Use of an Ecosystem-Based Approach to Shed Light on the Heterogeneity of the Contamination Pattern of Listeria monocytogenes on Conveyor Belt Surfaces in a Swine Slaughterhouse in the Province of Quebec, Canada
Abstract
:1. Introduction
2. Results
2.1. Listeria monocytogenes Detection
2.2. Listeria monocytogenes Classical and Molecular Characterization
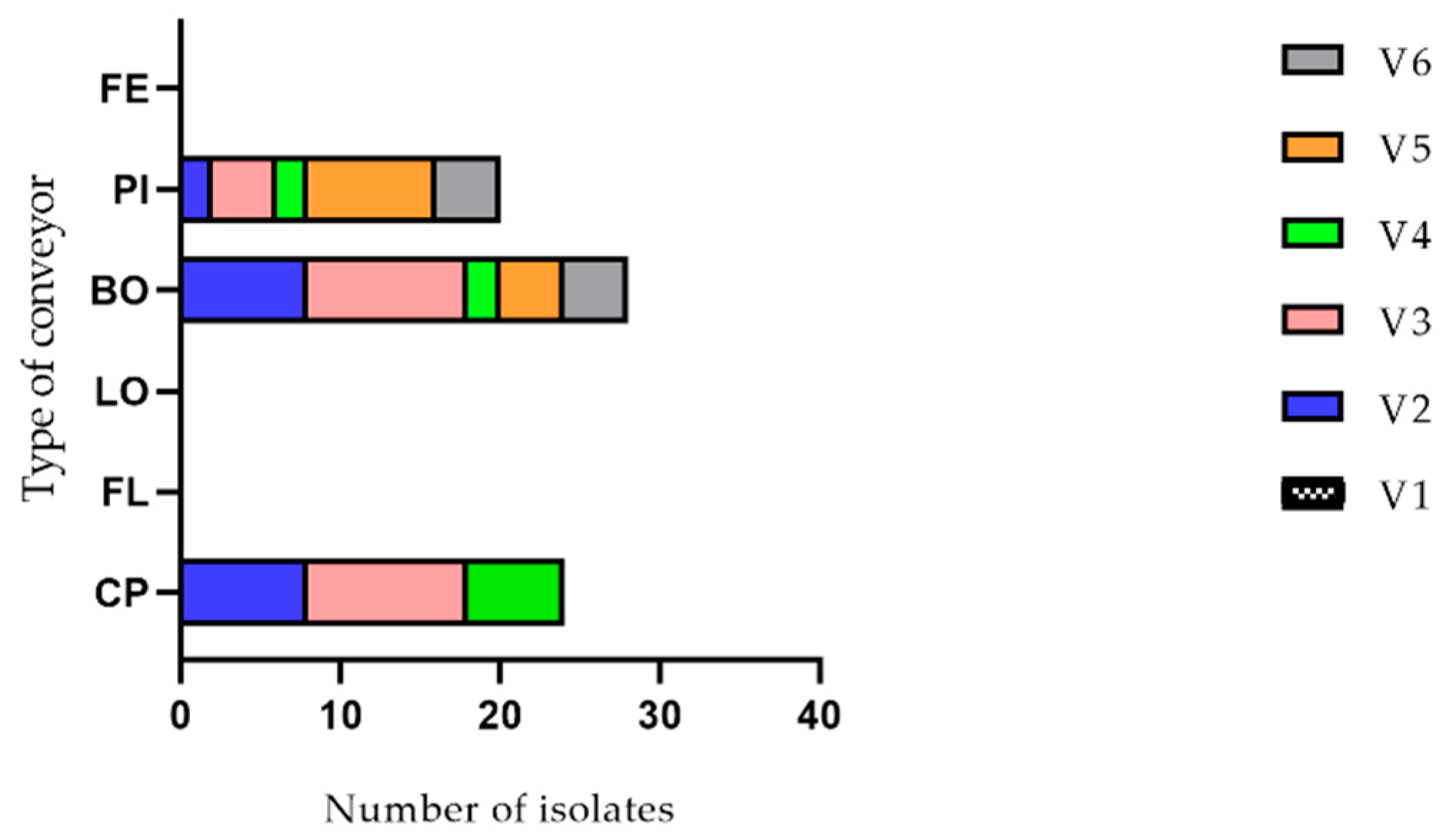
2.2.1. Listeria monocytogenes PCR-Serogroups
2.2.2. InlA Sequencing
2.2.3. Determination of the Ability to Form Biofilm
2.2.4. MLST and cgMLST Characterization, Virulence, Antimicrobial Resistance and Stress-Related Genes
2.3. Microbiota Analysis
2.3.1. Sequencing Data
2.3.2. Alpha Diversity
2.3.3. Beta Diversity
3. Discussion
4. Materials and Methods
4.1. Sampling
4.2. Listeria monocytogenes Detection
4.3. Total Microbiota Harvesting
4.4. Listeria monocytogenes Classical and Molecular Characterization
4.5. Selection, DNA Isolation, Library Preparation and Sequencing of the L. monocytogenes Isolates
4.6. MLST and cgMLST Characterization and Virulence, Antimicrobial Resistance and Stress-Related Genes
4.7. DNA Extraction and Purification of the Total Microbiota
4.8. Total Microbiota 16S Sequencing and Bio-Informatics Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ramaswamy, V.; Cresence, V.M.; Rejitha, J.S.; Lekshmi, M.U.; Dharsana, K.S.; Prasad, S.P.; Vijila, H.M. Listeria-review of epidemiology and pathogenesis. J. Microbiol. Immunol. Infect. 2007, 40, 4–13. [Google Scholar]
- Burall, L.S.; Grim, C.J.; Mammel, M.K.; Datta, A.R.A. Comprehensive Evaluation of the Genetic Relatedness of Listeria monocytogenes Serotype 4b Variant Strains. Front. Public Health 2017, 5, 241. [Google Scholar] [CrossRef] [Green Version]
- Norton, D.M.; McCamey, M.A.; Gall, K.L.; Scarlett, J.M.; Boor, K.J.; Wiedmann, M. Molecular studies on the ecology of Listeria monocytogenes in the smoked fish processing industry. Appl. Environ. Microbiol. 2001, 67, 198–205. [Google Scholar] [CrossRef] [Green Version]
- Laksanalamai, P.; Joseph, L.A.; Silk, B.J.; Burall, L.S.; Tarr, C.L.; Gerner-Smidt, P.; Datta, A.R. Genomic Characterization of Listeria monocytogenes Strains Involved in a Multistate Listeriosis Outbreak Associated with Cantaloupe in US. PLoS ONE 2012, 7, e42448. [Google Scholar] [CrossRef]
- Currie, A.; Farber, J.M.; Nadon, C.; Sharma, D.; Whitfield, Y.; Gaulin, C.; Galanis, E.; Bekal, S.; Flint, J.; Tschetter, L.; et al. Multi-Province Listeriosis Outbreak Linked to Contaminated Deli Meat Consumed Primarily in Institutional Settings, Canada, 2008. Foodborne Pathog. Dis. 2015, 12, 645–652. [Google Scholar] [CrossRef] [PubMed]
- Thomas, M.K.; Vriezen, R.; Farber, J.M.; Currie, A.; Schlech, W.; Fazil, A. Economic Cost of a Listeria monocytogenes Outbreak in Canada, 2008. Foodborne Pathog. Dis. 2015, 12, 966–971. [Google Scholar] [CrossRef] [Green Version]
- Simmons, C.K.; Wiedmann, M. Identification and classification of sampling sites for pathogen environmental monitoring programs for Listeria monocytogenes: Results from an expert elicitation. Food Microbiol. 2018, 75, 2–17. [Google Scholar] [CrossRef] [PubMed]
- Nastasijevic, I.; Milanov, D.; Velebit, B.; Djordjevic, V.; Swift, C.; Painset, A.; Lakicevic, B. Tracking of Listeria monocytogenes in meat establishment using Whole Genome Sequencing as a food safety management tool: A proof of concept. Int. J. Food Microbiol. 2017, 257, 157–164. [Google Scholar] [CrossRef] [PubMed]
- Bolocan, A.S.; Nicolau, A.I.; Alvarez-Ordóñez, A.; Borda, D.; Oniciuc, E.A.; Stessl, B.; Gurgu, L.; Wagner, M.; Jordan, K. Dynamics of Listeria monocytogenes colonisation in a newly-opened meat processing facility. Meat Sci. 2016, 113, 26–34. [Google Scholar] [CrossRef]
- Bergholz, T.M.; Shah, M.K.; Burall, L.S.; Rakic-Martinez, M.; Datta, A.R. Genomic and phenotypic diversity of Listeria monocytogenes clonal complexes associated with human listeriosis. Appl. Microbiol. Biotechnol. 2018, 102, 3475–3485. [Google Scholar] [CrossRef] [PubMed]
- Martín, B.; Perich, A.; Gómez, D.; Yangüela, J.; Rodríguez, A.; Garriga, M.; Aymerich, T. Diversity and distribution of Listeria monocytogenes in meat processing plants. Food Microbiol. 2014, 44, 119–127. [Google Scholar] [CrossRef]
- Kurpas, M.; Osek, J.; Moura, A.; Leclercq, A.; Lecuit, M.; Wieczorek, K. Genomic Characterization of Listeria monocytogenes Isolated From Ready-to-Eat Meat and Meat Processing Environments in Poland. Front. Microbiol. 2020, 11, 1412. [Google Scholar] [CrossRef]
- Moura, A.; Criscuolo, A.; Pouseele, H.; Maury, M.M.; Leclercq, A.; Tarr, C.; Björkman, J.T.; Dallman, T.; Reimer, A.; Enouf, V.; et al. Whole genome-based population biology and epidemiological surveillance of Listeria monocytogenes. Nat. Microbiol. 2016, 2, 16185. [Google Scholar] [CrossRef] [PubMed]
- De las Heras, A.; Cain, R.J.; Bielecka, M.K.; Vázquez-Boland, J.A. Regulation of Listeria virulence: PrfA master and commander. Curr. Opin. Microbiol. 2011, 14, 118–127. [Google Scholar] [CrossRef] [PubMed]
- Maury, M.M.; Tsai, Y.H.; Charlier, C.; Touchon, M.; Chenal-Francisque, V.; Leclercq, A.; Criscuolo, A.; Gaultier, C.; Roussel, S.; Brisabois, A.; et al. Uncovering Listeria monocytogenes hypervirulence by harnessing its biodiversity. Nat. Genet. 2016, 48, 308–313. [Google Scholar] [CrossRef] [Green Version]
- Jacquet, C.; Doumith, M.; Gordon, J.I.; Martin, P.M.; Cossart, P.; Lecuit, M. A molecular marker for evaluating the pathogenic potential of foodborne Listeria monocytogenes. J. Infect. Dis. 2004, 189, 2094–2100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nightingale, K.K.; Windham, K.; Martin, K.E.; Yeung, M.; Wiedmann, M. Select Listeria monocytogenes subtypes commonly found in foods carry distinct nonsense mutations in inlA, leading to expression of truncated and secreted internalin A, and are associated with a reduced invasion phenotype for human intestinal epithelial cells. Appl. Environ. Microbiol. 2005, 71, 8764–8772. [Google Scholar]
- Fravalo, P.; Cherifi, T.; Feliciano, K.D.N.; Letellier, A.; Fairbrother, J.-H.; Bekal, S. Characterisation of InlA truncation in Listeria monocytogenes isolates from farm animals and human cases in the province of Quebec. Vet. Rec. Open 2017, 4, e000199. [Google Scholar] [CrossRef] [Green Version]
- Hein, I.; Klinger, S.; Dooms, M.; Flekna, G.; Stessl, B.; Leclercq, A.; Hill, C.; Allerberger, F.; Wagner, M. Stress survival islet 1 (SSI-1) survey in Listeria monocytogenes reveals an insert common to listeria innocua in sequence type 121 L. monocytogenes strains. Appl. Environ. Microbiol. 2011, 77, 2169–2173. [Google Scholar] [CrossRef] [Green Version]
- Dutta, V.; Elhanafi, D.; Kathariou, S. Conservation and distribution of the benzalkonium chloride resistance cassette bcrABC in Listeria monocytogenes. Appl. Environ. Microbiol. 2013, 79, 6067–6074. [Google Scholar] [CrossRef] [Green Version]
- Cherifi, T.; Carrillo, C.; Lambert, D.; Miniaï, I.; Quessy, S.; Lariviere-Gauthier, G.; Blais, B.; Fravalo, P. Genomic characterization of Listeria monocytogenes isolates reveals that their persistence in a pig slaughterhouse is linked to the presence of benzalkonium chloride resistance genes. BMC Microbiol. 2018, 18, 220. [Google Scholar] [CrossRef]
- Kremer, P.H.C.; Lees, J.A.; Koopmans, M.M.; Ferwerda, B.; Arends, A.W.A.; Feller, M.M.; Schipper, K.; Valls Seron, M.; van der Ende, A.; Brouwer, M.C.; et al. Benzalkonium tolerance genes and outcome in Listeria monocytogenes meningitis. Clin. Microbiol. Infect. 2017, 23, 265.e1–265.e7. [Google Scholar] [CrossRef] [Green Version]
- Carpentier, B.; Cerf, O. Review—Persistence of Listeria monocytogenes in food industry equipment and premises. Int. J. Food Microbiol. 2011, 145, 1–8. [Google Scholar] [CrossRef]
- Stasiewicz, M.J.; Oliver, H.F.; Wiedmann, M.; den Bakker, H.C. Whole-Genome Sequencing Allows for Improved Identification of Persistent Listeria monocytogenes in Food-Associated Environments. Appl. Environ. Microbiol. 2015, 81, 6024–6037. [Google Scholar] [CrossRef] [Green Version]
- Zilelidou, E.A.; Skandamis, P.N. Growth, detection and virulence of Listeria monocytogenes in the presence of other microorganisms: Microbial interactions from species to strain level. Int. J. Food Microbiol. 2018, 277, 10–25. [Google Scholar] [CrossRef]
- Chasseignaux, E.; Gérault, P.; Toquin, M.T.; Salvat, G.; Colin, P.; Ermel, G. Ecology of Listeria monocytogenes in the environment of raw poultry meat and raw pork meat processing plants. FEMS Microbiol. Lett. 2002, 210, 271–275. [Google Scholar] [CrossRef]
- Carpentier, B.; Chassaing, D. Interactions in biofilms between Listeria monocytogenes and resident microorganisms from food industry premises. Int. J. Food Microbiol. 2004, 97, 111–122. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.; Chung, T.; Chen, Y.; Macarisin, D.; LaBorde, L.; Kovac, J. The occurrence of Listeria monocytogenes is associated with built environment microbiota in three tree fruit processing facilities. Microbiome 2019, 7, 115. [Google Scholar] [CrossRef]
- Giaouris, E.; Heir, E.; Hébraud, M.; Chorianopoulos, N.; Langsrud, S.; Møretrø, T.; Habimana, O.; Desvaux, M.; Renier, S.; Nychas, G.J. Attachment and biofilm formation by foodborne bacteria in meat processing environments: Causes, implications, role of bacterial interactions and control by alternative novel methods. Meat Sci. 2014, 97, 298–309. [Google Scholar] [CrossRef] [PubMed]
- Pang, X.; Yuk, H.G. Effects of the colonization sequence of Listeria monocytogenes and Pseudomonas fluorescens on survival of biofilm cells under food-related stresses and transfer to salmon. Food Microbiol. 2019, 82, 142–150. [Google Scholar] [CrossRef] [PubMed]
- Papaioannou, E.; Giaouris, E.D.; Berillis, P.; Boziaris, I.S. Dynamics of biofilm formation by Listeria monocytogenes on stainless steel under mono-species and mixed-culture simulated fish processing conditions and chemical disinfection challenges. Int. J. Food Microbiol. 2018, 267, 9–19. [Google Scholar] [CrossRef]
- Mitri, S.; Xavier, J.B.; Foster, K.R. Social evolution in multispecies biofilms. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. 2), 10839. [Google Scholar] [CrossRef] [Green Version]
- Rendueles, O.; Ghigo, J.M. Multi-species biofilms: How to avoid unfriendly neighbors. FEMS Microbiol. Rev. 2012, 36, 972–989. [Google Scholar] [CrossRef]
- Ripolles-Avila, C.; Hascoët, A.S.; Martínez-Suárez, J.V.; Capita, R.; Rodríguez-Jerez, J.J. Evaluation of the microbiological contamination of food processing environments through implementing surface sensors in an iberian pork processing plant: An approach towards the control of Listeria monocytogenes. Food Control 2019, 99, 40–47. [Google Scholar] [CrossRef]
- Ferreira, V.; Wiedmann, M.; Teixeira, P.; Stasiewicz, M.J. Listeria monocytogenes persistence in food-associated environments: Epidemiology, strain characteristics, and implications for public health. J. Food Prot. 2014, 77, 150–170. [Google Scholar] [CrossRef] [PubMed]
- Langsrud, S.; Moen, B.; Møretrø, T.; Løype, M.; Heir, E. Microbial dynamics in mixed culture biofilms of bacteria surviving sanitation of conveyor belts in salmon-processing plants. J. Appl. Microbiol. 2016, 120, 366–378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stephan, R.; Althaus, D.; Kiefer, S.; Lehner, A.; Hatz, C.; Schmutz, C.; Jost, M.; Gerber, N.; Baumgartner, A.; Hächler, H.; et al. Foodborne transmission of Listeria monocytogenes via ready-to-eat salad: A nationwide outbreak in Switzerland, 2013–2014. Food Control 2015, 57, 14–17. [Google Scholar] [CrossRef]
- Colagiorgi, A.; Bruini, I.; Di Ciccio, P.A.; Zanardi, E.; Ghidini, S.; Ianieri, A. Listeria monocytogenes Biofilms in the Wonderland of Food Industry. Pathogens 2017, 6, 41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fagerlund, A.; Møretrø, T.; Heir, E.; Briandet, R.; Langsrud, S. Cleaning and Disinfection of Biofilms Composed of Listeria monocytogenes and Background Microbiota from Meat Processing Surfaces. Appl. Environ. Microbiol. 2017, 83, e01046-e17. [Google Scholar] [CrossRef] [Green Version]
- Ciccio, P.D.; Conter, M.; Zanardi, E.; Ghidini, S.; Vergara, A.; Paludi, D.; Festino, A.R.; Ianieri, A. Listeria monocytogenes: Biofilms in Food Processing. Ital. J. Food Sci. 2012, 24, 203–213. [Google Scholar]
- Fagerlund, A.; Langsrud, S.; Møretrø, T. Microbial diversity and ecology of biofilms in food industry environments associated with Listeria monocytogenes persistence. Curr. Opin. Food Sci. 2021, 37, 171–178. [Google Scholar] [CrossRef]
- Bremer, P.J.; Monk, I.; Osborne, C.M. Survival of Listeria monocytogenes attached to stainless steel surfaces in the presence or absence of Flavobacterium spp. J. Food Prot. 2001, 64, 1369–1376. [Google Scholar] [CrossRef]
- Heir, E.; Møretrø, T.; Simensen, A.; Langsrud, S. Listeria monocytogenes strains show large variations in competitive growth in mixed culture biofilms and suspensions with bacteria from food processing environments. Int. J. Food Microbiol. 2018, 275, 46–55. [Google Scholar] [CrossRef]
- Møretrø, T.; Langsrud, S.; Heir, E. Bacteria on Meat Abattoir Process Surfaces after Sanitation: Characterisation of Survival Properties of Listeria monocytogenes and the Commensal Bacterial Flora. Adv. Microbiol. 2013, 3, 10. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez-López, P.; Bernárdez, M.; Rodríguez-Herrera, J.J.; Comesaña, Á.S.; Cabo, M.L. Identification and metagenetic characterisation of Listeria monocytogenes-harbouring communities present in food-related industrial environments. Food Control 2019, 95, 6–17. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Zhang, H.; Wu, C.; Deng, W.; Wang, D.; Zhao, G.; Song, J.; Jiang, Y. Molecular analysis of dominant species in Listeria monocytogenes-positive biofilms in the drains of food processing facilities. Appl. Microbiol. Biotechnol. 2016, 100, 3165–3175. [Google Scholar] [CrossRef] [PubMed]
- Dzieciol, M.; Schornsteiner, E.; Muhterem-Uyar, M.; Stessl, B.; Wagner, M.; Schmitz-Esser, S. Bacterial diversity of floor drain biofilms and drain waters in a Listeria monocytogenes contaminated food processing environment. Int. J. Food Microbiol. 2016, 223, 33–40. [Google Scholar] [CrossRef]
- Fox, E.M.; Solomon, K.; Moore, J.E.; Wall, P.G.; Fanning, S. Phylogenetic profiles of in-house microflora in drains at a food production facility: Comparison and biocontrol implications of Listeria-positive and-negative bacterial populations. Appl. Microbiol. Biotechnol. 2014, 80, 3369–3374. [Google Scholar] [CrossRef] [Green Version]
- Calasso, M.; Ercolini, D.; Mancini, L.; Stellato, G.; Minervini, F.; Di Cagno, R.; De Angelis, M.; Gobbetti, M. Relationships among house, rind and core microbiotas during manufacture of traditional Italian cheeses at the same dairy plant. Food Microbiol. 2016, 54, 115–126. [Google Scholar] [CrossRef]
- Doyle, C.J.; O’Toole, P.W.; Cotter, P.D. Metagenome-based surveillance and diagnostic approaches to studying the microbial ecology of food production and processing environments. Environ. Microbiol. 2017, 19, 4382–4391. [Google Scholar] [CrossRef] [Green Version]
- Røder, H.L.; Sørensen, S.J.; Burmølle, M. Studying Bacterial Multispecies Biofilms: Where to Start? Trends Microbiol. 2016, 24, 503–513. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-López, P.; Rodríguez-Herrera, J.J.; Cabo, M.L. Tracking bacteriome variation over time in Listeria monocytogenes-positive foci in food industry. Int. J. Food Microbiol. 2020, 315, 108439. [Google Scholar] [CrossRef] [PubMed]
- SALA, C.; Morar, A.; Tîrziu, E.; Nichita, I.; Imre, M.; Imre, K. Environmental Occurrence and Antibiotic Susceptibility Profile of Listeria monocytogenes at a Slaughterhouse Raw Processing Plant in Romania. J. Food Prot. 2016, 79, 1794–1797. [Google Scholar] [CrossRef] [PubMed]
- AUTIO, T.; Säteri, T.; Fredriksson-Ahomaa, M.; Rahkio, M.; Lundén, J.; Korkeala, H. Listeria monocytogenes Contamination Pattern in Pig Slaughterhouses. J. Food Prot. 2000, 63, 1438–1442. [Google Scholar] [CrossRef] [PubMed]
- Muhterem-Uyar, M.; Dalmasso, M.; Bolocan, A.S.; Hernandez, M.; Kapetanakou, A.E.; Kuchta, T.; Manios, S.G.; Melero, B.; Minarovičová, J.; Nicolau, A.I.; et al. Environmental sampling for Listeria monocytogenes control in food processing facilities reveals three contamination scenarios. Food Control 2015, 51, 94–107. [Google Scholar] [CrossRef]
- Bolocan, A.S.; Oniciuc, E.A.; Alvarez-Ordóñez, A.; Wagner, M.; Rychli, K.; Jordan, K.; Nicolau, A.I. Putative Cross-Contamination Routes of Listeria monocytogenes in a Meat Processing Facility in Romania. J. Food Prot. 2015, 78, 1664–1674. [Google Scholar] [CrossRef] [PubMed]
- Pagadala, S.; Parveen, S.; Rippen, T.; Luchansky, J.B.; Call, J.E.; Tamplin, M.L.; Porto-Fett, A.C. Prevalence, characterization and sources of Listeria monocytogenes in blue crab (Callinectus sapidus) meat and blue crab processing plants. Food Microbiol. 2012, 31, 263–270. [Google Scholar] [CrossRef]
- De Filippis, F.; La Storia, A.; Villani, F.; Ercolini, D. Exploring the sources of bacterial spoilers in beefsteaks by culture-independent high-throughput sequencing. PLoS ONE 2013, 8, e70222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orsi, R.H.; den Bakker, H.C.; Wiedmann, M. Listeria monocytogenes lineages: Genomics, evolution, ecology, and phenotypic characteristics. Int. J. Med. Microbiol. 2011, 301, 79–96. [Google Scholar] [CrossRef]
- Ebner, R.; Stephan, R.; Althaus, D.; Brisse, S.; Maury, T.; Tasara, M. Phenotypic and genotypic characteristics of Listeria monocytogenes strains isolated during 2011–2014 from different food matrices in Switzerland. Food Control 2015, 57, 321–326. [Google Scholar] [CrossRef] [Green Version]
- Matereke, L.T.; Okoh, A.I. Listeria monocytogenes Virulence, Antimicrobial Resistance and Environmental Persistence: A Review. Pathogens 2020, 9, 528. [Google Scholar] [CrossRef] [PubMed]
- Muhterem-Uyar, M.; Ciolacu, L.; Wagner, K.-H.; Wagner, M.; Schmitz-Esser, S.; Stessl, B. New Aspects on Listeria monocytogenes ST5-ECVI Predominance in a Heavily Contaminated Cheese Processing Environment. Front. Microbiol. 2018, 9, 64. [Google Scholar] [CrossRef] [Green Version]
- Maury, M.M.; Bracq-Dieye, H.; Huang, L.; Vales, G.; Lavina, M.; Thouvenot, P.; Disson, O.; Leclercq, A.; Brisse, S.; Lecuit, M. Hypervirulent Listeria monocytogenes clones’ adaption to mammalian gut accounts for their association with dairy products. Nat. Commun. 2019, 10, 2488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hingston, P.; Chen, J.; Dhillon, B.K.; Laing, C.; Bertelli, C.; Gannon, V.; Tasara, T.; Allen, K.; Brinkman, F.S.L.; Truelstrup Hansen, L.; et al. Genotypes Associated with Listeria monocytogenes Isolates Displaying Impaired or Enhanced Tolerances to Cold, Salt, Acid, or Desiccation Stress. Front. Microbiol. 2017, 8, 369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chakraborty, T.; Leimeister-Wächter, M.; Domann, E.; Hartl, M.; Goebel, W.; Nichterlein, T.; Notermans, S. Coordinate regulation of virulence genes in Listeria monocytogenes requires the product of the prfA gene. J. Bacteriol. 1992, 174, 568–574. [Google Scholar] [CrossRef] [Green Version]
- Maury, M.M.; Chenal-Francisque, V.; Bracq-Dieye, H.; Han, L.; Leclercq, A.; Vales, G.; Moura, A.; Gouin, E.; Scortti, M.; Disson, O.; et al. Spontaneous Loss of Virulence in Natural Populations of Listeria monocytogenes. Infect. Immun. 2017, 85, e00541-e17. [Google Scholar] [CrossRef] [Green Version]
- Ryan, S.; Begley, M.; Hill, C.; Gahan, C.G.M. A five-gene stress survival islet (SSI-1) that contributes to the growth of Listeria monocytogenes in suboptimal conditions. J. Appl. Microbiol. 2010, 109, 984–995. [Google Scholar] [CrossRef]
- Crespo-Piazuelo, D.; Estellé, J.; Revilla, M.; Criado-Mesas, L.; Ramayo-Caldas, Y.; Óvilo, C.; Fernández, A.I.; Ballester, M.; Folch, J.M. Characterization of bacterial microbiota compositions along the intestinal tract in pigs and their interactions and functions. Sci. Rep. 2018, 8, 12727. [Google Scholar] [CrossRef]
- Huang, A.; Cai, R.; Wang, Q.; Shi, L.; Li, C.; Yan, H. Dynamic Change of Gut Microbiota During Porcine Epidemic Diarrhea Virus Infection in Suckling Piglets. Front. Microbiol. 2019, 10, 322. [Google Scholar] [CrossRef] [Green Version]
- Hinton, A., Jr.; Hume, M.E.; Deloach, J.R. Role of Metabolic Intermediates in the Inhibition of Salmonella typhimurium and Salmonella enteritidis by Veillonella. J. Food Prot. 1993, 56, 932–937. [Google Scholar] [CrossRef]
- Hinton, A., Jr.; Hume, M.E. Antibacterial activity of the metabolic by-products of a Veillonella species and Bacteroides fragilis. Anaerobe 1995, 1, 121–127. [Google Scholar] [CrossRef]
- Durant, J.A.; Nisbet, D.J.; Ricke, S.C. Comparison of batch culture growth and fermentation of a poultry Veillonella isolate and selected Veillonella species grown in a defined medium. Anaerobe 1997, 3, 391–397. [Google Scholar] [CrossRef] [PubMed]
- Kérouanton, A.; Marault, M.; Petit, L.; Grout, J.; Dao, T.T.; Brisabois, A. Evaluation of a multiplex PCR assay as an alternative method for Listeria monocytogenes serotyping. J. Microbiol. Methods 2010, 80, 134–137. [Google Scholar] [CrossRef] [PubMed]
- Burall, L.S.; Simpson, A.C.; Datta, A.R. Evaluation of a serotyping scheme using a combination of an antibody-based serogrouping method and a multiplex PCR assay for identifying the major serotypes of Listeria monocytogenes. J. Food Prot. 2011, 74, 403–409. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Ragon, M.; Wirth, T.; Hollandt, F.; Lavenir, R.; Lecuit, M.; Le Monnier, A.; Brisse, S. A New Perspective on Listeria monocytogenes Evolution. PLOS Pathog. 2008, 4, e1000146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larivière-Gauthier, G.; Thibodeau, A.; Letellier, A.; Yergeau, É.; Fravalo, P. Reduction of Salmonella Shedding by Sows during Gestation in Relation to Its Fecal Microbiome. Front. Microbiol. 2017, 8, 2219. [Google Scholar] [CrossRef] [Green Version]
- Thibodeau, A.; Fravalo, P.; Yergeau, É.; Arsenault, J.; Lahaye, L.; Letellier, A. Chicken Caecal Microbiome Modifications Induced by Campylobacter jejuni Colonization and by a Non-Antibiotic Feed Additive. PLoS ONE 2015, 10, e0131978. [Google Scholar] [CrossRef] [Green Version]
- Apprill, A.; McNally, S.; Parsons, R.; Weber, L. Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat. Microb. Ecol. 2015, 75, 129–137. [Google Scholar] [CrossRef] [Green Version]
- Parada, A.E.; Needham, D.M.; Fuhrman, J.A. Every base matters: Assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 2016, 18, 1403–1414. [Google Scholar] [CrossRef]





| Table. | Presence | Absence |
|---|---|---|
| Observed | 230.88 | 223.07 |
| Shannon | 3.13 | 3.24 |
| Inv. Simpson | 11.63 | 12.74 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shedleur-Bourguignon, F.; Thériault, W.P.; Longpré, J.; Thibodeau, A.; Fravalo, P. Use of an Ecosystem-Based Approach to Shed Light on the Heterogeneity of the Contamination Pattern of Listeria monocytogenes on Conveyor Belt Surfaces in a Swine Slaughterhouse in the Province of Quebec, Canada. Pathogens 2021, 10, 1368. https://doi.org/10.3390/pathogens10111368
Shedleur-Bourguignon F, Thériault WP, Longpré J, Thibodeau A, Fravalo P. Use of an Ecosystem-Based Approach to Shed Light on the Heterogeneity of the Contamination Pattern of Listeria monocytogenes on Conveyor Belt Surfaces in a Swine Slaughterhouse in the Province of Quebec, Canada. Pathogens. 2021; 10(11):1368. https://doi.org/10.3390/pathogens10111368
Chicago/Turabian StyleShedleur-Bourguignon, Fanie, William P. Thériault, Jessie Longpré, Alexandre Thibodeau, and Philippe Fravalo. 2021. "Use of an Ecosystem-Based Approach to Shed Light on the Heterogeneity of the Contamination Pattern of Listeria monocytogenes on Conveyor Belt Surfaces in a Swine Slaughterhouse in the Province of Quebec, Canada" Pathogens 10, no. 11: 1368. https://doi.org/10.3390/pathogens10111368
APA StyleShedleur-Bourguignon, F., Thériault, W. P., Longpré, J., Thibodeau, A., & Fravalo, P. (2021). Use of an Ecosystem-Based Approach to Shed Light on the Heterogeneity of the Contamination Pattern of Listeria monocytogenes on Conveyor Belt Surfaces in a Swine Slaughterhouse in the Province of Quebec, Canada. Pathogens, 10(11), 1368. https://doi.org/10.3390/pathogens10111368






