Identification of cbiO Gene Critical for Biofilm Formation by MRSA CFSa36 Strain Isolated from Pediatric Patient with Cystic Fibrosis
Abstract
:1. Introduction
2. Results
2.1. Characterization of the Capacity of Biofilm Formation by S. aureus Strains Isolated from Pediatric Patients with Cystic Fibrosis
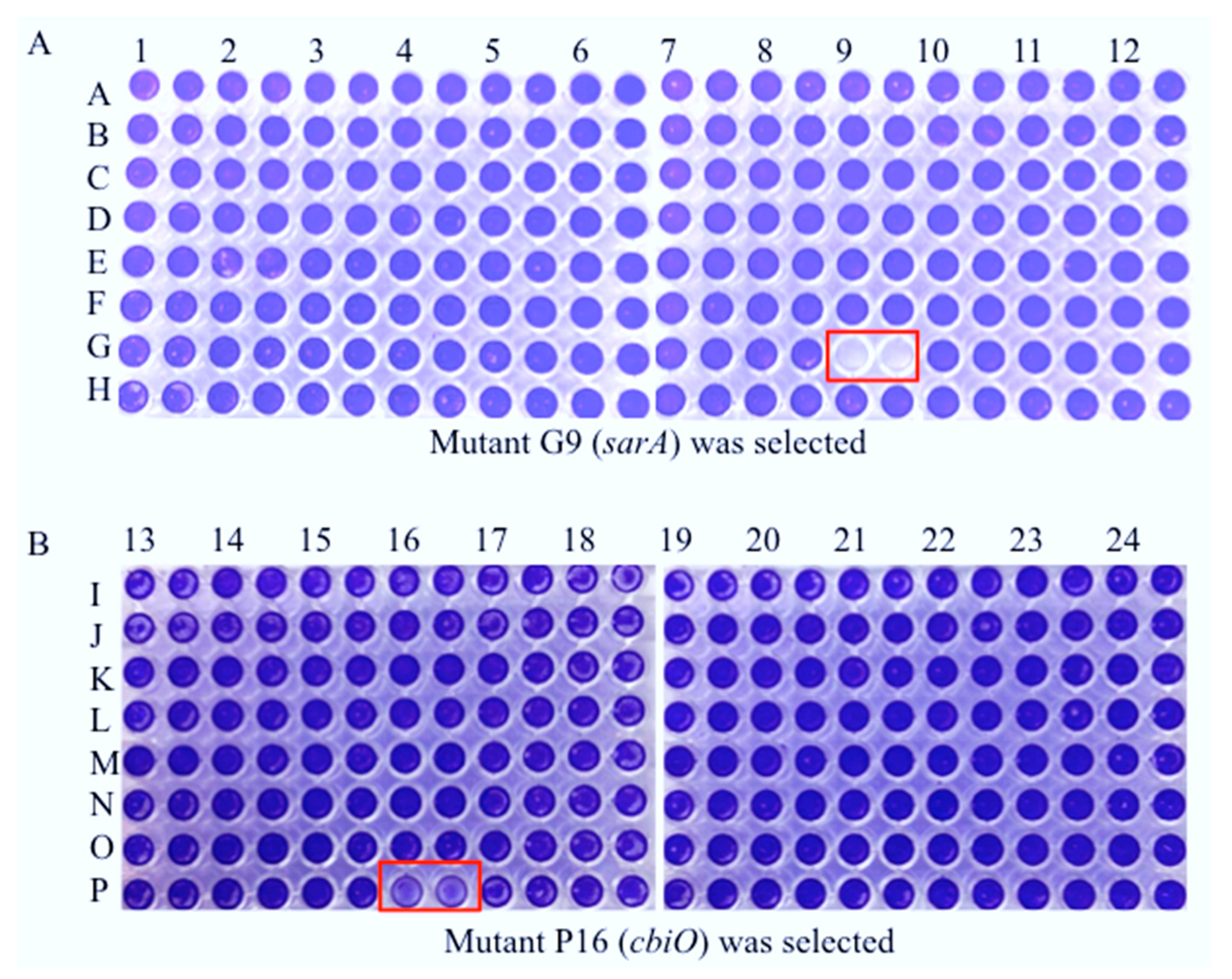
2.2. Random Identification of the cbiO Gene Required for Biofilm Formation by CA-MRSA on Plastic Surface In Vitro
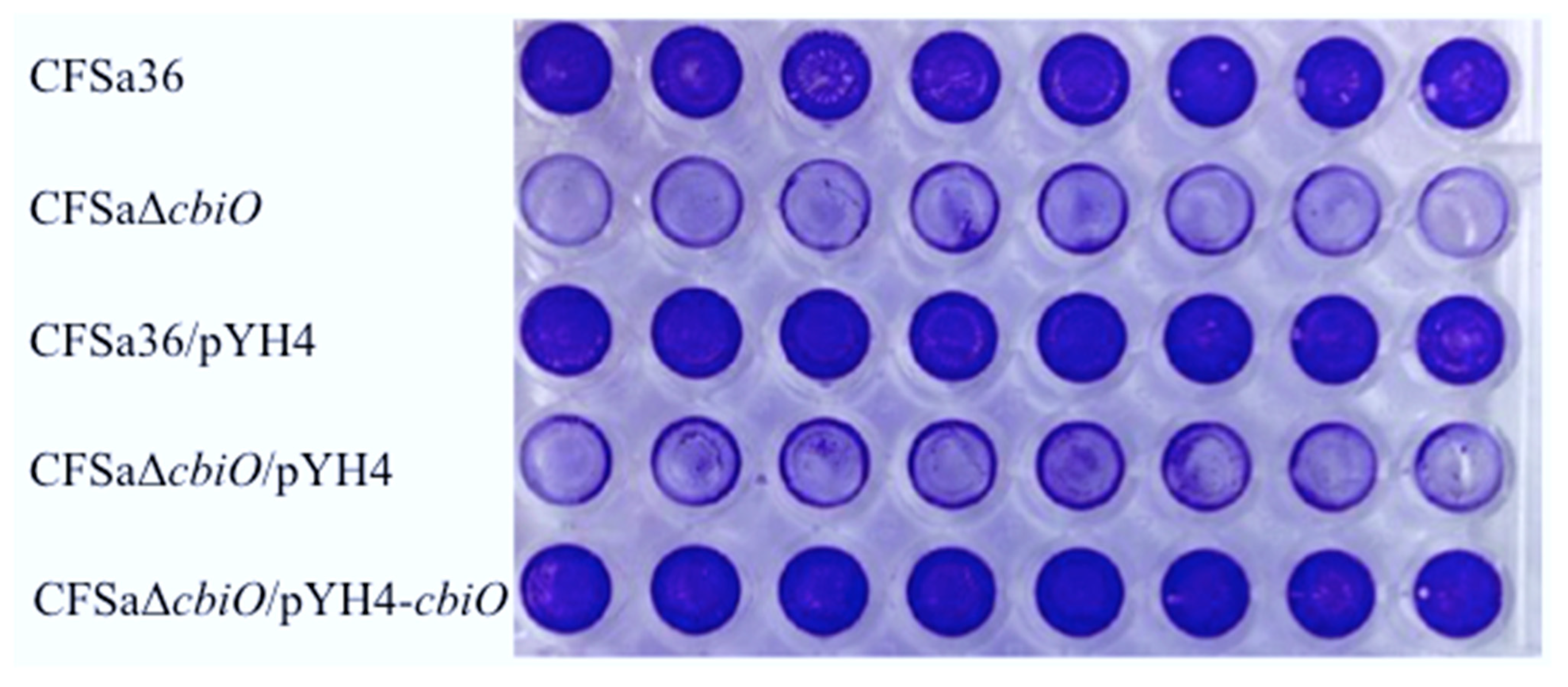
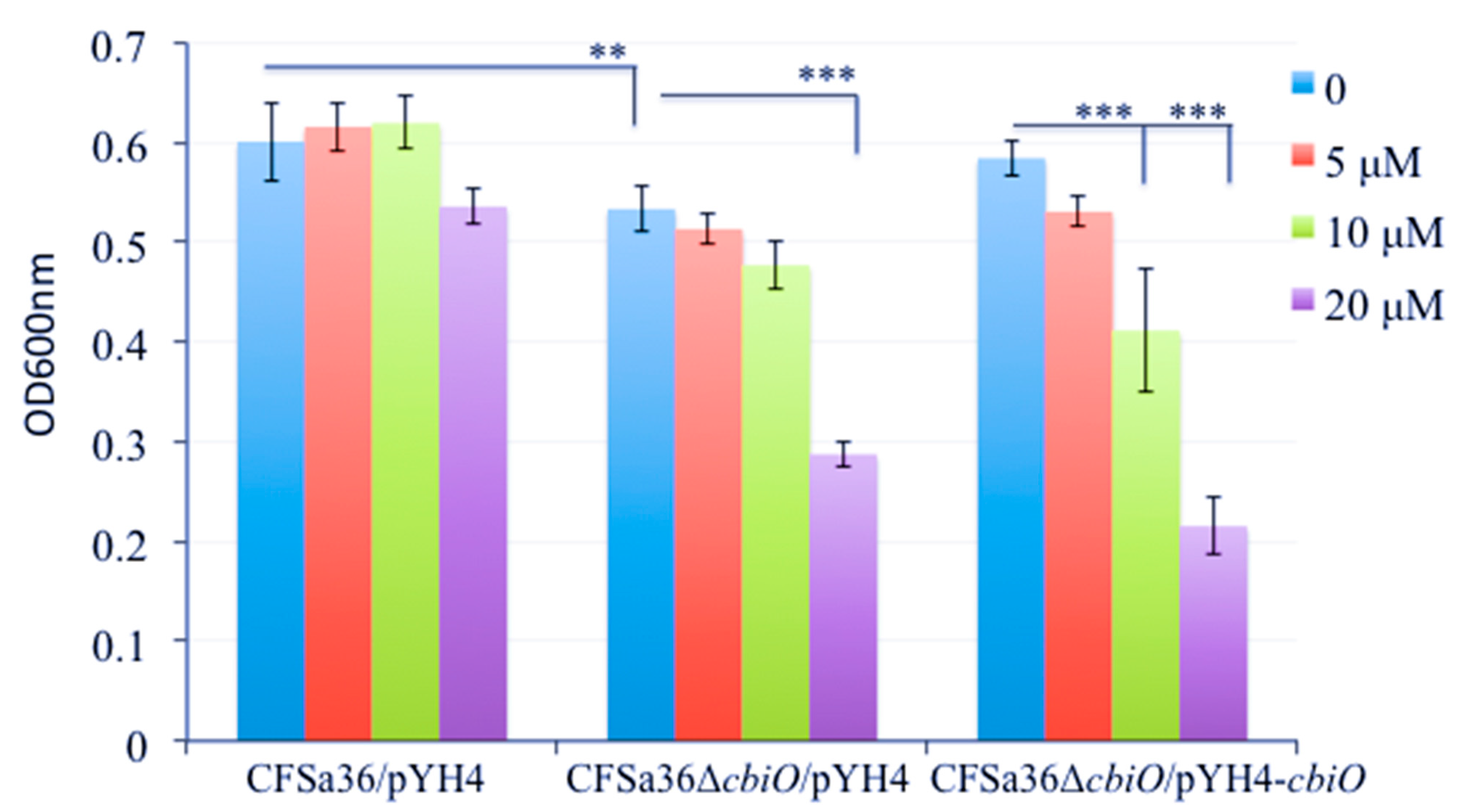
2.3. The Deletion Mutation of cbiO Dramatically Affected the Capacity of Biofilm Formation by an MRSA Strain, CFSa36, Isolated from a Pediatric Patient with Cystic Fibrosis
2.4. The Introduction of cbiO Expression Plasmid Complemented the Biofilm Formation Capacity of the cbiO Null Mutant
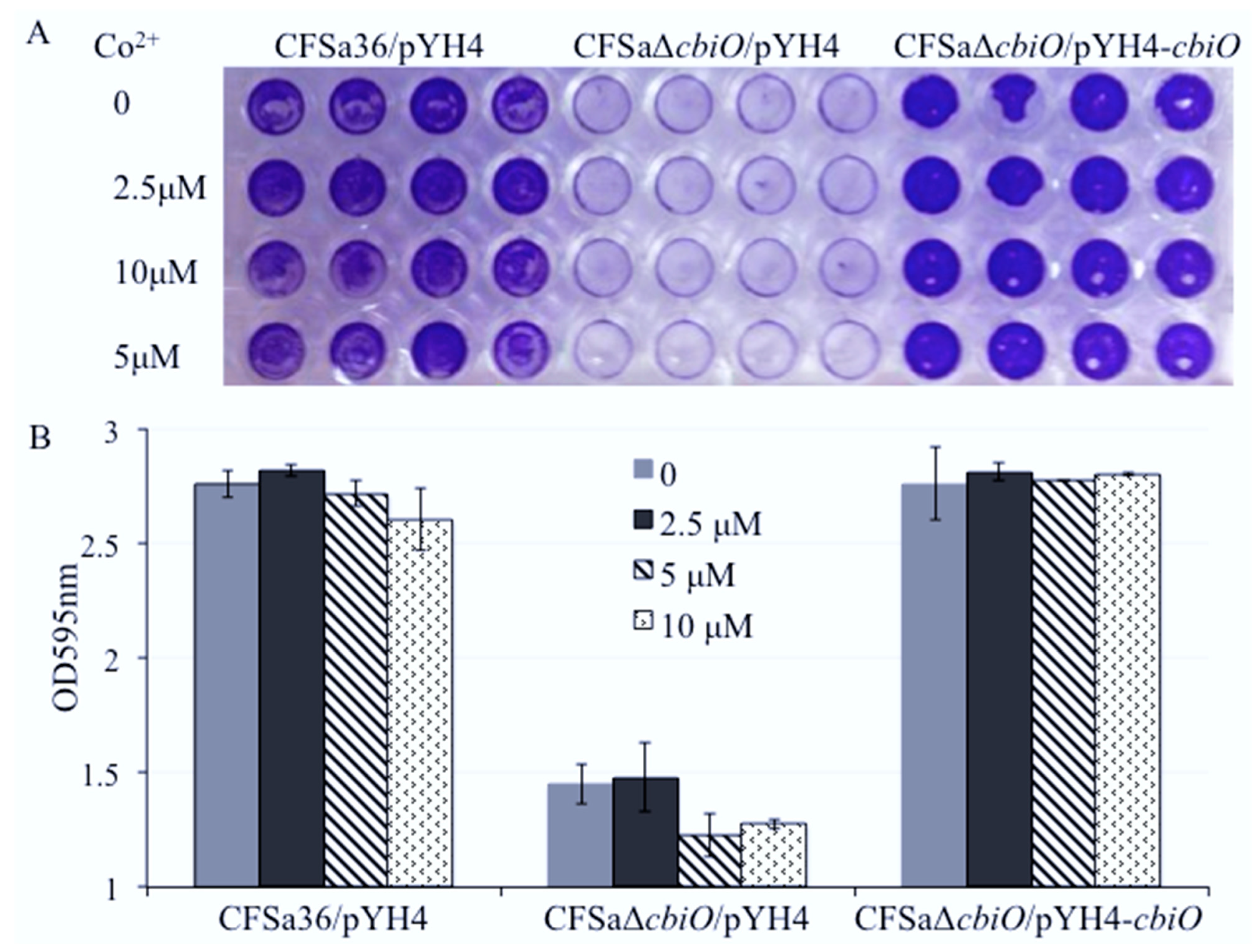
2.5. The Addition of Extra Copper Ion (CuSO4) in a Chemically Defined Medium (CDM) Had No Influence on Bacterial Growth, but It Complemented the Biofilm Formation Capacity of the cbiO Knockout Mutant
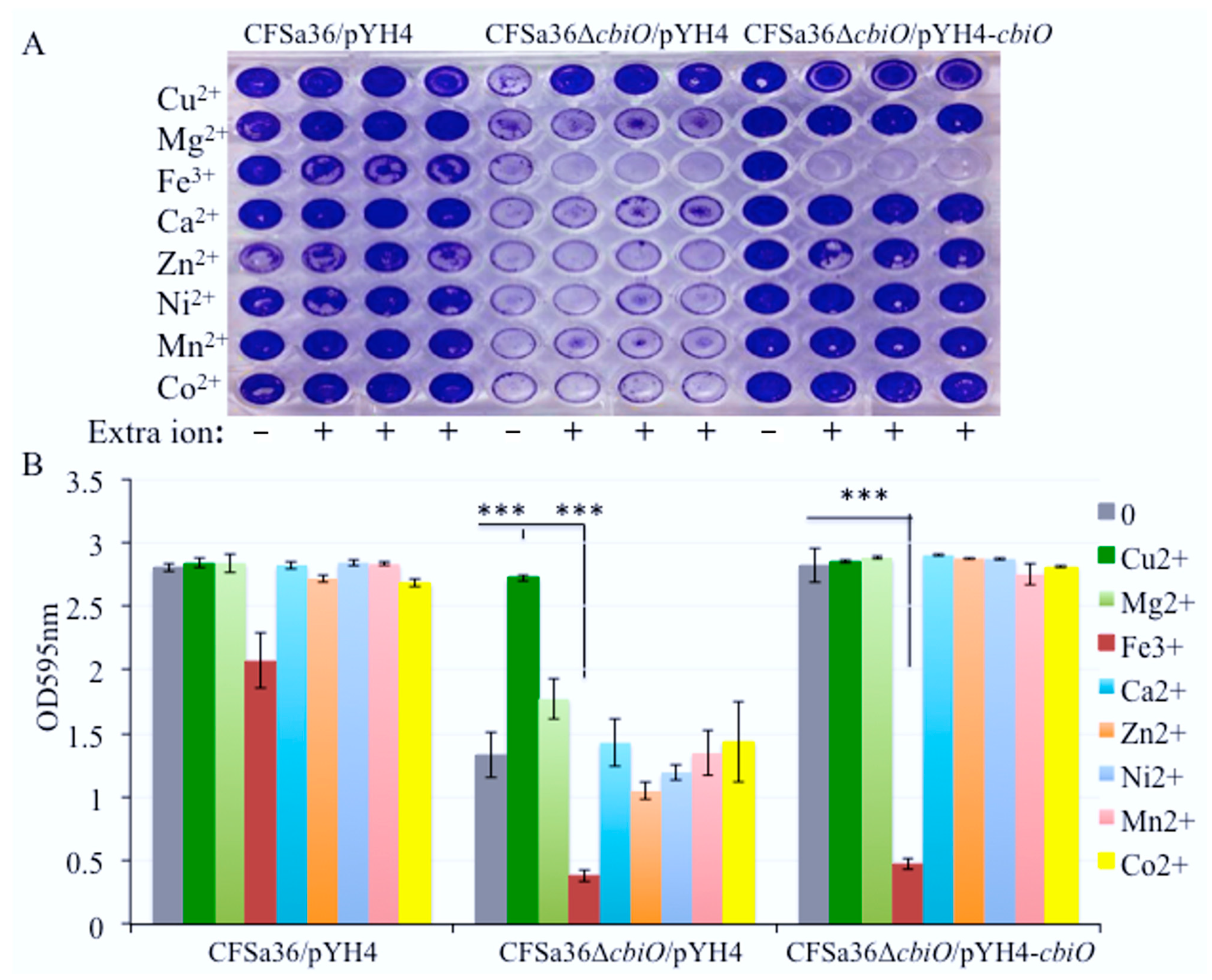
2.6. The Addition of Extra Fe3+ in CDM Enhanced the Effect of cbiO Null Mutation on Biofilm Formation
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains, Plasmids and Growth Media
4.2. Construction of the cbiO Gene Deletion Mutant and the cbiO Gene Complemented Strains
4.3. Static Biofilm Formation Assays
4.4. Data Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Li, C.; Wu, Y.; Riehle, A.; Ma, J.; Kamler, M.; Gulbins, E.; Grassmé, H. Staphylococcus aureus Survives in Cystic Fibrosis Macro-phages, Forming a Reservoir for Chronic Pneumonia. Infect. Immun. 2017, 85, e00883-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kahl, B.C. Impact of Staphylococcus aureus on the pathogenesis of chronic cystic fibrosis lung disease. Int. J. Med. Microbiol. 2010, 300, 514–519. [Google Scholar] [CrossRef]
- Cascioferro, S.; Carbone, D.; Parrino, B.; Pecoraro, C.; Giovannetti, E.; Cirrincione, G.; Diana, P. Therapeutic Strategies to Counteract Antibi-otic Resistance in MRSA Biofilm-Associated Infections. ChemMedChem 2021, 16, 65–80. [Google Scholar] [CrossRef]
- Dasenbrook, E.C.; Merlo, C.A.; Diener-West, M.; Lechtzin, N.; Boyle, M.P. Persistent methicillin-resistant Staphylococcus aure-us and rate of FEV1 decline in cystic fibrosis. Am. J. Respir. Crit. Care Med. 2008, 178, 814–821. [Google Scholar] [CrossRef] [PubMed]
- Boles, B.R.; Horswill, A.R. Staphylococcal biofilm disassembly. Trends Microbiol. 2011, 19, 449–455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Zhang, J.; Ji, Y. Environmental factors modulate biofilm formation by Staphylococcus aureus. Sci. Prog. 2020, 103, 0036850419898659. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Zhang, J.; Zhong, D.; Ji, L.; Yang, J.; Phillips, J.; Ji, Y. Characterization of Staphylococcus aureus isolates from pediatric patients with cystic fibrosis. World J. Microbiol. Biotechnol. 2016, 32, 162. [Google Scholar] [CrossRef] [PubMed]
- Valle, J.; Toledo-Arana, A.; Berasain, C.; Ghigo, J.-M.; Amorena, B.; Penadés, J.R.; Lasa, I. SarA and not σB is essential for biofilm development by Staphylococcus aureus. Mol. Microbiol. 2003, 48, 1075–1087. [Google Scholar] [CrossRef]
- O’Neill, E.; Humphreys, H.; O’Gara, J.P. Carriage of both the fnbA and fnbB genes and growth at 37 degrees C promote FnBP-mediated biofilm development in meticillin-resistant Staphylococcus aureus clinical isolates. J. Med. Microbiol. 2009, 58 Pt 4, 399–402. [Google Scholar] [CrossRef] [Green Version]
- O’Neill, E.; Pozzi, C.; Houston, P.; Humphreys, H.; Robinson, D.A.; Loughman, A.; Foster, T.J.; O’Gara, J.P. A novel Staphylococ-cus aureus biofilm phenotype mediated by the fibronectin-binding proteins, FnBPA and FnBPB. J. Bacteriol. 2008, 190, 3835–3850. [Google Scholar] [CrossRef] [Green Version]
- Beenken, K.E.; Blevins, J.S.; Smeltzer, M.S. Mutation of sarA in Staphylococcus aureus Limits Biofilm Formation. Infect. Immun. 2003, 71, 4206–4211. [Google Scholar] [CrossRef] [Green Version]
- Jefferson, K.K.; Pier, D.B.; Goldmann, D.A.; Pier, G.B. The Teicoplanin-Associated Locus Regulator (TcaR) and the Intercellular Adhesin Locus Regulator (IcaR) Are Transcriptional Inhibitors of the ica Locus in Staphylococcus aureus. J. Bacteriol. 2004, 186, 2449–2456. [Google Scholar] [CrossRef] [Green Version]
- Mlynek, K.D.; Callahan, M.T.; Shimkevitch, A.V.; Farmer, J.T.; Endres, J.L.; Marchand, M.; Bayles, K.W.; Horswill, A.R.; Kaplan, J.B. Effects of Low-Dose Amoxicillin on Staphylococcus aureus USA300 Biofilms. Antimicrob. Agents Chemother. 2016, 60, 2639–2651. [Google Scholar] [CrossRef] [Green Version]
- Lyczak, J.B.; Cannon, C.L.; Pier, G.B. Lung Infections Associated with Cystic Fibrosis. Clin. Microbiol. Rev. 2002, 15, 194–222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- LiPuma, J.J. The Changing Microbial Epidemiology in Cystic Fibrosis. Clin. Microbiol. Rev. 2010, 23, 299–323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phillips, J.R.; Tripp, T.J.; Regelmann, W.E.; Schlievert, P.M.; Wangensteen, O.D. Staphylococcal alpha-toxin causes increased tracheal epithelial permeability. Pediatr. Pulmonol. 2006, 41, 1146–1152. [Google Scholar] [CrossRef] [PubMed]
- Andersen, D.H. The Present Diagnosis and Therapy of Cystic Fibrosis of the Pancreas. Proc. R. Soc. Med. 1949, 42, 25–32. [Google Scholar] [CrossRef] [Green Version]
- Yagci, S.; Hascelik, G.; Dogru, D.; Ozcelik, U.; Sener, B. Prevalence and genetic diversity of Staphylococcus aureus small-colony variants in cystic fibrosis patients. Clin. Microbiol. Infect. 2013, 19, 77–84. [Google Scholar] [CrossRef] [Green Version]
- Emerson, J.; McNamara, S.; Buccat, A.M.; Worrell, K.; Burns, J.L. Changes in cystic fibrosis sputum microbiology in the United States between 1995 and 2008. Pediatr. Pulmonol. 2010, 45, 363–370. [Google Scholar] [CrossRef]
- Vu-Thien, H.; Hormigos, K.; Corbineau, G.; Fauroux, B.; Corvol, H.; Moissenet, D.; Vergnaud, G.; Pourcel, C. Longitudinal survey of Staphylococcus aureus in cystic fibrosis patients using a multiple-locus variable-number of tandem-repeats analysis method. BMC Microbiol. 2010, 10, 24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Costerton, J.W.; Stewart, P.; Greenberg, E. Bacterial Biofilms: A Common Cause of Persistent Infections. Science 1999, 284, 1318–1322. [Google Scholar] [CrossRef] [Green Version]
- O’Grady, N.P.; Alexander, M.; Dellinger, E.P.; Gerberding, J.L.; Heard, S.O.; Maki, D.G.; Masur, H.; McCormick, R.D.; Mermel, L.A.; Pearson, M.L.; et al. Guidelines for the prevention of intravascular catheter-related infections. Centers for Disease Control and Prevention. MMWR Recomm. Rep. 2002, 51, 1–29. [Google Scholar]
- Lemire, J.A.; Harrison, J.; Turner, R.J. Antimicrobial activity of metals: Mechanisms, molecular targets and applications. Nat. Rev. Genet. 2013, 11, 371–384. [Google Scholar] [CrossRef]
- Dinh, Т.L.; Akhmetova, G.R.; Martykanova, D.S.; Rudakova, N.L.; Sharipova, М.R. Influence of Divalent Metal Ions on Biofilm Formation by Bacillus subtilis. BioNanoScience 2019, 9, 521–527. [Google Scholar] [CrossRef]
- Sirelkhatim, A.; Mahmud, S.; Seeni, A.; Kaus, N.H.M.; Ann, L.C.; Bakhori, S.K.M.; Hasan, H.; Mohamad, D. Review on Zinc Oxide Nanoparticles: Antibacterial Activity and Toxicity Mechanism. Nano-Micro Lett. 2015, 7, 219–242. [Google Scholar] [CrossRef] [Green Version]
- Dupont, C.L.; Grass, G.; Rensing, C. Copper toxicity and the origin of bacterial resistance—new insights and applications. Metallomics 2011, 3, 1109–1118. [Google Scholar] [CrossRef] [PubMed]
- Hsueh, Y.-H.; Ke, W.-J.; Hsieh, C.-T.; Lin, K.-S.; Tzou, D.-Y.; Chiang, C.-L. ZnO Nanoparticles Affect Bacillus subtilis Cell Growth and Biofilm Formation. PLoS ONE 2015, 10, e0128457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahire, J.J.; Dicks, L.M.T. Nisin Incorporated With 2,3-Dihydroxybenzoic Acid in Nanofibers Inhibits Biofilm Formation by a Methicillin-Resistant Strain of Staphylococcus aureus. Probiotics Antimicrob. Proteins 2014, 7, 52–59. [Google Scholar] [CrossRef] [PubMed]
- García, C.F.; Kretschmer, M.; Lozano-Andrade, C.N.; Schönleitner, M.; Dragos, A.; Kovács, Á.T.; Lieleg, O. Metal ions weaken the hydrophobicity and antibiotic resistance of Bacillus subtilis NCIB 3610 biofilms. NPJ Biofilms Microbiomes 2020, 6 Pt 8, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Abraham, N.M.; Lamlertthon, S.; Fowler, V.G.; Jefferson, K.K. Chelating agents exert distinct effects on biofilm formation in Staphylococcus aureus depending on strain background: Role for clumping factor B. J. Med. Microbiol. 2012, 61, 1062–1070. [Google Scholar] [CrossRef] [Green Version]
- Zielinska, A.K.; Beenken, K.E.; Mrak, L.N.; Spencer, H.J.; Post, G.R.; Skinner, R.A.; Tackett, A.J.; Horswill, A.R.; Smeltzer, M.S. sarA-mediated repression of protease production plays a key role in the pathogenesis of Staphylococcus aureus USA300 isolates. Mol. Microbiol. 2012, 86, 1183–1196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abd El-Hamid, M.I.; YEl-Naenaeey, E.S.; MKandeel, T.; Hegazy, W.A.H.; Mosbah, R.A.; Nassar, M.S.; Bakhrebah, M.A.; Abdulaal, W.H.; Alhakamy, N.A.; Bendary, M.M. Promising Antibiofilm Agents: Recent Breakthrough against Biofilm Producing Methi-cillin-Resistant Staphylococcus aureus. Antibiotics 2020, 9, 667. [Google Scholar] [CrossRef] [PubMed]
- Carbone, D.; Parrino, B.; Cascioferro, S.; Pecoraro, C.; Giovannetti, E.; Di Sarno, V.; Musella, S.; Auriemma, G.; Cirrincione, G.; Diana, P. 1,2,4-Oxadiazole Topsentin Analogs with Antiproliferative Activity against Pancreatic Cancer Cells, Targeting GSK3β Kinase. ChemMedChem 2021, 16, 537–554. [Google Scholar] [CrossRef]
- Monk, I.R.; Shah, I.M.; Xu, M.; Tan, M.-W.; Foster, T.J. Transforming the Untransformable: Application of Direct Transformation to Manipulate Genetically Staphylococcus aureus and Staphylococcus epidermidis. mBio 2012, 3, e00277-11. [Google Scholar] [CrossRef] [Green Version]
- Fey, P.D.; Endres, J.L.; Yajjala, V.K.; Widhelm, T.J.; Boissy, R.J.; Bose, J.L.; Bayles, K.W. A genetic resource for rapid and compre-hensive phenotype screening of nonessential Staphylococcus aureus genes. mBio 2013, 4, e00537-12. [Google Scholar] [CrossRef] [Green Version]
- Bae, T.; Schneewind, O. Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid 2006, 55, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; O’Toole, P.; Shen, W.; Amrine-Madsen, H.; Jiang, X.; Lobo, N.; Palmer, L.M.; Voelker, L.; Fan, F.; Gwynn, M.N.; et al. Novel Chromosomally Encoded Multidrug Efflux Transporter MdeA in Staphylococcus aureus. Antimicrob. Agents Chemother. 2004, 48, 909–917. [Google Scholar] [CrossRef] [Green Version]
- Yan, M.; Yu, C.; Yang, J.; Ji, Y. The essential two-component system YhcSR is involved in regulation of the nitrate respira-tory pathway of Staphylococcus aureus. J. Bacteriol. 2011, 193, 1799–1805. [Google Scholar] [CrossRef]
- Cassat, J.E.; Smeltzer, M.S.; Lee, C.Y. Investigation of Biofilm Formation in Clinical Isolates of Staphylococcus aureus. Methods Mol. Biol. 2014, 1085, 195–211. [Google Scholar] [CrossRef] [Green Version]



| Strain, Plasmid or Primer | Relevant Characteristics | Reference |
|---|---|---|
| Strain DC10B | Dam-E. coli | [34] |
| Nebraska Tn-mutant library | 1920 genes transposon mutants, ErmR | [35] |
| JE2 | USA300 CA-MRSA isolate | [35] |
| 15983 | Clinical S. aureus biofilm formation strain | [8] |
| CFSa isolates (49) CFSa36 | Human clinical MRSA isolate Human clinical MRSA isolate, sigB+/rsbU+ | [7] [7] |
| CFSa36ΔcbiO | CFSa36 cbiO deletion mutant | This study |
| CFSa36ΔcbiO/pYH4 | CFSa36 cbiO deletion mutant with pYH4; ErmR | This study |
| CFSa36ΔcbiO/pYH4-cbiO | CFSa36 cbiO deletion mutant with pYH4-cbiO; ErmR | This study |
| Plasmid | ||
| pKOR1 | Temperature sensitive inducible allelic exchange plasmid for S. aureus; CmR | [36] |
| pKOR1-cbiO | pKOR1 with in-frame cbiO upstream/downstream deletion region; CmR | This study |
| pYH4 | Shuttle vector with Tc inducible promoter; ErmR | [37] |
| pYH4-cbiO | cbiO cloned downstream of pYH4 tet promoter; ErmR | This study |
| Primer | ||
| cbiO_pKOR1_L_F | 5′GGGGACAAGTTTGTACAAAAAAGCAGGCTGTGCAAACAC CCAAAGATATG3′ | |
| cbiO_pKOR1_R_R | 5′GGGGACCACTTTGTACAAGAAAGCTGGGTGCTGACATGA TGAAAGTGCG3′ | |
| cbiO_L_R | 5′GAAGGGCTGGTGGATCAAC3′ | |
| cbiO_R_F | 5′Phos/CACTTGTCTCTCTCCTTTAC3′ | |
| cbiO_F | 5′AGCTTTGTTTAAACGTGGAGGATAAGAATTCAG5′ | |
| cbiO_R | 5′AGGCGCGCCTCATAGTTGATCCACCAG CC3′ | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Yang, J.; Ji, M.; Phillips, J.; Wylam, M.; Ji, Y. Identification of cbiO Gene Critical for Biofilm Formation by MRSA CFSa36 Strain Isolated from Pediatric Patient with Cystic Fibrosis. Pathogens 2021, 10, 1363. https://doi.org/10.3390/pathogens10111363
Liu Y, Yang J, Ji M, Phillips J, Wylam M, Ji Y. Identification of cbiO Gene Critical for Biofilm Formation by MRSA CFSa36 Strain Isolated from Pediatric Patient with Cystic Fibrosis. Pathogens. 2021; 10(11):1363. https://doi.org/10.3390/pathogens10111363
Chicago/Turabian StyleLiu, Ying, Junshu Yang, Michelle Ji, James Phillips, Mark Wylam, and Yinduo Ji. 2021. "Identification of cbiO Gene Critical for Biofilm Formation by MRSA CFSa36 Strain Isolated from Pediatric Patient with Cystic Fibrosis" Pathogens 10, no. 11: 1363. https://doi.org/10.3390/pathogens10111363
APA StyleLiu, Y., Yang, J., Ji, M., Phillips, J., Wylam, M., & Ji, Y. (2021). Identification of cbiO Gene Critical for Biofilm Formation by MRSA CFSa36 Strain Isolated from Pediatric Patient with Cystic Fibrosis. Pathogens, 10(11), 1363. https://doi.org/10.3390/pathogens10111363







