Imperi, F.; Massai, F.; Facchini, M.; Frangipani, E.; Visaggio, D.; Leoni, L.; Bragonzi, A.; Visca, P. Repurposing the antimycotic drug flucytosine for suppression of Pseudomonas aeruginosa pathogenicity. Proc. Natl. Acad. Sci. USA 2013, 110, 7458–7463. [Google Scholar] [CrossRef]
- Costabile, G.; d’Angelo, I.; d’Emmanuele di Villa Bianca, R.; Mitidieri, E.; Pompili, B.; Del Porto, P.; Leoni, L.; Visca, P.; Miro, A.; Quaglia, F.; et al. Development of inhalable hyaluronan/mannitol composite dry powders for flucytosine repositioning in local therapy of lung infections. J. Control Release 2016, 238, 80–91. [Google Scholar] [CrossRef] [PubMed]
- Ochsner, U.A.; Snyder, A.; Vasil, A.I.; Vasil, M.L. Effects of the twin-arginine translocase on secretion of virulence factors, stress response, and pathogenesis. Proc. Natl. Acad. Sci. USA 2002, 99, 8312–8317. [Google Scholar] [CrossRef] [PubMed]
- Vasil, M.L.; Tomaras, A.P.; Pritchard, A.E. Identification and evaluation of twin-arginine translocase inhibitors. Antimicrob. Agents Chemother. 2012, 56, 6223–6234. [Google Scholar] [CrossRef]
- Massai, F.; Saleeb, M.; Doruk, T.; Elofsson, M.; Forsberg, Å. Development, Optimization, and Validation of a High Throughput Screening Assay for Identification of Tat and Type II Secretion Inhibitors of Pseudomonas aeruginosa. Front. Cell Infect. Microbiol. 2019, 9, 250. [Google Scholar] [CrossRef]
- Lee, J.H.; Kim, Y.G.; Cho, M.H.; Kim, J.A.; Lee, J. 7-fluoroindole as an antivirulence compound against Pseudomonas aeruginosa. FEMS Microbiol. Lett. 2012, 329, 36–44. [Google Scholar] [CrossRef]
- Peppoloni, S.; Pericolini, E.; Colombari, B.; Pinetti, D.; Cermelli, C.; Fini, F.; Prati, F.; Caselli, E.; Blasi, E. The beta-Lactamase Inhibitor Boronic Acid Derivative SM23 as a New Anti-Pseudomonas aeruginosa Biofilm. Front. Microbiol. 2020, 11, 35. [Google Scholar] [CrossRef]
- Zhou, J.W.; Luo, H.Z.; Jiang, H.; Jian, T.K.; Chen, Z.Q.; Jia, A.Q. Hordenine: A Novel Quorum Sensing Inhibitor and Antibiofilm Agent against Pseudomonas aeruginosa. J. Agric. Food Chem. 2018, 66, 1620–1628. [Google Scholar] [CrossRef]
- Kang, D.; Kirienko, N.V. High-Throughput Genetic Screen Reveals that Early Attachment and Biofilm Formation Are Necessary for Full Pyoverdine Production by Pseudomonas aeruginosa. Front. Microbiol. 2017, 8, 1707. [Google Scholar] [CrossRef]
- Braud, A.; Hoegy, F.; Jezequel, K.; Lebeau, T.; Schalk, I.J. New insights into the metal specificity of the Pseudomonas aeruginosa pyoverdine-iron uptake pathway. Environ. Microbiol. 2009, 11, 1079–1091. [Google Scholar] [CrossRef] [PubMed]
- Ross-Gillespie, A.; Weigert, M.; Brown, S.P.; Kummerli, R. Gallium-mediated siderophore quenching as an evolutionarily robust antibacterial treatment. Evol. Med. Public Health 2014, 2014, 18–29. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Kleerekoper, Q.; Revtovich, A.V.; Kang, D.; Kirienko, N.V. Identification and validation of a novel anti-virulent that binds to pyoverdine and inhibits its function. Virulence 2020, 11, 1293–1309. [Google Scholar] [CrossRef] [PubMed]
- Kirienko, D.R.; Kang, D.; Kirienko, N.V. Novel Pyoverdine Inhibitors Mitigate Pseudomonas aeruginosa Pathogenesis. Front. Microbiol. 2019, 9, 3317. [Google Scholar] [CrossRef] [PubMed]
- Mahajan-Miklos, S.; Tan, M.W.; Rahme, L.G.; Ausubel, F.M. Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa-Caenorhabditis elegans pathogenesis model. Cell 1999, 96, 47–56. [Google Scholar] [CrossRef]
- Usher, L.R.; Lawson, R.A.; Geary, I.; Taylor, C.J.; Bingle, C.D.; Taylor, G.W.; Whyte, M.K. Induction of neutrophil apoptosis by the Pseudomonas aeruginosa exotoxin pyocyanin: A potential mechanism of persistent infection. J. Immunol. 2002, 168, 1861–1868. [Google Scholar] [CrossRef]
- Eraso, A.J.; Albesa, I. Potential Toxicity of Pyoverdin From Pseudomonas on Mouse Liver. J. Food Process. Preserv. 1996, 20, 12. [Google Scholar] [CrossRef]
- Folschweiller, N.; Gallay, J.; Vincent, M.; Abdallah, M.A.; Pattus, F.; Schalk, I.J. The interaction between pyoverdin and its outer membrane receptor in Pseudomonas aeruginosa leads to different conformers: A time-resolved fluorescence study. Biochemistry 2002, 41, 14591–14601. [Google Scholar] [CrossRef]
- Oliver, J.M.; Berlin, R.D.; Davis, B.H. Use of horseradish peroxidase and fluorescent dextrans to study fluid pinocytosis in leukocytes. Methods Enzymol. 1984, 108, 336–347. [Google Scholar] [CrossRef]
- Lee, D.G.; Urbach, J.M.; Wu, G.; Liberati, N.T.; Feinbaum, R.L.; Miyata, S.; Diggins, L.T.; He, J.; Saucier, M.; Déziel, E.; et al. Genomic analysis reveals that Pseudomonas aeruginosa virulence is combinatorial. Genome Biol. 2006, 7, R90. [Google Scholar] [CrossRef]
- Visca, P.; Imperi, F.; Lamont, I.L. Pyoverdine siderophores: From biogenesis to biosignificance. Trends Microbiol. 2007, 15, 22–30. [Google Scholar] [CrossRef] [PubMed]
- Aiello, D.; Williams, J.D.; Majgier-Baranowska, H.; Patel, I.; Peet, N.P.; Huang, J.; Lory, S.; Bowlin, T.L.; Moir, D.T. Discovery and characterization of inhibitors of Pseudomonas aeruginosa type III secretion. Antimicrob. Agents Chemother. 2010, 54, 1988–1999. [Google Scholar] [CrossRef] [PubMed]
- Starkey, M.; Lepine, F.; Maura, D.; Bandyopadhaya, A.; Lesic, B.; He, J.; Kitao, T.; Righi, V.; Milot, S.; Tzika, A.; et al. Identification of anti-virulence compounds that disrupt quorum-sensing regulated acute and persistent pathogenicity. PLoS Pathog. 2014, 10, e1004321. [Google Scholar] [CrossRef]
- Zhu, J.; Cai, X.; Harris, T.L.; Gooyit, M.; Wood, M.; Lardy, M.; Janda, K.D. Disarming Pseudomonas aeruginosa virulence factor LasB by leveraging a Caenorhabditis elegans infection model. Chem. Biol. 2015, 22, 483–491. [Google Scholar] [CrossRef] [PubMed]
- Kany, A.M.; Sikandar, A.; Haupenthal, J.; Yahiaoui, S.; Maurer, C.K.; Proschak, E.; Köhnke, J.; Hartmann, R.W. Binding Mode Characterization and Early in Vivo Evaluation of Fragment-Like Thiols as Inhibitors of the Virulence Factor LasB from Pseudomonas aeruginosa. ACS Infect. Dis. 2018, 4, 988–997. [Google Scholar] [CrossRef]
- Moy, T.I.; Conery, A.L.; Larkins-Ford, J.; Wu, G.; Mazitschek, R.; Casadei, G.; Lewis, K.; Carpenter, A.E.; Ausubel, F.M. High-throughput screen for novel antimicrobials using a whole animal infection model. ACS Chem. Biol. 2009, 4, 527–533. [Google Scholar] [CrossRef]
- Breger, J.; Fuchs, B.B.; Aperis, G.; Moy, T.I.; Ausubel, F.M.; Mylonakis, E. Antifungal chemical compounds identified using a C. elegans pathogenicity assay. PLoS Pathog. 2007, 3, e18. [Google Scholar] [CrossRef]
- Kim, W.; Zhu, W.; Hendricks, G.L.; Van Tyne, D.; Steele, A.D.; Keohane, C.E.; Fricke, N.; Conery, A.L.; Shen, S.; Pan, W.; et al. A new class of synthetic retinoid antibiotics effective against bacterial persisters. Nature 2018, 556, 103–107. [Google Scholar] [CrossRef]
- Wareham, D.W.; Papakonstantinopoulou, A.; Curtis, M.A. The Pseudomonas aeruginosa PA14 type III secretion system is expressed but not essential to virulence in the Caenorhabditis elegans-P. aeruginosa pathogenicity model. FEMS Microbiol. Lett. 2005, 242, 209–216. [Google Scholar] [CrossRef]
- Woodworth, B.A.; Tamashiro, E.; Bhargave, G.; Cohen, N.A.; Palmer, J.N. An in vitro model of Pseudomonas aeruginosa biofilms on viable airway epithelial cell monolayers. Am. J. Rhinol. 2008, 22, 235–238. [Google Scholar] [CrossRef]
- Carterson, A.J.; Honer zu Bentrup, K.; Ott, C.M.; Clarke, M.S.; Pierson, D.L.; Vanderburg, C.R.; Buchanan, K.L.; Nickerson, C.A.; Schurr, M.J. A549 lung epithelial cells grown as three-dimensional aggregates: Alternative tissue culture model for Pseudomonas aeruginosa pathogenesis. Infect. Immun. 2005, 73, 1129–1140. [Google Scholar] [CrossRef]
- Sato, H.; Frank, D.W. ExoU is a potent intracellular phospholipase. Mol. Microbiol. 2004, 53, 1279–1290. [Google Scholar] [CrossRef]
- Shafikhani, S.H.; Morales, C.; Engel, J. The Pseudomonas aeruginosa type III secreted toxin ExoT is necessary and sufficient to induce apoptosis in epithelial cells. Cell Microbiol. 2008, 10, 994–1007. [Google Scholar] [CrossRef] [PubMed]
- Sutterwala, F.S.; Mijares, L.A.; Li, L.; Ogura, Y.; Kazmierczak, B.I.; Flavell, R.A. Immune recognition of Pseudomonas aeruginosa mediated by the IPAF/NLRC4 inflammasome. J. Exp. Med. 2007, 204, 3235–3245. [Google Scholar] [CrossRef]
- Miao, E.A.; Ernst, R.K.; Dors, M.; Mao, D.P.; Aderem, A. Pseudomonas aeruginosa activates caspase 1 through Ipaf. Proc. Natl. Acad. Sci. USA 2008, 105, 2562–2567. [Google Scholar] [CrossRef] [PubMed]
- Franchi, L.; Stoolman, J.; Kanneganti, T.D.; Verma, A.; Ramphal, R.; Nunez, G. Critical role for Ipaf in Pseudomonas aeruginosa-induced caspase-1 activation. Eur. J. Immunol. 2007, 37, 3030–3039. [Google Scholar] [CrossRef] [PubMed]
- Deng, Q.; Wang, Y.; Zhang, Y.; Li, M.; Li, D.; Huang, X.; Wu, Y.; Pu, J.; Wu, M. Pseudomonas aeruginosa Triggers Macrophage Autophagy to Escape Intracellular Killing by Activation of the NLRP3 Inflammasome. Infect. Immun. 2016, 84, 56–66. [Google Scholar] [CrossRef] [PubMed]
- Golovkine, G.; Faudry, E.; Bouillot, S.; Voulhoux, R.; Attree, I.; Huber, P. VE-cadherin cleavage by LasB protease from Pseudomonas aeruginosa facilitates type III secretion system toxicity in endothelial cells. PLoS Pathog. 2014, 10, e1003939. [Google Scholar] [CrossRef] [PubMed]
- Huber, P.; Bouillot, S.; Elsen, S.; Attree, I. Sequential inactivation of Rho GTPases and Lim kinase by Pseudomonas aeruginosa toxins ExoS and ExoT leads to endothelial monolayer breakdown. Cell Mol. Life Sci. 2014, 71, 1927–1941. [Google Scholar] [CrossRef] [PubMed]
- Pukatzki, S.; Kessin, R.H.; Mekalanos, J.J. The human pathogen Pseudomonas aeruginosa utilizes conserved virulence pathways to infect the social amoeba Dictyostelium discoideum. Proc. Natl. Acad. Sci. USA 2002, 99, 3159–3164. [Google Scholar] [CrossRef]
- Leong, W.; Lutz, C.; Williams, J.; Poh, Y.H.; Yee, B.Y.K.; Chua, C.; Rice, S.A.; Givskov, M.; Sanderson-Smith, M.; McDougald, D. Pseudomonas aeruginosa isolates co-incubated with Acanthamoeba castellanii exhibit phenotypes similar to chronic cystic fibrosis isolates. bioRxiv 2020. [Google Scholar] [CrossRef]
- Conery, A.L.; Larkins-Ford, J.; Ausubel, F.M.; Kirienko, N.V. High-throughput screening for novel anti-infectives using a C. elegans pathogenesis model. Curr. Protoc. Chem. Biol. 2014, 6, 25–37. [Google Scholar] [CrossRef] [PubMed]
- Anderson, Q.L.; Revtovich, A.V.; Kirienko, N.V. A High-throughput, High-content, Liquid-based C. elegans Pathosystem. J. Vis. Exp. 2018, 137, e58068. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kang, D.; Kirienko, N.V. An In Vitro Cell Culture Model for Pyoverdine-Mediated Virulence. Pathogens 2021, 10, 9. https://doi.org/10.3390/pathogens10010009
Kang D, Kirienko NV. An In Vitro Cell Culture Model for Pyoverdine-Mediated Virulence. Pathogens. 2021; 10(1):9. https://doi.org/10.3390/pathogens10010009
Chicago/Turabian StyleKang, Donghoon, and Natalia V. Kirienko. 2021. "An In Vitro Cell Culture Model for Pyoverdine-Mediated Virulence" Pathogens 10, no. 1: 9. https://doi.org/10.3390/pathogens10010009
APA StyleKang, D., & Kirienko, N. V. (2021). An In Vitro Cell Culture Model for Pyoverdine-Mediated Virulence. Pathogens, 10(1), 9. https://doi.org/10.3390/pathogens10010009




