The Flo Adhesin Family
Abstract
1. Introduction
2. Redefinition of the Flo Adhesin Family Based on the Protein Architecture
3. Structure and Function of Flocculation Adhesins
3.1. PA14/GLEYA Flo Type Adhesin Structure
3.2. Flo11 Type Adhesin Structure
4. Yeasts Expressing Flo Proteins Involved in Human Infections
4.1. Pathogenic Candida Species
4.1.1. Candida glabrata
4.1.2. Candida lusitaniae (Clavispora lusitaniae)
4.1.3. Candida parapsilosis and Candida tropicalis
4.1.4. Candida auris
4.1.5. Candida Species That Rarely Cause Infections
4.2. Non-Pathogenic S. cerevisiae Encountered in Rare Infections
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bongomin, F.; Gago, S.; Oladele, R.O.; Denning, D.W. Global and multi-national prevalence of fungal diseases—Estimate precision. J. Fungi 2017, 3, 57. [Google Scholar] [CrossRef]
- Denning, D.W. Calling upon all public health mycologists: To accompany the country burden papers from 14 countries. Eur. J. Clin. Microbiol. Infect. Dis. 2017, 36, 923–924. [Google Scholar] [CrossRef] [PubMed]
- Geddes-McAlister, J.; Shapiro, R.S. New pathogens, new tricks: Emerging, drug-resistant fungal pathogens and future prospects for antifungal therapeutics. Ann. N. Y. Acad. Sci. 2019, 1435, 57–78. [Google Scholar] [CrossRef] [PubMed]
- Richardson, M.D. Changing patterns and trends in systemic fungal infections. J. Antimicrob. Chemother. 2005, 56, i5–i11. [Google Scholar] [CrossRef]
- Huttunen, R.; Syrjänen, J. Obesity and the risk and outcome of infection. Int. J. Obes. 2013, 37, 333–340. [Google Scholar] [CrossRef]
- Rytter, M.J.H.; Kolte, L.; Briend, A.; Friis, H.; Christensen, V.B. The immune system in children with malnutrition—A systematic review. PLoS ONE 2014, 9, e105017. [Google Scholar]
- Ibrahim, M.K.; Zambruni, M.; Melby, C.L.; Melby, P.C. Impact of childhood malnutrition on host defense and infection. Clin. Microbiol. Rev. 2017, 30, 919–971. [Google Scholar] [CrossRef] [PubMed]
- Koehler, P.; Cornely, O.A.; Böttiger, B.W.; Dusse, F.; Eichenauer, D.A.; Fuchs, F.; Hallek, M.; Jung, N.; Klein, F.; Persigehl, T.; et al. COVID-19 associated pulmonary aspergillosis. Mycoses 2020, 63, 528–534. [Google Scholar] [CrossRef] [PubMed]
- Alanio, A.; Dellière, S.; Fodil, S.; Bretagne, S.; Mégarbane, B. Prevalence of putative invasive pulmonary aspergillosis in critically ill patients with COVID-19. Lancet Respir. Med. 2020, 8, e48–e49. [Google Scholar] [CrossRef]
- van Arkel, A.L.E.; Rijpstra, T.A.; Belderbos, H.N.A.; van Wijngaarden, P.; Verweij, P.E.; Bentvelsen, R.G. COVID-19-associated pulmonary aspergillosis. Am. J. Respir. Crit. Care Med. 2020, 202, 132–135. [Google Scholar] [CrossRef] [PubMed]
- Arastehfar, A.; Carvalho, A.; van de Veerdonk, F.L.; Jenks, J.D.; Koehler, P.; Krause, R.; Cornely, O.A.; Perlin, D.S.; Lass-Flörl, C.; Hoenigl, M. COVID-19 associated pulmonary aspergillosis (CAPA)—From immunology to treatment. J. Fungi 2020, 6, 91. [Google Scholar] [CrossRef]
- Koehler, P.; Bassetti, M.; Chakrabarti, A.; Chen, S.C.A.; Colombo, A.L.; Hoenigl, M.; Klimko, N.; Lass-Flörl, C.; Oladele, R.O.; Vinh, D.C.; et al. Defining and managing COVID-19-associated pulmonary aspergillosis: The 2020 ECMM/ISHAM consensus criteria for research and clinical guidance. Lancet Infect. Dis. 2021, 21, e149–e162. [Google Scholar] [CrossRef]
- Vos, T.; Flaxman, A.D.; Naghavi, M.; Lozano, R.; Michaud, C.; Ezzati, M.; Shibuya, K.; Salomon, J.A.; Abdalla, S.; Aboyans, V.; et al. Years lived with disability (YLDs) for 1160 sequelae of 289 diseases and injuries 1990–2010: A systematic analysis for the Global Burden of Disease Study 2010. Lancet 2012, 380, 2163–2196. [Google Scholar] [CrossRef]
- Brown, G.D.; Denning, D.W.; Levitz, S.M. Tackling human fungal infections. Science 2012, 336, 647. [Google Scholar] [CrossRef] [PubMed]
- Brown, G.D.; Denning, D.W.; Gow, N.A.R.; Levitz, S.M.; Netea, M.G.; White, T.C. Hidden killers: Human fungal infections. Sci. Transl. Med. 2012, 4, 165rv13. [Google Scholar] [CrossRef]
- Havlickova, B.; Czaika, V.A.; Friedrich, M. Epidemiological trends in skin mycoses worldwide. Mycoses 2008, 51, 2–15. [Google Scholar] [CrossRef] [PubMed]
- Sobel, J.D. Vulvovaginal candidosis. Lancet 2007, 369, 1961–1971. [Google Scholar] [CrossRef]
- Castón-Osorio, J.J.; Rivero, A.; Torre-Cisneros, J. Epidemiology of invasive fungal infection. Int. J. Antimicrob. Agents 2008, 32, S103–S109. [Google Scholar] [CrossRef]
- Miceli, M.H.; Díaz, J.A.; Lee, S.A. Emerging opportunistic yeast infections. Lancet Infect. Dis. 2011, 11, 142–151. [Google Scholar] [CrossRef]
- Galimberti, R.; Torre, A.C.; Baztán, M.C.; Rodriguez-Chiappetta, F. Emerging systemic fungal infections. Clin. Dermatol. 2012, 30, 633–650. [Google Scholar] [CrossRef] [PubMed]
- Willaert, R. Micro- and Nanoscale Approaches in Antifungal Drug Discovery. Fermentation 2018, 4, 43. [Google Scholar] [CrossRef]
- Lemke, A.; Kiderlen, A.F.; Kayser, O. Amphotericin, B. Appl. Microbiol. Biotechnol. 2005, 68, 151–162. [Google Scholar] [CrossRef]
- Bellmann, R. Pharmacodynamics and Pharmacokinetics of Antifungals for Treatment of Invasive Aspergillosis. Curr. Pharm. Des. 2013, 19, 3629–3647. [Google Scholar] [CrossRef] [PubMed]
- Pfaller, M.A. Antifungal Drug Resistance: Mechanisms, Epidemiology, and Consequences for Treatment. Am. J. Med. 2012, 125, S3–S13. [Google Scholar] [CrossRef] [PubMed]
- Pianalto, K.M.; Alspaugh, J.A. New horizons in antifungal therapy. J. Fungi 2016, 2, 26. [Google Scholar] [CrossRef] [PubMed]
- Nivoix, Y.; Ubeaud-Sequier, G.; Engel, P.; Leveque, D.; Herbrecht, R. Drug-Drug Interactions of Triazole Antifungal Agents in Multimorbid Patients and Implications for Patient Care. Curr. Drug Metab. 2009, 10, 395–409. [Google Scholar] [CrossRef]
- Lewis, R.E. Current concepts in antifungal pharmacology. In Proceedings of the Mayo Clinic Proceedings; Elsevier Ltd.: Amsterdam, The Netherlands, 2011; Volume 86, pp. 805–817. [Google Scholar]
- McCarthy, M.W.; Walsh, T.J. Drugs currently under investigation for the treatment of invasive candidiasis. Expert Opin. Investig. Drugs 2017, 26, 825–831. [Google Scholar] [CrossRef] [PubMed]
- Perfect, J.R. The antifungal pipeline: A reality check. Nat. Rev. Drug Discov. 2017, 16, 603–616. [Google Scholar] [CrossRef] [PubMed]
- Fuentefria, A.M.; Pippi, B.; Dalla Lana, D.F.; Donato, K.K.; de Andrade, S.F. Antifungals discovery: An insight into new strategies to combat antifungal resistance. Lett. Appl. Microbiol. 2018, 66, 2–13. [Google Scholar] [CrossRef]
- Silva, L.N.; de Mello, T.P.; de Souza Ramos, L.; Branquinha, M.H.; dos Santos, A.L.S. New and Promising Chemotherapeutics for Emerging Infections Involving Drug-resistant Non-albicans Candida Species. Curr. Top. Med. Chem. 2019, 19, 2527–2553. [Google Scholar] [CrossRef] [PubMed]
- Sousa, F.; Ferreira, D.; Reis, S.; Costa, P. Current insights on antifungal therapy: Novel nanotechnology approaches for drug delivery systems and new drugs from natural sources. Pharmaceuticals 2020, 13, 248. [Google Scholar] [CrossRef] [PubMed]
- Mota Fernandes, C.; Dasilva, D.; Haranahalli, K.; McCarthy, J.B.; Mallamo, J.; Ojima, I.; Del Poeta, M. The Future of Antifungal Drug Therapy: Novel Compounds and Targets. Antimicrob. Agents Chemother. 2021, 65, 65. [Google Scholar] [CrossRef]
- Scorzoni, L.; Fuchs, B.B.; Junqueira, J.C.; Mylonakis, E. Current and promising pharmacotherapeutic options for candidiasis. Expert Opin. Pharmacother. 2021, 22, 867–887. [Google Scholar] [CrossRef] [PubMed]
- Dranginis, A.M.; Rauceo, J.M.; Coronado, J.E.; Lipke, P.N. A Biochemical Guide to Yeast Adhesins: Glycoproteins for Social and Antisocial Occasions. Microbiol. Mol. Biol. Rev. 2007, 71, 282–294. [Google Scholar] [CrossRef]
- de Groot, P.W.J.; Bader, O.; de Boer, A.D.; Weig, M.; Chauhan, N. Adhesins in Human Fungal Pathogens: Glue with Plenty of Stick. Eukaryot. Cell 2013, 12, 470–481. [Google Scholar] [CrossRef]
- Teunissen, A.W.R.H.; Steensma, H.Y. The dominant flocculation genes of Saccharomyces cerevisiae constitute a new subtelomeric gene family. Yeast 1995, 11, 1001–1013. [Google Scholar] [CrossRef]
- Guo, B.; Styles, C.A.; Feng, Q.; Fink, G.R. A Saccharomyces gene family involved in invasive growth, cell-cell adhesion, and mating. Proc. Natl. Acad. Sci. USA 2000, 97, 12158–12163. [Google Scholar] [CrossRef]
- Hoyer, L.L. The ALS gene family of Candida albicans. Trends Microbiol. 2001, 9, 176–180. [Google Scholar] [CrossRef]
- Lo, W.-S.; Dranginis, A.M. The Cell Surface Flocculin Flo11 Is Required for Pseudohyphae Formation and Invasion by. Mol. Biol. Cell 1998, 9, 161–171. [Google Scholar] [CrossRef] [PubMed]
- Vyas, V.K.; Kuchin, S.; Berkey, C.D.; Carlson, M. Snf1 Kinases with Different β-Subunit Isoforms Play Distinct Roles in Regulating Haploid Invasive Growth. Mol. Cell. Biol. 2003, 23, 1341–1348. [Google Scholar] [CrossRef]
- Gaur, N.K.; Klotz, S.A. Expression, cloning, and characterization of a Candida albicans gene, ALA1, that confers adherence properties upon Saccharomyces cerevisiae for extracellular matrix proteins. Infect. Immun. 1997, 65, 5289–5294. [Google Scholar] [CrossRef]
- Reynolds, T.B.; Fink, G.R.; TB, R.; GR, F. Bakers’ yeast, a model for fungal biofilm formation. Science 2001, 291, 878–881. [Google Scholar] [CrossRef]
- Van Mulders, S.E.; Christianen, E.; Saerens, S.M.G.G.; Daenen, L.; Verbelen, P.J.; Willaert, R.; Verstrepen, K.J.; Delvaux, F.R. Phenotypic diversity of Flo protein family-mediated adhesion in Saccharomyces cerevisiae. FEMS Yeast Res. 2009, 9, 178–190. [Google Scholar] [CrossRef] [PubMed]
- Cormack, B.P. An Adhesin of the Yeast Pathogen Candida glabrata Mediating Adherence to Human Epithelial Cells. Science 1999, 285, 578–582. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Palecek, S.P. EAP1, a Candida albicans Gene Involved in Binding Human Epithelial Cells. Eukaryot. Cell 2003, 2, 1266–1273. [Google Scholar] [CrossRef]
- Zupancic, M.L.; Frieman, M.; Smith, D.; Alvarez, R.A.; Cummings, R.D.; Cormack, B.P. Glycan microarray analysis of Candida glabrata adhesin ligand specificity. Mol. Microbiol. 2008, 68, 547–559. [Google Scholar] [CrossRef]
- Verstrepen, K.J.; Fink, G.R. Genetic and epigenetic mechanisms underlying cell-surface variability in protozoa and fungi. Annu. Rev. Genet. 2009, 43, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Goossens, K.; Willaert, R. Flocculation protein structure and cell-cell adhesion mechanism in Saccharomyces cerevisiae. Biotechnol. Lett. 2010, 32, 1571–1585. [Google Scholar] [CrossRef] [PubMed]
- Reitsma, S.; Slaaf, D.W.; Vink, H.; Van Zandvoort, M.A.M.J.M.J.; Oude Egbrink, M.G.A.A.; Zandvoort, M.A.M.J.; Oude Egbrink, M.G.A.A. The endothelial glycocalyx: Composition, functions, and visualization. Pflügers Arch.—Eur. J. Physiol. 2007, 454, 345–359. [Google Scholar] [CrossRef]
- Critchley, I.A.; Douglas, L.J. Role of glycosides as epithelial cell receptors for Candida albicans. J. Gen. Microbiol. 1987, 133, 637–643. [Google Scholar] [CrossRef] [PubMed]
- Vardar-Ünlü, G.; McSharry, C.; Douglas, L.J. Fucose-specific adhesins on germ tubes of Candida albicans. FEMS Immunol. Med. Microbiol. 1998, 20, 55–67. [Google Scholar] [CrossRef]
- Ielasi, F.S.; Alioscha-Perez, M.; Donohue, D.; Claes, S.; Sahli, H.; Schols, D.; Willaert, R.G. Lectin-glycan interaction network-based identification of host receptors of microbial pathogenic adhesins. MBio 2016, 7. [Google Scholar] [CrossRef]
- Perlroth, J.; Choi, B.; Spellberg, B. Nosocomial fungal infections: Epidemiology, diagnosis, and treatment. Med. Mycol. 2007, 45, 321–346. [Google Scholar] [CrossRef] [PubMed]
- Ramage, G.; Martínez, J.P.; López-Ribot, J.L. Candida biofilms on implanted biomaterials: A clinically significant problem. FEMS Yeast Res. 2006, 6, 979–986. [Google Scholar] [CrossRef] [PubMed]
- Busscher, H.J.; Rinastiti, M.; Siswomihardjo, W.; Van Der Mei, H.C. Biofilm formation on dental restorative and implant materials. J. Dent. Res. 2010, 89, 657–665. [Google Scholar] [CrossRef] [PubMed]
- Ramage, G.; Culshaw, S.; Jones, B.; Williams, C. Are we any closer to beating the biofilm: Novel methods of biofilm control. Curr. Opin. Infect. Dis. 2010, 23, 560–566. [Google Scholar] [CrossRef] [PubMed]
- Pfaller, M.A. Epidemiology of candidiasis. J. Hosp. Infect. 1995, 30, 329–338. [Google Scholar] [CrossRef]
- Pfaller, M.A.; Diekema, D.J. Epidemiology of invasive candidiasis: A persistent public health problem. Clin. Microbiol. Rev. 2007, 20, 133–163. [Google Scholar] [CrossRef]
- Pfaller, M.A.; Diekema, D.J.; Steele-Moore, L.; Denys, G.; Staley, C.; Dipersio, J.R.; Saubolle, M.; Wilson, M.L.; Overturf, G.D.; Peterson, L.R.; et al. Twelve years of fluconazole in clinical practice: Global-trends in species distribution and fluconazole susceptibility of bloodstream isolates of Candida. Clin. Microbiol. Infect. 2004, 10, 11–23. [Google Scholar] [CrossRef]
- García-Martos, P.; Domínguez, I.; Marín, P.; García-Agudo, R.; Aoufi, S.; Maria, J. Antifungal susceptibility of emerging yeast pathogens. Enferm. Infecc. Microbiol. Clin. 2001, 19, 249–256. [Google Scholar] [CrossRef]
- Coles, M.; Cox, K.; Chao, A. Candida haemulonii: An emerging opportunistic pathogen in the United States? IDCases 2020, 21, e00900. [Google Scholar] [CrossRef]
- Willaert, R.; Section, V. Beverages: The Beer Brewing Process: Wort Production and Beer Fermentation. In Handbook of Food Products Manufacturing; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2006; Volume 1, pp. 443–506. ISBN 9780470049648. [Google Scholar]
- Brückner, S.; Mösch, H.-U.U. Choosing the right lifestyle: Adhesion and development in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 2011, 36, 25–58. [Google Scholar] [CrossRef] [PubMed]
- Gimeno, C.J.; Ljungdahl, P.O.; Styles, C.A.; Fink, G.R. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: Regulation by starvation and RAS. Cell 1992, 68, 1077–1090. [Google Scholar] [CrossRef]
- Cullen, P.J.; Sprague, G.F. Glucose depletion causes haploid invasive growth in yeast. Proc. Natl. Acad. Sci. USA 2000, 97, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Palková, Z. Multicellular microorganisms: Laboratory versus nature. EMBO Rep. 2004, 5, 470–476. [Google Scholar] [CrossRef] [PubMed]
- Stratford, M.; Assinder, S. Yeast flocculation: Flo1 and NewFlo phenotypes and receptor structure. Yeast 1991, 7, 559–574. [Google Scholar] [CrossRef] [PubMed]
- Kraushaar, T.; Brückner, S.; Veelders, M.; Rhinow, D.; Schreiner, F.; Birke, R.; Pagenstecher, A.; Mösch, H.U.; Essen, L.O. Interactions by the fungal Flo11 adhesin depend on a fibronectin type III-like adhesin domain girdled by aromatic bands. Structure 2015, 23, 1005–1017. [Google Scholar] [CrossRef]
- Lambrechts, M.G.; Bauer, F.F.; Marmur, J.; Pretorius, I.S. MUC1, a mucin-like protein that is regulated by MSS10, is critical for pseudohyphal differentiation in yeast. Proc. Natl. Acad. Sci. USA 1996, 93, 8419–8424. [Google Scholar] [CrossRef]
- Reynolds, T.B. Going with the flo: The role of flo11-dependent and independent interactions in yeast mat formation. J. Fungi 2018, 4, 132. [Google Scholar] [CrossRef] [PubMed]
- Purevdorj-Gage, B.; Orr, M.E.; Stoodley, P.; Sheehan, K.B.; Hyman, L.E. The role of FLO11 in Saccharomyces cerevisiae biofilm development in a laboratory based flow-cell system. FEMS Yeast Res. 2007, 7, 372–379. [Google Scholar] [CrossRef] [PubMed]
- Bojsen, R.K.; Andersen, K.S.; Regenberg, B. Saccharomyces cerevisiae—A model to uncover molecular mechanisms for yeast biofilm biology. FEMS Immunol. Med. Microbiol. 2012, 65, 169–182. [Google Scholar] [CrossRef]
- Douglas, L.M.; Li, L.; Yang, Y.; Dranginis, A.M. Expression and Characterization of the Flocculin Flo11/Muc1, a Saccharomyces cerevisiae Mannoprotein with Homotypic Properties of Adhesion. Eukaryot. Cell 2007, 6, 2214–2221. [Google Scholar] [CrossRef]
- Goossens, K.V.Y.; Willaert, R.G. The N-terminal domain of the Flo11 protein from Saccharomyces cerevisiae is an adhesin without mannose-binding activity. FEMS Yeast Res. 2012, 12, 78–87. [Google Scholar] [CrossRef] [PubMed]
- Erdman, S.; Lin, L.; Malczynski, M.; Snyder, M. Pheromone-regulated genes required for yeast mating differentiation. J. Cell Biol. 1998, 140, 461–483. [Google Scholar] [CrossRef] [PubMed]
- Roy, A.; Lu, C.F.; Marykwas, D.L.; Lipke, P.N.; Kurjan, J. The AGA1 product is involved in cell surface attachment of the Saccharomyces cerevisiae cell adhesion glycoprotein a-agglutinin. Mol. Cell. Biol. 1991, 11, 4196–4206. [Google Scholar] [CrossRef] [PubMed]
- Cappellaro, C.; Hauser, K.; Mrśa, V.; Watzele, M.; Watzele, G.; Gruber, C.; Tanner, W. Saccharomyces cerevisiae a- and alpha-agglutinin: Characterization of their molecular interaction. EMBO J. 1991, 10, 4081–4088. [Google Scholar] [CrossRef] [PubMed]
- Lipke, P.N.; Wojciechowicz, D.; Kurjan, J. AG alpha 1 is the structural gene for the Saccharomyces cerevisiae alpha-agglutinin, a cell surface glycoprotein involved in cell-cell interactions during mating. Mol. Cell. Biol. 1989, 9, 3155–3165. [Google Scholar] [CrossRef]
- Caro, L.H.P.; Tettelin, H.; Vossen, J.H.; Ram, A.F.J.; Van Den Ende, H.; Klis, F.M. In silicio identification of glycosyl-phosphatidylinositol-anchored plansma-membrane and cell wall proteins of Saccharomyces cerevisiae. Yeast 1997, 13, 1477–1489. [Google Scholar] [CrossRef]
- Jentoft, N. Why are proteins O-glycosylated? Trends Biochem. Sci. 1990, 15, 291–294. [Google Scholar] [CrossRef]
- Verstrepen, K.J.; Jansen, A.; Lewitter, F.; Fink, G.R. Intragenic tandem repeats generate functional variability. Nat. Genet. 2005, 37, 986–990. [Google Scholar] [CrossRef]
- Liu, N.; Wang, D.; Wang, Z.Y.; He, X.P.; Zhang, B. Genetic basis of flocculation phenotype conversion in Saccharomyces cerevisiae. FEMS Yeast Res. 2007, 7, 1362–1370. [Google Scholar] [CrossRef][Green Version]
- Westman, J.O.; Mapelli, V.; Taherzadeh, M.J.; Franzén, C.J. Flocculation causes inhibitor tolerance in Saccharomyces cerevisiae for second-generation bioethanol production. Appl. Environ. Microbiol. 2014, 80, 6908–6918. [Google Scholar] [CrossRef] [PubMed]
- Verstrepen, K.J.; Klis, F.M. Flocculation, adhesion and biofilm formation in yeasts. Mol. Microbiol. 2006, 60, 5–15. [Google Scholar] [CrossRef]
- Willaert, R.G. Adhesins of yeasts: Protein structure and interactions. J. Fungi 2018, 4, 119. [Google Scholar] [CrossRef]
- Rigden, D.J.; Mello, L.V.; Galperin, M.Y. The PA14 domain, a conserved all-β domain in bacterial toxins, enzymes, adhesins and signaling molecules. Trends Biochem. Sci. 2004, 29, 335–339. [Google Scholar] [CrossRef] [PubMed]
- Petosa, C.; Collier, R.J.; Klimpel, K.R.; Leppla, S.H.; Liddington, R.C. Crystal structure of the anthrax toxin protective antigen. Nature 1997, 385, 833–838. [Google Scholar] [CrossRef]
- Yoshida, E.; Hidaka, M.; Fushinobu, S.; Koyanagi, T.; Minami, H.; Tamaki, H.; Kitaoka, M.; Katayama, T.; Kumagai, H. Role of a PA14 domain in determining substrate specificity of a glycoside hydrolase family 3 β-glucosidase from Kluyveromyces marxianus. Biochem. J. 2010, 431, 39–49. [Google Scholar] [CrossRef]
- Silipo, A.; Larsbrink, J.; Marchetti, R.; Lanzetta, R.; Brumer, H.; Molinaro, A. NMR spectroscopic analysis reveals extensive binding interactions of complex xyloglucan oligosaccharides with the Cellvibrio japonicus glycoside hydrolase family 31 α-xylosidase. Chem.—Eur. J. 2012, 18, 13395–13404. [Google Scholar] [CrossRef]
- Linder, T.; Gustafsson, C.M. Molecular phylogenetics of ascomycotal adhesins—A novel family of putative cell-surface adhesive proteins in fission yeasts. Fungal Genet. Biol. 2008, 45, 485–497. [Google Scholar] [CrossRef] [PubMed]
- Ielasi, F.S.; Decanniere, K.; Willaert, R.G. The epithelial adhesin 1 (Epa1p) from the human-pathogenic yeast Candida glabrata: Structural and functional study of the carbohydrate-binding domain. Acta Crystallogr. Sect. D Biol. Crystallogr. 2012, 68, 210–217. [Google Scholar] [CrossRef] [PubMed]
- Goossens, K.V.Y.; Ielasi, F.S.; Nookaew, I.; Stals, I.; Alonso-sarduy, L.; Daenen, L.; Nielsen, J.; Devreese, B.; Willaert, G. Molecular Mechanism of Flocculation Self-Recognition in Yeast and. MBio 2015, 6, 1–16. [Google Scholar] [CrossRef]
- Frieman, M.B.; McCaffery, J.M.; Cormack, B.P. Modular domain structure in the Candida glabrata adhesin Epa1p, a β1,6 glucan-cross-linked cell wall protein. Mol. Microbiol. 2002, 46, 479–492. [Google Scholar] [CrossRef] [PubMed]
- Maestre-Reyna, M.; Diderrich, R.; Veelders, M.S.; Eulenburg, G.; Kalugin, V.; Brückner, S.; Keller, P.; Rupp, S.; Mösch, H.U.; Essen, L.O. Structural basis for promiscuity and specificity during Candida glabrata invasion of host epithelia. Proc. Natl. Acad. Sci. USA 2012, 109, 16864–16869. [Google Scholar] [CrossRef] [PubMed]
- Sim, L.; Groes, M.; Olesen, K.; Henriksen, A. Structural and biochemical characterization of the N-terminal domain of flocculin Lg-Flo1p from Saccharomyces pastorianusreveals a unique specificity for phosphorylated mannose. FEBS J. 2013, 280, 1073–1083. [Google Scholar] [CrossRef] [PubMed]
- Kock, M.; Brückner, S.; Wozniak, N.; Maestre-Reyna, M.; Veelders, M.; Schlereth, J.; Mösch, H.-U.; Essen, L.-O. Structural and Functional Characterization of PA14/Flo5-Like Adhesins From Komagataella pastoris. Front. Microbiol. 2018, 9, 2581. [Google Scholar] [CrossRef] [PubMed]
- Diderrich, R.; Kock, M.; Maestre-Reyna, M.; Keller, P.; Steuber, H.; Rupp, S.; Essen, L.O.; Mösch, H.U. Structural Hot Spots Determine Functional Diversity of the Candida glabrata Epithelial Adhesin Family. J. Biol. Chem. 2015, 290, 19597–19613. [Google Scholar] [CrossRef]
- Brückner, S.; Schubert, R.; Kraushaar, T.; Hartmann, R.; Hoffmann, D.; Jelli, E.; Drescher, K.; Müller, D.J.; Essen, L.O.; Mösch, H.U. Kin discrimination in social yeast is mediated by cell surface receptors of the flo11 adhesin family. Elife 2020, 9. [Google Scholar] [CrossRef] [PubMed]
- Veelders, M.; Brückner, S.; Ott, D.; Unverzagt, C.; Mösch, H.U.; Essen, L.O. Structural basis of flocculin-mediated social behavior in yeast. Proc. Natl. Acad. Sci. USA 2010, 107, 22511–22516. [Google Scholar] [CrossRef] [PubMed]
- Goossens, K.V.Y.Y.; Stassen, C.; Stals, I.; Donohue, D.S.; Devreese, B.; de Greve, H.; Willaert, R.G. The N-terminal domain of the flo1 flocculation protein from Saccharomyces cerevisiae binds specifically to mannose carbohydrates. Eukaryot. Cell 2011, 10, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Bony, M.; Thines-Sempoux, D.; Barre, P.; Blondin, B. Localization and Cell Surface Anchoring of the Saccharomyces cerevisiae Flocculation Protein Flo1p. J. Bacteriol. 2011, 179, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Bucior, I.; Burger, M.M. Carbohydrate–carbohydrate interactions in cell recognition. Curr. Opin. Struct. Biol. 2004, 14, 631–637. [Google Scholar] [CrossRef] [PubMed]
- Fernàndez-Busquets, X.; Körnig, A.; Bucior, I.; Burger, M.M.; Anselmetti, D. Self-Recognition and Ca2+-Dependent Carbohydrate–Carbohydrate Cell Adhesion Provide Clues to the Cambrian Explosion. Mol. Biol. Evol. 2009, 26, 2551–2561. [Google Scholar] [CrossRef] [PubMed]
- Glaser, F.; Pupko, T.; Paz, I.; Bell, R.E.; Bechor-Shental, D.; Martz, E.; Ben-Tal, N. ConSurf: Identification of functional regions in proteins by surface-mapping of phylogenetic information. Bioinformatics 2003, 19, 163–164. [Google Scholar] [CrossRef] [PubMed]
- Landau, M.; Mayrose, I.; Rosenberg, Y.; Glaser, F.; Martz, E.; Pupko, T.; Ben-Tal, N. ConSurf 2005: The projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005, 33. [Google Scholar] [CrossRef] [PubMed]
- Dehullu, J.; Vorholt, J.A.; Lipke, P.N.; Dufrêne, Y.F. Fluidic Force Microscopy Captures Amyloid Bonds between Microbial Cells. Trends Microbiol. 2019, 27, 728–730. [Google Scholar] [CrossRef] [PubMed]
- Dehullu, J.; Valotteau, C.; Herman-Bausier, P.; Garcia-Sherman, M.; Mittelviefhaus, M.; Vorholt, J.A.; Lipke, P.N.; Dufrêne, Y.F.; Jérô Me Dehullu, J.; Valotteau, C.; et al. Fluidic Force Microscopy Demonstrates That Homophilic Adhesion by Candida albicans Als Proteins Is Mediated by Amyloid Bonds between Cells. Nano Lett. 2019, 19, 3846–3853. [Google Scholar] [CrossRef]
- Lipke, P.N.; Mathelié-Guinlet, M.; Viljoen, A.; Dufrêne, Y.F. A New Function for Amyloid-Like Interactions: Cross-Beta Aggregates of Adhesins form Cell-to-Cell Bonds. Pathogens 2021, 10, 1013. [Google Scholar] [CrossRef] [PubMed]
- Lipke, P.N.; Klotz, S.A.; Dufrene, Y.F.; Jackson, D.N.; Garcia-Sherman, M.C. Amyloid-Like β-Aggregates as Force-Sensitive Switches in Fungal Biofilms and Infections. Microbiol. Mol. Biol. Rev. 2018, 82. [Google Scholar] [CrossRef] [PubMed]
- El-Kirat-Chatel, S.; Beaussart, A.; Vincent, S.P.; Flos, M.A.; Hols, P.; Lipke, P.N.; Dufrêne, Y.F. Forces in yeast flocculation. Nanoscale 2015, 7, 1760–1767. [Google Scholar] [CrossRef]
- Lo, W.S.; Dranginis, A.M. FLO11, a yeast gene related to the STA genes, encodes a novel cell surface flocculin. J. Bacteriol. 1996, 178, 7144–7151. [Google Scholar] [CrossRef]
- Rupp, S. MAP kinase and cAMP filamentation signaling pathways converge on the unusually large promoter of the yeast FLO11 gene. EMBO J. 1999, 18, 1257–1269. [Google Scholar] [CrossRef] [PubMed]
- Roberts, R.L.; Fink, G.R. Elements of a single map kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: Mating and invasive growth. Genes Dev. 1994, 8, 2974–2985. [Google Scholar] [CrossRef] [PubMed]
- Kron, S.J.; Styles, C.A.; Fink, G.R. Symmetric cell division in pseudohyphae of the yeast Saccharomyces cerevisiae. Mol. Biol. Cell 1994, 5, 1003–1022. [Google Scholar] [CrossRef]
- Mösch, H.U. Pseudohyphal development of Saccharomyces cerevisiae. Contrib. Microbiol. 2000, 5, 185–200. [Google Scholar] [PubMed]
- Strittmatter, A.W.; Fischer, C.; Kleinschmidt, M.; Braus, G.H. FLO11 mediated filamentous growth of the yeast Saccharomyces cerevisiae depends on the expression of the ribosomal RPS26 genes. Mol. Genet. Genomics 2006, 276, 113–125. [Google Scholar] [CrossRef] [PubMed]
- Pitoniak, A.; Birkaya, B.; Dionne, H.M.; Vadaie, N.; Cullen, P.J. The signaling mucins Msb2 and Hkr1 differentially regulate the filamentation mitogen-activated protein kinase pathway and contribute to a multimodal response. Mol. Biol. Cell 2009, 20, 3101–3114. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, T.B.; Jansen, A.; Peng, X.; Fink, G.R. Mat formation in Saccharomyces cerevisiae requires nutrient and pH gradients. Eukaryot. Cell 2008, 7, 122–130. [Google Scholar] [CrossRef] [PubMed]
- Sarode, N.; Davis, S.E.; Tams, R.N.; Reynolds, T.B. The Wsc1p cell wall signaling protein controls biofilm (Mat) formation independently of Flo11p in Saccharomyces cerevisiae. G3 Genes Genomes Genet. 2014, 4, 199–207. [Google Scholar] [CrossRef] [PubMed]
- Ishigami, M.; Nakagawa, Y.; Hayakawa, M.; Iimura, Y. FLO11 is the primary factor in flor formation caused by cell surface hydrophobicity in wild-type flor yeast. Biosci. Biotechnol. Biochem. 2006, 70, 660–666. [Google Scholar] [CrossRef]
- Zara, S.; Bakalinsky, A.T.; Zara, G.; Pirino, G.; Demontis, M.A.; Budroni, M. FLO11-Based Model for Air-Liquid Interfacial Biofilm Formation by Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2005, 71, 2934–2939. [Google Scholar] [CrossRef]
- Ishigami, M.; Nakagawa, Y.; Hayakawa, M.; Iimura, Y. FLO11 is essential for flor formation caused by the C-terminal deletion of NRG1 in Saccharomyces cerevisiae. FEMS Microbiol. Lett. 2004, 237, 425–430. [Google Scholar] [CrossRef] [PubMed]
- Alexandre, H. Flor yeasts of Saccharomyces cerevisiae-Their ecology, genetics and metabolism. Int. J. Food Microbiol. 2013, 167, 269–275. [Google Scholar] [CrossRef] [PubMed]
- Mortensen, H.D.; Dupont, K.; Jespersen, L.; Arneborg, N. The Flo11p-deficient Saccharomyces cerevisiae strain background S288c can adhere to plastic surfaces. Colloids Surf. B Biointerfaces 2007, 60, 131–134. [Google Scholar] [CrossRef]
- Mortensen, H.D.; Dupont, K.; Jespersen, L.; Willats, W.G.T.; Arneborg, N. Identification of amino acids involved in the Flo11p-mediated adhesion of Saccharomyces cerevisiae to a polystyrene surface using phage display with competitive elution. J. Appl. Microbiol. 2007, 103, 1041–1047. [Google Scholar] [CrossRef] [PubMed]
- Bayly, J.C.; Douglas, L.M.; Pretorius, I.S.; Bauer, F.F.; Dranginis, A.M. Characteristics of Flo11-dependent flocculation in Saccharomyces cerevisiae. FEMS Yeast Res. 2005, 5, 1151–1156. [Google Scholar] [CrossRef] [PubMed]
- Smit, G.; Straver, M.H.; Lugtenberg, B.J.J.; Kijne, J.W. Flocculence of Saccharomyces cerevisiae cells is induced by nutrient limitation, with cell surface hydrophobicity as a major determinant. Appl. Environ. Microbiol. 1992, 58, 3709–3714. [Google Scholar] [CrossRef] [PubMed]
- Straver, M.H.; Aar, P.C.V.D.; Smit, G.; Kijne, J.W. Determinants of flocculence of brewer’s yeast during fermentation in wort. Yeast 1993, 9, 527–532. [Google Scholar] [CrossRef] [PubMed]
- Wilcocks, K.L.; Smart, K.A. The importance of surface charge and hydrophobicity for the flocculation of chain-forming brewing yeast strains and resistance of these parameters to acid washing. FEMS Microbiol. Lett. 1995, 134, 293–297. [Google Scholar] [CrossRef] [PubMed]
- Barua, S.; Li, L.; Lipke, P.N.; Dranginis, A.M. Molecular Basis for Strain Variation in the Saccharomyces cerevisiae Adhesin Flo11p. mSphere 2016, 1. [Google Scholar] [CrossRef]
- Koide, A.; Bailey, C.W.; Huang, X.; Koide, S. The fibronectin type III domain as a scaffold for novel binding proteins. J. Mol. Biol. 1998, 284, 1141–1151. [Google Scholar] [CrossRef]
- Ramsook, C.B.; Tan, C.; Garcia, M.C.; Fung, R.; Soybelman, G.; Henry, R.; Litewka, A.; O’Meally, S.; Otoo, H.N.; Khalaf, R.A.; et al. Yeast Cell Adhesion Molecules Have Functional Amyloid-Forming Sequences. Eukaryot. Cell 2010, 9, 393–404. [Google Scholar] [CrossRef]
- Fidel, P.L.; Vazquez, J.A.; Sobel, J.D. Candida glabrata: Review of Epidemiology, Pathogenesis, and Clinical Disease with Comparison to C. albicans. Clin. Microbiol. Rev. 1999, 12, 80–96. [Google Scholar] [CrossRef]
- Kumar, K.; Askari, F.; Sahu, M.S.; Kaur, R. Candida glabrata: A Lot More Than Meets the Eye. Microorganisms 2019, 7, 39. [Google Scholar] [CrossRef] [PubMed]
- Silva, S.S.; Negri, M.; Henriques, M.; Oliveira, R.; Williams, D.W.; Azeredo, J. Adherence and biofilm formation of non-Candida albicans Candida species. Trends Microbiol. 2011, 19, 241–247. [Google Scholar] [CrossRef] [PubMed]
- Brunke, S.; Hube, B. Two unlike cousins: Candida albicans and C.glabrata infection strategies. Cell. Microbiol. 2013, 15, 701–708. [Google Scholar] [CrossRef] [PubMed]
- Kaur, R.; Domergue, R.; Zupancic, M.L.; Cormack, B.P. A yeast by any other name: Candida glabrata and its interaction with the host. Curr. Opin. Microbiol. 2005, 8, 378–384. [Google Scholar] [CrossRef]
- Roetzer, A.; Gabaldón, T.; Schüller, C. From Saccharomyces cerevisiae to Candida glabrata in a few easy steps: Important adaptations for an opportunistic pathogen. FEMS Microbiol. Lett. 2010, 314, 1–9. [Google Scholar] [CrossRef]
- Pfaller, M.A.; Moet, G.J.; Messer, S.A.; Jones, R.N.; Castanheira, M. Candida bloodstream infections: Comparison of species distributions and antifungal resistance patterns in community-onset and nosocomial isolates in the SENTRY Antimicrobial Surveillance Program, 2008–2009. Antimicrob. Agents Chemother. 2011, 55, 561–566. [Google Scholar] [CrossRef] [PubMed]
- Guinea, J. Global trends in the distribution of Candida species causing candidemia. Clin. Microbiol. Infect. 2014, 20, 5–10. [Google Scholar] [CrossRef] [PubMed]
- Hassan, Y.; Chew, S.Y.; Than, L.T.L. Candida glabrata: Pathogenicity and Resistance Mechanisms for Adaptation and Survival. J. Fungi 2021, 7, 667. [Google Scholar] [CrossRef] [PubMed]
- Zarb, P.; Coignard, B.; Griskeviciene, J.; Muller, A.; Vankerckhoven, V.; Weist, K.; Goossens, M.M.; Vaerenberg, S.; Hopkins, S.; Catry, B.; et al. The European Centre for Disease Prevention and Control (ECDC) pilot point prevalence survey of healthcare-associated infections and antimicrobial use. Eurosurveillance 2012, 17, 20316. [Google Scholar] [CrossRef] [PubMed]
- Mota, S.; Alves, R.; Carneiro, C.; Silva, S.; Brown, A.J.; Istel, F.; Kuchler, K.; Sampaio, P.; Casal, M.; Henriques, M.; et al. Candida glabrata susceptibility to antifungals and phagocytosis is modulated by acetate. Front. Microbiol. 2015, 6, 919. [Google Scholar] [CrossRef] [PubMed]
- Kale, P.; Johnson, L.B. Second-generation azole antifungal agents. Drugs Today 2005, 41, 91–105. [Google Scholar] [CrossRef] [PubMed]
- Richardson, M.; Lass-Flörl, C. Changing epidemiology of systemic fungal infections. Clin. Microbiol. Infect. 2008, 14, 5–24. [Google Scholar] [CrossRef]
- Cohen, Y.; Karoubi, P.; Adrie, C.; Gauzit, R.; Marsepoil, T.; Zarka, D.; Clec’H, C. Early prediction of Candida glabrata fungemia in nonneutropenic critically ill patients. Crit. Care Med. 2010, 38, 826–830. [Google Scholar] [CrossRef]
- Nobile, C.J.; Schneider, H.A.; Nett, J.E.; Sheppard, D.C.; Filler, S.G.; Andes, D.R.; Mitchell, A.P. Complementary Adhesin Function in C. albicans Biofilm Formation. Curr. Biol. 2008, 18, 1017–1024. [Google Scholar] [CrossRef] [PubMed]
- Silva, S.; Henriques, M.; Martins, A.; Oliveira, R.; Williams, D.; Azeredo, J. Biofilms of non-Candida albicans Candida species: Quantification, structure and matrix composition. Med. Mycol. 2009, 47, 681–689. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, C.F.; Rodrigues, M.E.; Silva, S.; Henriques, M. Candida glabrata Biofilms: How Far Have We Come? J. Fungi 2017, 3, 11. [Google Scholar] [CrossRef] [PubMed]
- Norimatsu, Y.; Morii, D.; Kogure, A.; Hamanaka, T.; Kuwano, Y.; Yokozawa, T.; Oda, T. A case of breakthrough Candida parapsilosis fungemia during micafungin therapy for a Candida glabrata bloodstream infection. Med. Mycol. Case Rep. 2017, 16, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Timmermans, B.; Las Peñas, A.D.; Castaño, I.; Van Dijck, P. Adhesins in candida glabrata. J. Fungi 2018, 4, 60. [Google Scholar] [CrossRef] [PubMed]
- Douglas, L.J. Candida biofilms and their role in infection. Trends Microbiol. 2003, 11, 30–36. [Google Scholar] [CrossRef]
- Kojic, E.M.; Darouiche, R.O. Candida Infections of Medical Devices. Clin. Microbiol. Rev. 2004, 17, 255–267. [Google Scholar] [CrossRef]
- Ramage, G.; Saville, S.P.; Thomas, D.P.; López-Ribot, J.L. Candida biofilms: An update. Eukaryot. Cell 2005, 4, 633–638. [Google Scholar] [CrossRef]
- Chaudhary, S.; Gupta, C.; Das, S.; Saha, R.; Rani, M.; Ramachandran, V.G. Biofilm formation by Candida species on intrauretheral catheter and its antifungal susceptibility profile. Indian J. Med. Microbiol. 2014, 32, 467–468. [Google Scholar] [CrossRef] [PubMed]
- Chandra, J.; Mukherjee, P.K. Candida Biofilms: Development, Architecture, and Resistance. Microbiol. Spectr. 2015, 3. [Google Scholar] [CrossRef]
- Weig, M.; Jänsch, L.; Groß, U.; De Koster, C.G.; Klis, F.M.; Groot, P.W.J. De Systematic identification in silico of covalently bound cell wall proteins and analysis of protein–polysaccharide linkages of the human pathogen Candida glabrata. Microbiology 2004, 150, 3129–3144. [Google Scholar] [CrossRef]
- de Groot, P.W.J.J.; Kraneveld, E.A.; Yin, Q.Y.; Dekker, H.L.; Gross, U.; Crielaard, W.; de Koster, C.G.; Bader, O.; Klis, F.M.; Weig, M.; et al. The Cell Wall of the Human Pathogen Candida glabrata: Differential Incorporation of Novel Adhesin-Like Wall Proteins. Eukaryot. Cell 2008, 7, 1951–1964. [Google Scholar] [CrossRef]
- Ielasi, F.S.; Verhaeghe, T.; Desmet, T.; Willaert, R.G. Engineering the carbohydrate-binding site of Epa1p from Candida glabrata: Generation of adhesin mutants with different carbohydrate specificity. Glycobiology 2014, 24, 1312–1322. [Google Scholar] [CrossRef] [PubMed]
- Gabaldón, T.; Martin, T.; Marcet-Houben, M.; Durrens, P.; Bolotin-Fukuhara, M.; Lespinet, O.; Arnaise, S.; Boisnard, S.; Aguileta, G.; Atanasova, R.; et al. Comparative genomics of emerging pathogens in the Candida glabrata clade. BMC Genom. 2013, 14, 623. [Google Scholar] [CrossRef] [PubMed]
- Hadfield, T.L.; Smith, M.B.; Winn, R.E.; Rinaldi, M.G.; Guerra, C. Mycoses caused by Candida lusitaniae. Rev. Infect. Dis. 1987, 9, 1006–1012. [Google Scholar] [CrossRef]
- Raja, A.; Park, J. Disseminated Candida lusitaniae: Nosocomial Acquisition Secondary to an Indwelling Urinary Catheter. Case Rep. Infect. Dis. 2021, 2021, 6632730. [Google Scholar] [CrossRef]
- Apsemidou, A.; Füller, M.A.; Idelevich, E.A.; Kurzai, O.; Tragiannidis, A.; Groll, A.H. Candida lusitaniae Breakthrough Fungemia in an Immuno-Compromised Adolescent: Case Report and Review of the Literature. J. Fungi 2020, 6, 380. [Google Scholar] [CrossRef]
- Hawkins, J.L.; Baddour, L.M. Candida lusitaniae Infections in the Era of Fluconazole Availability. Clin. Infect. Dis. 2003, 36, e14–e18. [Google Scholar] [CrossRef] [PubMed]
- Perpetuini, G.; Rossetti, A.P.; Battistelli, N.; Arfelli, G.; Tofalo, R. Adhesion Properties, Biofilm Forming Potential, and Susceptibility to Disinfectants of Contaminant Wine Yeasts. Microorganisms 2021, 9, 654. [Google Scholar] [CrossRef]
- Trofa, D.; Gácser, A.; Nosanchuk, J.D. Candida parapsilosis, an emerging fungal pathogen. Clin. Microbiol. Rev. 2008, 21, 606–625. [Google Scholar] [CrossRef] [PubMed]
- Silva, S.; Negri, M.; Henriques, M.; Oliveira, R.; Williams, D.W.; Azeredo, J. Candida glabrata, Candida parapsilosis and Candida tropicalis: Biology, epidemiology, pathogenicity and antifungal resistance. FEMS Microbiol. Rev. 2012, 36, 288–305. [Google Scholar] [CrossRef]
- Tavanti, A.; Davidson, A.D.; Gow, N.A.R.; Maiden, M.C.J.; Odds, F.C. Candida orthopsilosis and Candida metapsilosis spp. nov. to replace Candida parapsilosis groups II and III. J. Clin. Microbiol. 2005, 43, 284–292. [Google Scholar] [CrossRef]
- Kossoff, E.H.; Buescher, E.S.; Karlowicz, M.G. Candidemia in a neonatal intensive care unit: Trends during fifteen years and clinical features of 111 cases. Pediatr. Infect. Dis. J. 1998, 17, 504–508. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, D.; Almirante, B.; Park, B.J.; Cuenca-Estrella, M.; Planes, A.M.; Sanchez, F.; Gene, A.; Xercavins, M.; Fontanals, D.; Rodriguez-Tudela, J.L.; et al. Candidemia in neonatal intensive care units: Barcelona, Spain. Pediatr. Infect. Dis. J. 2006, 25, 224–229. [Google Scholar] [CrossRef] [PubMed]
- Diekema, D.J.; Messer, S.A.; Boyken, L.B.; Hollis, R.J.; Kroeger, J.; Tendolkar, S.; Pfaller, M.A. In vitro activity of seven systemically active antifungal agents against a large global collection of rare Candida species as determined by CLSI broth microdilution methods. J. Clin. Microbiol. 2009, 47, 3170–3177. [Google Scholar] [CrossRef]
- Liu, Y.; Kang, M.; Ye, H.; Zong, Z.; Lv, X. Analysis on clinical characteristics and drug resistance of Candida parapsilosis bloodstream infections in West China Hospital, China, from 2012 to 2015. J. Mycol. Med. 2018, 28, 222–226. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Wei, D.; Gong, X.; Shen, Y.; Zhu, Y.; Wang, J.; Gao, Z. Initial use of voriconazole positively affects outcome of Candida parapsilosis bloodstream infection: A retrospective analysis. Transl. Pediatr. 2020, 9, 48086–48486. [Google Scholar] [CrossRef] [PubMed]
- Pfaller, M.A.; Boyken, L.; Hollis, R.J.; Kroeger, J.; Messer, S.A.; Tendolkar, S.; Diekema, D.J. In vitro susceptibility of invasive isolates of Candida spp. to anidulafungin, caspofungin, and micafungin: Six years of global surveillance. J. Clin. Microbiol. 2008, 46, 150–156. [Google Scholar] [CrossRef] [PubMed]
- Al-Sweih, N.; Khan, Z.; Khan, S.; Devarajan, L.V. Neonatal candidaemia in Kuwait: A 12-year study of risk factors, species spectrum and antifungal susceptibility. Mycoses 2009, 52, 518–523. [Google Scholar] [CrossRef] [PubMed]
- Blyth, C.C.; Chen, S.C.A.; Slavin, M.A.; Serena, C.; Nguyen, Q.; Marriott, D.; Ellis, D.; Meyer, W.; Sorrell, T.C. Not Just Little Adults: Candidemia Epidemiology, Molecular Characterization, and Antifungal Susceptibility in Neonatal and Pediatric Patients. Pediatrics 2009, 123, 1360–1368. [Google Scholar] [CrossRef]
- Nucci, M.; Queiroz-Telles, F.; Tobón, A.M.; Restrepo, A.; Colombo, A.L. Epidemiology of Opportunistic Fungal Infections in Latin America. Clin. Infect. Dis. 2010, 51, 561–570. [Google Scholar] [CrossRef]
- Lockhart, S.R.; Iqbal, N.; Cleveland, A.A.; Farley, M.M.; Harrison, L.H.; Bolden, C.B.; Baughman, W.; Stein, B.; Hollick, R.; Park, B.J.; et al. Species identification and antifungal susceptibility testing of Candida bloodstream isolates from population-based surveillance studies in two U.S. cities from 2008 to 2011. J. Clin. Microbiol. 2012, 50, 3435–3442. [Google Scholar] [CrossRef]
- das Miranda, L.N.; Rodrigues, E.C.A.; Costa, S.F.; van derHeijden, I.M.; Dantas, K.C.; Lobo, R.D.; Basso, M.; Varkulja, G.F.; Krebs, V.L.J.; Gibelli, M.A.B.C.; et al. Candida parapsilosis candidaemia in a neonatal unit over 7 years: A case series study. BMJ Open 2012, 2, e000992. [Google Scholar] [CrossRef]
- Ziccardi, M.; Souza, L.O.P.; Gandra, R.M.; Galdino, A.C.M.; Baptista, A.R.S.; Nunes, A.P.F.; Ribeiro, M.A.; Branquinha, M.H.; Santos, A.L.S. Candida parapsilosis (sensu lato) isolated from hospitals located in the Southeast of Brazil: Species distribution, antifungal susceptibility and virulence attributes. Int. J. Med. Microbiol. 2015, 305, 848–859. [Google Scholar] [CrossRef] [PubMed]
- de Paula Menezes, R.; de Oliveira Melo, S.G.; Bessa, M.A.S.; Silva, F.F.; Alves, P.G.V.; Araújo, L.B.; Penatti, M.P.A.; Abdallah, V.O.S.; von Dollinger de Brito Röder, D.; dos Santos Pedroso, R. Candidemia by Candida parapsilosis in a neonatal intensive care unit: Human and environmental reservoirs, virulence factors, and antifungal susceptibility. Braz. J. Microbiol. 2020, 51, 851–860. [Google Scholar] [CrossRef]
- Abi-chacra, É.A.; Souza, L.O.P.; Cruz, L.P.; Braga-Silva, L.A.; Gonçalves, D.S.; Sodré, C.L.; Ribeiro, M.D.; Seabra, S.H.; Figueiredo-Carvalho, M.H.G.; Barbedo, L.S.; et al. Phenotypical properties associated with virulence from clinical isolates belonging to the Candida parapsilosis complex. FEMS Yeast Res. 2013, 13, 831–848. [Google Scholar] [CrossRef] [PubMed]
- Panagoda, G.J.; Ellepola, A.N.B.; Samaranayake, L.P. Adhesion of Candida parapsilosis to epithelial and acrylic surfaces correlates with cell surface hydrophobicity. Mycoses 2001, 44, 29–35. [Google Scholar] [CrossRef] [PubMed]
- Klotz, S.A.; Drutz, D.J.; Zajic, J.E. Factors governing adherence of Candida species to plastic surfaces. Infect. Immun. 1985, 50, 97–101. [Google Scholar] [CrossRef] [PubMed]
- Hazen, K.C.; Plotkin, B.J.; Klimas, D.M. Influence of growth conditions on cell surface hydrophobicity of Candida albicans and Candida glabrata. Infect. Immun. 1986, 54, 269–271. [Google Scholar] [CrossRef]
- Panagoda, G.J.; Samaranayake, L.P. The relationship between the cell length, adhesion to acrylic and relative cell surface hydrophobicity of Candida parapsilosis. Med. Mycol. 1998, 36, 373–378. [Google Scholar] [CrossRef]
- Suzuki, T.; Miyamae, Y.; Ishida, I. Variation of colony morphology and chromosomal rearrangement in Candida tropicalis pK233. J. Gen. Microbiol. 1991, 137, 161–167. [Google Scholar] [CrossRef][Green Version]
- Okawa, Y.; Goto, K. Antigenicity of Candida tropicalis strain cells cultured at 27 and 37 °C. FEMS Immunol. Med. Microbiol. 2006, 46, 438–443. [Google Scholar] [CrossRef]
- Chai, L.Y.A.; Denning, D.W.; Warn, P. Candida tropicalis in human disease. Crit. Rev. Microbiol. 2010, 36, 282–298. [Google Scholar] [CrossRef]
- Eggimann, P.; Garbino, J.; Pittet, D. Epidemiology of Candida species infections in critically ill non-immunosuppressed patients. Lancet Infect. Dis. 2003, 3, 685–702. [Google Scholar] [CrossRef]
- Álvarez-Lerma, F.; Nolla-Salas, J.; León, C.; Palomar, M.; Jordá, R.; Carrasco, N.; Bobillo, F. Candiduria in critically ill patients admitted to intensive care medical units. Intensive Care Med. 2003, 29, 1069–1076. [Google Scholar] [CrossRef]
- Binelli, C.A.; Moretti, M.L.; Assis, R.S.; Sauaia, N.; Menezes, P.R.; Ribeiro, E.; Geiger, D.C.P.; Mikami, Y.; Miyaji, M.; Oliveira, M.S.; et al. Investigation of the possible association between nosocomial candiduria and candidaemia. Clin. Microbiol. Infect. 2006, 12, 538–543. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Colombo, A.L.; Nucci, M.; Park, B.J.; Nouér, S.A.; Arthington-Skaggs, B.; Da Matta, D.A.; Warnock, D.; Morgan, J. Epidemiology of candidemia in Brazil: A nationwide sentinel surveillance of candidemia in eleven medical centers. J. Clin. Microbiol. 2006, 44, 2816–2823. [Google Scholar] [CrossRef] [PubMed]
- Colombo, A.L.; Guimarães, T.; Silva, L.R.B.F.; de Monfardini, L.P.A.; Cunha, A.K.B.; Rady, P.; Alves, T.; Rosas, R.C. Prospective Observational Study of Candidemia in São Paulo, Brazil: Incidence Rate, Epidemiology, and Predictors of Mortality. Infect. Control Hosp. Epidemiol. 2007, 28, 570–576. [Google Scholar] [CrossRef] [PubMed]
- Hasan, F.; Xess, I.; Wang, X.; Jain, N.; Fries, B.C. Biofilm formation in clinical Candida isolates and its association with virulence. Microbes Infect. 2009, 11, 753–761. [Google Scholar] [CrossRef]
- Miranda, L.N.; van der Heijden, I.M.; Costa, S.F.; Sousa, A.P.I.; Sienra, R.A.; Gobara, S.; Santos, C.R.; Lobo, R.D.; Pessoa, V.P.; Levin, A.S. Candida colonisation as a source for candidaemia. J. Hosp. Infect. 2009, 72, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Kauffman, C.A.; Vazquez, J.A.; Sobel, J.D.; Gallis, H.A.; McKinsey, D.S.; Karchmer, A.W.; Sugar, A.M.; Sharkey, P.K.; Wise, G.J.; Mangi, R.; et al. Prospective Multicenter Surveillance Study of Funguria in Hospitalized Patients. Clin. Infect. Dis. 2000, 30, 14–18. [Google Scholar] [CrossRef] [PubMed]
- Almirante, B.; Rodríguez, D.; Park, B.J.; Cuenca-Estrella, M.; Planes, A.M.; Almela, M.; Mensa, J.; Sanchez, F.; Ayats, J.; Gimenez, M.; et al. Epidemiology and predictors of mortality in cases of Candida bloodstream infection: Results from population-based surveillance, Barcelona, Spain, from 2002 to 2003. J. Clin. Microbiol. 2005, 43, 1829–1835. [Google Scholar] [CrossRef] [PubMed]
- Kontoyiannis, D.P.; Vaziri, I.; Hanna, H.A.; Boktour, M.; Thornby, J.; Hachem, R.; Bodey, G.P.; Raad, I.I. Risk Factors for Candida tropicalis Fungemia in Patients with Cancer. Clin. Infect. Dis. 2001, 33, 1676–1681. [Google Scholar] [CrossRef]
- Ásmundsdóttir, L.R.; Erlendsdóttir, H.; Gottfredsson, M. Increasing incidence of candidemia: Results from a 20-year nationwide study in Iceland. J. Clin. Microbiol. 2002, 40, 3489–3492. [Google Scholar] [CrossRef][Green Version]
- Nucci, M.; Colombo, A.L. Candidemia due to Candida tropicalis: Clinical, epidemiologic, and microbiologic characteristics of 188 episodes occurring in tertiary care hospitals. Diagn. Microbiol. Infect. Dis. 2007, 58, 77–82. [Google Scholar] [CrossRef]
- Butler, G.; Rasmussen, M.D.; Lin, M.F.; Santos, M.A.S.; Sakthikumar, S.; Munro, C.A.; Rheinbay, E.; Grabherr, M.; Forche, A.; Reedy, J.L.; et al. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature 2009, 459, 657–662. [Google Scholar] [CrossRef] [PubMed]
- Kozik, A.; Karkowska-Kuleta, J.; Zajac, D.; Bochenska, O.; Kedracka-Krok, S.; Jankowska, U.; Rapala-Kozik, M. Fibronectin-, vitronectin- and laminin-binding proteins at the cell walls of Candida parapsilosis and Candida tropicalis pathogenic yeasts. BMC Microbiol. 2015, 15, 197. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.-H.; Smith, B.; Miller, A.N.; Staker, B.; Fields, C.; Hernandez, A.; Hoyer, L.L. Agglutinin-Like Sequence (ALS) Genes in the Candida parapsilosis Species Complex: Blurring the Boundaries Between Gene Families That Encode Cell-Wall Proteins. Front. Microbiol. 2019, 10, 781. [Google Scholar] [CrossRef] [PubMed]
- Neale, M.N.; Glass, K.A.; Longley, S.J.; Kim, D.J.; Laforce-Nesbitt, S.S.; Wortzel, J.D.; Shaw, S.K.; Bliss, J.M. Role of the inducible adhesin CpAls7 in binding of Candida parapsilosis to the extracellular matrix under fluid shear. Infect. Immun. 2018, 86, e00892-17. [Google Scholar] [CrossRef] [PubMed]
- Bertini, A.; Zoppo, M.; Lombardi, L.; Rizzato, C.; De Carolis, E.; Vella, A.; Torelli, R.; Sanguinetti, M.; Tavanti, A. Targeted gene disruption in Candida parapsilosis demonstrates a role for CPAR2_404800 in adhesion to a biotic surface and in a murine model of ascending urinary tract infection. Virulence 2016, 7, 85–97. [Google Scholar] [CrossRef]
- Zuo, X.; Liu, Y.; Cai, X.; Zhan, L.; Hu, K. Association of different Candida species with catheter-related candidemia, and the potential antifungal treatments against their adhesion properties and biofilm-forming capabilities. J. Clin. Lab. Anal. 2021, 35, e23738. [Google Scholar] [CrossRef]
- Satoh, K.; Makimura, K.; Hasumi, Y.; Nishiyama, Y.; Uchida, K.; Yamaguchi, H. Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol. Immunol. 2009, 53, 41–44. [Google Scholar] [CrossRef]
- Navalkele, B.D.; Revankar, S.; Chandrasekar, P. Candida auris: A worrisome, globally emerging pathogen. Expert Rev. Anti Infect. Ther. 2017, 15, 819–827. [Google Scholar] [CrossRef]
- de Cássia Orlandi Sardi, J.; Silva, D.R.; Soares Mendes-Giannini, M.J.; Rosalen, P.L. Candida auris: Epidemiology, risk factors, virulence, resistance, and therapeutic options. Microb. Pathog. 2018, 125, 116–121. [Google Scholar] [CrossRef]
- Nett, J.E. Candida auris: An emerging pathogen “incognito”? PLoS Pathog. 2019, 15, e1007638. [Google Scholar] [CrossRef]
- Spivak, E.S.; Hanson, K.E. Candida auris: An emerging fungal pathogen. J. Clin. Microbiol. 2018, 56, e01588-17. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Bing, J.; Hu, T.; Ennis, C.L.; Nobile, C.J.; Huang, G. Candida auris: Epidemiology, biology, antifungal resistance, and virulence. PLoS Pathog. 2020, 16, e1008921. [Google Scholar] [CrossRef] [PubMed]
- Lone, S.A.; Ahmad, A. Candida auris—The growing menace to global health. Mycoses 2019, 62, 620–637. [Google Scholar] [CrossRef] [PubMed]
- Muñoz, J.F.; Welsh, R.M.; Shea, T.; Batra, D.; Gade, L.; Howard, D.; Rowe, L.A.; Meis, J.F.; Litvintseva, A.P.; Cuomo, C.A. Clade-specific chromosomal rearrangements and loss of subtelomeric adhesins in Candida auris. Genetics 2021, 218, iyab029. [Google Scholar] [CrossRef]
- Ahmad, S.; Alfouzan, W. Candida auris: Epidemiology, Diagnosis, Pathogenesis, Antifungal Susceptibility, and Infection Control Measures to Combat the Spread of Infections in Healthcare Facilities. Microorganisms 2021, 9, 807. [Google Scholar] [CrossRef]
- Chybowska, A.D.; Childers, D.S.; Farrer, R.A. Nine Things Genomics Can Tell Us About Candida auris. Front. Genet. 2020, 11, 351. [Google Scholar] [CrossRef]
- Lockhart, S.R.; Etienne, K.A.; Vallabhaneni, S.; Farooqi, J.; Chowdhary, A.; Govender, N.P.; Colombo, A.L.; Calvo, B.; Cuomo, C.A.; Desjardins, C.A.; et al. Simultaneous Emergence of Multidrug-Resistant Candida auris on 3 Continents Confirmed by Whole-Genome Sequencing and Epidemiological Analyses. Clin. Infect. Dis. 2017, 64, 134–140. [Google Scholar] [CrossRef]
- Sharma, C.; Kumar, N.; Pandey, R.; Meis, J.F.; Chowdhary, A. Whole genome sequencing of emerging multidrug resistant Candida auris isolates in India demonstrates low genetic variation. New Microbes New Infect. 2016, 13, 77. [Google Scholar] [CrossRef]
- Chatterjee, S.; Alampalli, S.V.; Nageshan, R.K.; Chettiar, S.T.; Joshi, S.; Tatu, U.S. Draft genome of a commonly misdiagnosed multidrug resistant pathogen Candida auris. BMC Genom. 2015, 16, 686. [Google Scholar] [CrossRef]
- Larkin, E.; Hager, C.; Chandra, J.; Mukherjee, P.K.; Retuerto, M.; Salem, I.; Long, L.; Isham, N.; Kovanda, L.; Borroto-Esoda, K.; et al. The emerging pathogen Candida auris: Growth phenotype, virulence factors, activity of antifungals, and effect of SCY-078, a novel glucan synthesis inhibitor, on growth morphology and biofilm formation. Antimicrob. Agents Chemother. 2017, 61, e02396-16. [Google Scholar] [CrossRef]
- Borman, A.M.; Szekely, A.; Johnson, E.M. Comparative Pathogenicity of United Kingdom Isolates of the Emerging Pathogen Candida auris and Other Key Pathogenic Candida Species. mSphere 2016, 1, e00189-16. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Uppuluri, P.; Mamouei, Z.; Alqarihi, A.; Elhassan, H.; French, S.; Lockhart, S.R.; Chiller, T.; Edwards, J.E., Jr.; Ibrahim, A.S. The NDV-3A vaccine protects mice from multidrug resistant Candida auris infection. PLoS Pathog. 2019, 15, e1007460. [Google Scholar] [CrossRef]
- Ben-Ami, R.; Berman, J.; Novikov, A.; Bash, E.; Shachor-Meyouhas, Y.; Zakin, S.; Maor, Y.; Tarabia, J.; Schechner, V.; Adler, A.; et al. Multidrug-resistant candida haemulonii and C. auris, Tel Aviv, Israel. Emerg. Infect. Dis. 2017, 23, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Khatamzas, E.; Madder, H.; Jeffery, K. Neurosurgical device-associated infections due to Candida auris—Three cases from a single tertiary center. J. Infect. 2019, 78, 409–421. [Google Scholar] [CrossRef] [PubMed]
- Kean, R.; Delaney, C.; Sherry, L.; Borman, A.; Johnson, E.M.; Richardson, M.D.; Rautemaa-Richardson, R.; Williams, C.; Ramage, G. Transcriptome Assembly and Profiling of Candida auris Reveals Novel Insights into Biofilm-Mediated Resistance. mSphere 2018, 3. [Google Scholar] [CrossRef] [PubMed]
- Kean, R.; Ramage, G. Combined Antifungal Resistance and Biofilm Tolerance: The Global Threat of Candida auris. mSphere 2019, 4. [Google Scholar] [CrossRef] [PubMed]
- Kean, R.; Brown, J.; Gulmez, D.; Ware, A.; Ramage, G. Candida auris: A Decade of Understanding of an Enigmatic Pathogenic Yeast. J. Fungi 2020, 6, 30. [Google Scholar] [CrossRef] [PubMed]
- Dominguez, E.G.; Zarnowski, R.; Choy, H.L.; Zhao, M.; Sanchez, H.; Nett, J.E.; Andes, D.R. Conserved Role for Biofilm Matrix Polysaccharides in Candida auris Drug Resistance. mSphere 2019, 4. [Google Scholar] [CrossRef]
- Berisio, R.; Vitagliano, L.; Mazzarella, L.; Zagari, A. Crystal structure of the collagen triple helix model [(Pro-Pro-Gly)10]3. Protein Sci. 2002, 11, 262–270. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharjee, A.; Bansal, M. Collagen Structure: The Madras Triple Helix and the Current Scenario. IUBMB Life 2005, 57, 161–172. [Google Scholar] [CrossRef]
- Szekely, A.; Borman, A.M.; Johnsona, E.M. Candida auris isolates of the southern asian and south african lineages exhibit different phenotypic and antifungal susceptibility profiles in vitro. J. Clin. Microbiol. 2019, 57. [Google Scholar] [CrossRef] [PubMed]
- Abdolrasouli, A.; Armstrong-James, D.; Ryan, L.; Schelenz, S. In vitro efficacy of disinfectants utilised for skin decolonisation and environmental decontamination during a hospital outbreak with Candida auris. Mycoses 2017, 60, 758–763. [Google Scholar] [CrossRef]
- Singh, R.; Kaur, M.; Chakrabarti, A.; Shankarnarayan, S.A.; Rudramurthy, S.M. Biofilm formation by Candida auris isolated from colonising sites and candidemia cases. Mycoses 2019, 62, 706–709. [Google Scholar] [CrossRef]
- Xin, H.; Mohiuddin, F.; Tran, J.; Adams, A.; Eberle, K. Experimental Mouse Models of Disseminated Candida auris Infection. mSphere 2019, 4. [Google Scholar] [CrossRef] [PubMed]
- Savini, V.; Catavitello, C.; Onofrillo, D.; Masciarelli, G.; Astolfi, D.; Balbinot, A.; Febbo, F.; D’Amario, C.; D’Antonio, D. What do we know about Candida guilliermondii? A voyage throughout past and current literature about this emerging yeast. Mycoses 2011, 54, 434–441. [Google Scholar] [CrossRef]
- Cooper, C.R. Yeasts Pathogenic to Humans. Yeasts 2011, 1, 9–19. [Google Scholar] [CrossRef]
- Kurtzman, C.P.; Suzuki, M. Phylogenetic analysis of ascomycete yeasts that form coenzyme Q-9 and the proposal of the new genera Babjeviella, Meyerozyma, Millerozyma, Priceomyces, and Scheffersomyces. Mycoscience 2010, 51, 2–14. [Google Scholar] [CrossRef]
- Vaughan-Martini, A.; Kurtzman, C.P.; Meyer, S.A.; O’Neill, E.B. Two new species in the Pichia guilliermondii clade: Pichia caribbica sp. nov., the ascosporic state of Candida fermentati, and Candida carpophila comb. nov. FEMS Yeast Res. 2005, 5, 463–469. [Google Scholar] [CrossRef][Green Version]
- Lan, L.; Xu, J. Multiple gene genealogical analyses suggest divergence and recent clonal dispersal in the opportunistic human pathogen Candida guilliermondii. Microbiology 2006, 152, 1539–1549. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lockhart, S.R.; Messer, S.A.; Pfaller, M.A.; Diekema, D.J. Identification and susceptibility profile of Candida fermentati from a worldwide collection of Candida guilliermondii clinical isolates. J. Clin. Microbiol. 2009, 47, 242–244. [Google Scholar] [CrossRef] [PubMed]
- Yan, W.; Gao, H.; Qian, X.; Jiang, Y.; Zhou, J.; Dong, W.; Xin, F.; Zhang, W.; Jiang, M. Biotechnological applications of the non-conventional yeast Meyerozyma guilliermondii. Biotechnol. Adv. 2021, 46, 107674. [Google Scholar] [CrossRef] [PubMed]
- Pfaller, M.A.; Diekema, D.J.; Mendez, M.; Kibbler, C.; Erzsebet, P.; Chang, S.C.; Gibbs, D.L.; Newell, V.A.; Finquelievich, J.; Tiraboschi, N.; et al. Candida guilliermondii, an opportunistic fungal pathogen with decreased susceptibility to fluconazole: Geographic and temporal trends from the ARTEMIS DISK antifungal surveillance program. J. Clin. Microbiol. 2006, 44, 3551–3556. [Google Scholar] [CrossRef] [PubMed]
- Pfaller, M.; Neofytos, D.; Diekema, D.; Azie, N.; Meier-Kriesche, H.U.; Quan, S.P.; Horn, D. Epidemiology and outcomes of candidemia in 3648 patients: Data from the Prospective Antifungal Therapy (PATH Alliance®) registry, 2004–2008. Diagn. Microbiol. Infect. Dis. 2012, 74, 323–331. [Google Scholar] [CrossRef]
- Cheng, J.W.; Liao, K.; Kudinha, T.; Yu, S.Y.; Xiao, M.; Wang, H.; Kong, F.; Xu, Y.C. Molecular epidemiology and azole resistance mechanism study of Candida guilliermondii from a Chinese surveillance system. Sci. Rep. 2017, 7, 907. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-Y.; Huang, S.-Y.; Tang, J.-L.; Tsay, W.; Yao, M.; Ko, B.-S.; Chou, W.-C.; Tien, H.-F.; Hsueh, P.-R. Clinical features of patients with infections caused by Candida guilliermondii and Candida fermentati and antifungal susceptibility of the isolates at a medical centre in Taiwan, 2001–10. J. Antimicrob. Chemother. 2013, 68, 2632–2635. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Passos, X.S.; Sales, W.S.; Maciel, P.J.; Costa, C.R.; Miranda, K.C.; de Lemos, J.A.; de Batista, M.A.; do Silva, M.R.R. Candida colonization in intensive care unit patients’ urine. Mem. Inst. Oswaldo Cruz 2005, 100, 925–928. [Google Scholar] [CrossRef][Green Version]
- Tseng, T.Y.; Chen, T.C.; Ho, C.M.; Lin, P.C.; Chou, C.H.; Tsai, C.T.; Wang, J.H.; Chi, C.Y.; Ho, M.W. Clinical features, antifungal susceptibility, and outcome of Candida guilliermondii fungemia: An experience in a tertiary hospital in mid-Taiwan. J. Microbiol. Immunol. Infect. 2018, 51, 552–558. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Tian, S.; Li, F.; Sun, G.; Yun, K.; Cheng, S.; Chu, Y. Clinical Characteristics and Outcomes of Candidemia Caused by Meyerozyma guilliermondii Complex in Cancer Patients Undergoing Surgery. Mycopathologia 2020, 185, 975–982. [Google Scholar] [CrossRef] [PubMed]
- Guinea, J.; Zaragoza, Ó.; Escribano, P.; Martín-Mazuelos, E.; Pemán, J.; Sánchez-Reus, F.; Cuenca-Estrella, M. Molecular identification and antifungal susceptibility of yeast isolates causing fungemia collected in a population-based study in Spain in 2010 and 2011. Antimicrob. Agents Chemother. 2014, 58, 1529–1537. [Google Scholar] [CrossRef]
- Papon, N.; Savini, V.; Lanoue, A.; Simkin, A.J.; Crèche, J.; Giglioli-Guivarc’h, N.; Clastre, M.; Courdavault, V.; Sibirny, A.A. Candida guilliermondii: Biotechnological applications, perspectives for biological control, emerging clinical importance and recent advances in genetics. Curr. Genet. 2013, 59, 73–90. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.W.; Yu, S.Y.; Xiao, M.; Wang, H.; Kudinha, T.; Kong, F.; Xu, Y.C. Identification and antifungal susceptibility profile of candida guilliermondii and candida fermentati from a multicenter study in China. J. Clin. Microbiol. 2016, 54, 2187–2189. [Google Scholar] [CrossRef] [PubMed]
- Hirayama, T.; Miyazaki, T.; Yamagishi, Y.; Mikamo, H.; Ueda, T.; Nakajima, K.; Takesue, Y.; Higashi, Y.; Yamamoto, Y.; Kimura, M.; et al. Clinical and microbiological characteristics of candida guilliermondii and candida fermentati. Antimicrob. Agents Chemother. 2018, 62. [Google Scholar] [CrossRef] [PubMed]
- Ahangarkani, F.; Badali, H.; Rezai, M.S.; Shokohi, T.; Abtahian, Z.; Nesheli, H.M.; Karami, H.; Roilides, E.; Tamaddoni, A. Candidemia due to candida guilliermondii in an immunocompromised infant: A case report and review of literature. Curr. Med. Mycol. 2019, 5, 32–36. [Google Scholar] [CrossRef]
- Tietz, H.J.; Czaika, V.; Sterry, W. Case report. Osteomyelitis caused by high resistant Candida guilliermondii. Mycoses 1999, 42, 577–580. [Google Scholar] [CrossRef] [PubMed]
- Krcmery, V.; Barnes, A.J. Non-albicans Candida spp. causing fungaemia: Pathogenicity and antifungal resistance. J. Hosp. Infect. 2002, 50, 243–260. [Google Scholar] [CrossRef]
- Girmenia, C.; Pizzarelli, G.; Cristini, F.; Barchiesi, F.; Spreghini, E.; Scalise, G.; Martino, P. Candida guilliermondii fungemia in patients with hematologic malignancies. J. Clin. Microbiol. 2006, 44, 2458–2464. [Google Scholar] [CrossRef] [PubMed]
- Pemán, J.; Bosch, M.; Cantón, E.; Viudes, Á.; Jarque, I.; Gómez-García, M.; García-Martínez, J.M.; Gobernado, M. Fungemia due to Candida guilliermondii in a pediatric and adult population during a 12-year period. Diagn. Microbiol. Infect. Dis. 2008, 60, 109–112. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarti, A.; Chatterjee, S.S.; Rao, K.L.N.; Zameer, M.M.; Shivaprakash, M.R.; Singhi, S.; Singh, R.; Varma, S.C. Recent experience with fungaemia: Change in species distribution and azole resistance. Scand. J. Infect. Dis. 2009, 41, 275–284. [Google Scholar] [CrossRef]
- Zepelin, M.B.; Kunz, L.; Rüchel, R.; Reichard, U.; Weig, M.; Groß, U. Epidemiology and antifungal susceptibilities of Candida spp. to six antifungal agents: Results from a surveillance study on fungaemia in Germany from July 2004 to August 2005. J. Antimicrob. Chemother. 2007, 60, 424–428. [Google Scholar] [CrossRef]
- Barchiesi, F.; Spreghini, E.; Tomassetti, S.; Della Vittoria, A.; Arzeni, D.; Manso, E.; Scalise, G. Effects of caspofungin against Candida guilliermondii and Candida parapsilosis. Antimicrob. Agents Chemother. 2006, 50, 2719–2727. [Google Scholar] [CrossRef]
- Savini, V.; Catavitello, C.; Di Marzio, I.; Masciarelli, G.; Astolfi, D.; Balbinot, A.; Bianco, A.; Pompilio, A.; Di Bonaventura, G.; D’Amario, C.; et al. Pan-azole-Resistant Candida guilliermondii from a Leukemia Patient’s Silent Funguria. Mycopathologia 2010, 169, 457–459. [Google Scholar] [CrossRef]
- Marcos-Zambrano, L.J.; Puig-Asensio, M.; Pérez-García, F.; Escribano, P.; Sánchez-Carrillo, C.; Zaragoza, O.; Padilla, B.; Cuenca-Estrella, M.; Almirante, B.; Martín-Gómez, M.T.; et al. Candida guilliermondii complex is characterized by high antifungal resistance but low mortality in 22 cases of candidemia. Antimicrob. Agents Chemother. 2017, 61. [Google Scholar] [CrossRef] [PubMed]
- Ruan, S.Y.; Chien, J.Y.; Hou, Y.C.; Hsueh, P.R. Catheter-related fungemia caused by Candida intermedia. Int. J. Infect. Dis. 2010, 14, e147–e149. [Google Scholar] [CrossRef] [PubMed]
- Jurado-Martín, I.; Marcos-Arias, C.; Tamayo, E.; Guridi, A.; de Groot, P.W.J.; Quindós, G.; Eraso, E. Candida duobushaemulonii: An Old But Unreported Pathogen. J. Fungi 2020, 6, 374. [Google Scholar] [CrossRef] [PubMed]
- Viswanathan, R.; Randhawa, H. Candida viswanathii sp. novo Isolated from a Case of Meningitis. Available online: https://www.cabdirect.org/cabdirect/abstract/19601301642 (accessed on 18 June 2021).
- Sandhu, D.K.; Sandhu, R.S.; Misra, V.C. Isolation of candida viswanathii from cerebrospinal fluid. Med. Mycol. 1976, 14, 251–254. [Google Scholar] [CrossRef]
- Randhawa, H.S.; Mishra, S.K.; Damodaran, V.N.; Prakash, A.; Chowdhary, A.; Khan, Z.U. Pathogenicity of Candida viswanathii for normal and cortisone-treated mice. J. Mycol. Med. 2015, 25, 287–292. [Google Scholar] [CrossRef]
- Kurtzman, C.P.; Robnett, C.J.; Basehoar-Powers, E. Phylogenetic relationships among species of Pichia, Issatchenkia and Williopsis determined from multigene sequence analysis, and the proposal of Barnettozyma gen. nov., Lindnera gen. nov. and Wickerhamomyces gen. nov. FEMS Yeast Res. 2008, 8, 939–954. [Google Scholar] [CrossRef]
- Minter, D.W. Cyberlindnera, a replacement name for Lindnera Kurtzman et al., nom. illegit. Mycotaxon 2009, 110, 473–476. [Google Scholar] [CrossRef]
- Dooley, D.P.; Beckius, M.L.; McAllister, C.K.; Jeffery, B.S. Prostatitis Caused by Hansenula fabianii. J. Infect. Dis. 1990, 161, 1040–1041. [Google Scholar] [CrossRef]
- Bhally, H.S.; Jain, S.; Shields, C.; Halsey, N.; Cristofalo, E.; Merz, W.G. Infection in a neonate caused by Pichia fabianii: Importance of molecular identification. Med. Mycol. 2006, 44, 185–187. [Google Scholar] [CrossRef]
- Jindal, N.; Arora, S.; Dhuria, N.; Arora, D. Cyberlindnera (Pichia) fabianii infection in a neutropenic child: Importance of molecular identification. JMM Case Rep. 2015, 2. [Google Scholar] [CrossRef]
- Grenouillet, F.; Millon, L.; Chamouine, A.; Thiriez, G.; Schulze, O.; Leroy, J. Pichia fabianii fungemia in a neonate. Pediatr. Infect. Dis. J. 2010, 29, 191. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Wang, J.; Li, W.; Jia, H.; Che, J.; Lu, J.; Liu, L.; Cheng, Y. Pichia fabianii blood infection in a premature infant in China: Case report. BMC Res. Notes 2013, 6, 77. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Ruiz, M.; Guinea, J.; Puig-Asensio, M.; Zaragoza, Ó.; Almirante, B.; Cuenca-Estrella, M.; Aguado, J.M.; Project, C.; REIPI, G.-G. (SEIMC) and Fungemia due to rare opportunistic yeasts: Data from a population-based surveillance in Spain. Med. Mycol. 2017, 55, 125–136. [Google Scholar] [CrossRef]
- Hof, H.; Amann, V.; Tauber, C.; Paulun, A. Peritonitis in a neonate due to Cyberlindnera fabianii, an ascomycetic yeast. Infection 2017, 45, 921–924. [Google Scholar] [CrossRef]
- Katagiri, S.; Gotoh, M.; Tone, K.; Akahane, D.; Ito, Y.; Ohyashiki, K.; Makimura, K. Fatal Cyberlindnera fabianii fungemia in a patient with mixed phenotype acute leukemia after umbilical cord blood transplantation. Int. J. Hematol. 2016, 103, 592–595. [Google Scholar] [CrossRef] [PubMed]
- Desai, M.; Nitta, B.; Dhanani, H.; Djurkovic, S.; Katugaha, S. Multiple organ dysfunction syndrome and death secondary to Cyberlindnera fabianii. Med. Mycol. Case Rep. 2019, 26, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Valenza, G.; Valenza, R.; Brederlau, J.; Frosch, M.; Kurzai, O. Identification of Candida fabianii as a cause of lethal septicaemia. Mycoses 2006, 49, 331–334. [Google Scholar] [CrossRef] [PubMed]
- Hamal, P.; Ostransky, J.; Dendis, M.; Horváth, R.; Ruzicka, F.; Buchta, V.; Vejsova, M.; Sauer, P.; Hejnar, P.; Raclavsky, V. A case of endocarditis caused by the yeast Pichia fabianii with biofilm production and developed in vitro resistance to azoles in the course of antifungal treatment. Med. Mycol. 2008, 46, 601–605. [Google Scholar] [CrossRef]
- Yun, J.W.; Park, K.S.; Ki, C.S.; Lee, N.Y. Catheter-related bloodstream infection by Lindnera fabianii in a neutropenic patient. J. Med. Microbiol. 2013, 62, 922–925. [Google Scholar] [CrossRef]
- Gabriel, F.; Noel, T.; Accoceberry, I. Lindnera (Pichia) fabianii blood infection after mesenteric ischemia. Med. Mycol. 2012, 50, 310–314. [Google Scholar] [CrossRef] [PubMed]
- Al-Sweih, N.; Ahmad, S.; Khan, S.; Joseph, L.; Asadzadeh, M.; Khan, Z. Cyberlindnera fabianii fungaemia outbreak in preterm neonates in Kuwait and literature review. Mycoses 2019, 62, 51–61. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.I.; Yu, S.; Park, J.S.; Joo, E.-J.; Shin, J.H.; Kwon, M.-J. Successful Treatment of Fungemia Caused by Cyberlindnera fabianii with Anidulafungin: A Case Report. Ann. Clin. Microbiol. 2015, 18, 94. [Google Scholar] [CrossRef]
- Mlinarić-Missoni, E.; Hatvani, L.; Kocsubé, S.; Vágvölgyi, C.; Škarić, I.; Lukić-Grlić, A. Cyberlindnera fabianii in the neonatal and paediatric intensive care unit: Case reports. JMM Case Rep. 2015, 2. [Google Scholar] [CrossRef]
- Baghdadi, J.; Hemarajata, P.; Humphries, R.; Kelesidis, T. First Report of Ventriculoperitoneal Shunt Infection due to Cyberlindnera fabianii. Case Rep. Infect. Dis. 2015, 2015, 1–6. [Google Scholar] [CrossRef]
- Kumar, A.; Prakash, A.; Singh, A.; Kumar, H.; Hagen, F.; Meis, J.F.; Chowdhary, A. Candida haemulonii species complex: An emerging species in India and its genetic diversity assessed with multilocus sequence and amplified fragment-length polymorphism analyses. Emerg. Microbes Infect. 2016, 5, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Yarrow, D.; Meyer, S.A. Proposal for amendment of the diagnosis of the genus Candida Berkhout nom. cons. Int. J. Syst. Bacteriol. 1978, 28, 611–615. [Google Scholar] [CrossRef]
- Douglass, A.P.; Offei, B.; Braun-Galleani, S.; Coughlan, A.Y.; Martos, A.A.R.R.; Ortiz-Merino, R.A.; Byrne, K.P.; Wolfe, K.H. Population genomics shows no distinction between pathogenic Candida krusei and environmental Pichia kudriavzevii: One species, four names. PLoS Pathog. 2018, 14, e1007138. [Google Scholar] [CrossRef] [PubMed]
- Mixão, V.; Hansen, A.P.; Saus, E.; Boekhout, T.; Lass-Florl, C.; Gabaldón, T. Whole-genome sequencing of the opportunistic yeast pathogen candida inconspicua uncovers its hybrid origin. Front. Genet. 2019, 10, 383. [Google Scholar] [CrossRef]
- Guitard, J.; Atanasova, R.; Brossas, J.Y.; Meyer, I.; Gits, M.; Marinach, C.; Vellaissamy, S.; Angoulvant, A.; Mazier, D.; Hennequin, C. Candida inconspicua and candida norvegensis: New insights into identification in relation to sexual reproduction and genome organization. J. Clin. Microbiol. 2015, 53, 1655–1661. [Google Scholar] [CrossRef]
- Guitard, J.; Angoulvant, A.; Letscher-Bru, V.; L’Ollivier, C.; Cornet, M.; Dalle, F.; Grenouillet, F.; Lacroix, C.; Vekhoff, A.; Maury, E.; et al. Invasive infections due to Candida norvegensis and Candida inconspicua: Report of 12 cases and review of the literature. Med. Mycol. 2013, 51, 795–799. [Google Scholar] [CrossRef]
- Majoros, L.; Kardos, G.; Szabó, B.; Kovács, M.; Maráz, A. Fluconazole susceptibility testing of Candida inconspicua clinical isolates: Comparison of four methods. J. Antimicrob. Chemother. 2005, 55, 275–276. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pfaller, M.A.; Diekema, D.J.; Gibbs, D.L.; Newell, V.A.; Ellis, D.; Tullio, V.; Rodloff, A.; Fu, W.; Ling, T.A. Results from the artemis disk global antifungal surveillance study, 1997 to 2007: A 10.5-year analysis of susceptibilities of candida species to fluconazole and voriconazole as determined by CLSI standardized disk diffusion. J. Clin. Microbiol. 2010, 48, 1366–1377. [Google Scholar] [CrossRef] [PubMed]
- D’Antonio, D.; Violante, B.; Mazzoni, A.; Bonfini, T.; Capuani, M.A.; D’Aloia, F.; Iacone, A.; Schioppa, F.; Romano, F. A nosocomial cluster of Candida inconspicua infections in patients with hematological malignancies. J. Clin. Microbiol. 1998, 36, 792–795. [Google Scholar] [CrossRef] [PubMed]
- Baily, G.G.; Moore, C.B.; Essayag, S.M.; de Wit, S.; Burnie, J.P.; Denning, D.W. Candida inconspicua, a Fluconazole-Resistant Pathogen in Patients Infected with Human Immunodeficiency Virus. Clin. Infect. Dis. 1997, 25, 161–163. [Google Scholar] [CrossRef] [PubMed]
- Nho, S.; Anderson, M.J.; Moore, C.B.; Denning, D.W. Species differentiation by internally transcribed spacer PCR and HhaI digestion of fluconazole-resistant Candida krusei, Candida inconspicua, and Candida norvegensis strains. J. Clin. Microbiol. 1997, 35, 1036–1039. [Google Scholar] [CrossRef] [PubMed]
- Szabó, Z.; Sóczó, G.; Miszti, C.; Hermann, P.; Rozgonyi, F. In vitro activity of fluconazole and amphotericin B against Candida inconspicua clinical isolates as determined by the time-kill method. Acta Microbiol. Immunol. Hung. 2008, 55, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Hansen, A.; Lass-Flörl, C.; Lackner, M.; Group, R.Y.S.; Aigner, M.; Alastruey-Izquierdo, A.; Arikan-Akdagli, S.; Bader, O.; Becker, K.; Boekhout, T.; et al. Antifungal susceptibility profiles of rare ascomycetous yeasts. J. Antimicrob. Chemother. 2019, 74, 2649–2656. [Google Scholar] [CrossRef] [PubMed]
- Boraston, A.B.; Bolam, D.N.; Gilbert, H.J.; Davies, G.J. Carbohydrate-binding modules: Fine-tuning polysaccharide recognition. Biochem. J. 2004, 382, 769–781. [Google Scholar] [CrossRef]
- Guillén, D.; Sánchez, S.; Rodríguez-Sanoja, R. Carbohydrate-binding domains: Multiplicity of biological roles. Appl. Microbiol. Biotechnol. 2009, 85, 1241–1249. [Google Scholar] [CrossRef]
- Sidar, A.; Albuquerque, E.D.; Voshol, G.P.; Ram, A.F.J.; Vijgenboom, E.; Punt, P.J. Carbohydrate Binding Modules: Diversity of Domain Architecture in Amylases and Cellulases From Filamentous Microorganisms. Front. Bioeng. Biotechnol. 2020, 8, 871. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Li, S.; Cheng, L.; Luo, L. Analyzing the relation between the microbial diversity of DaQu and the turbidity spoilage of traditional Chinese vinegar. Appl. Microbiol. Biotechnol. 2014, 98, 6073–6084. [Google Scholar] [CrossRef] [PubMed]
- Bourdichon, F.; Casaregola, S.; Farrokh, C.; Frisvad, J.C.; Gerds, M.L.; Hammes, W.P.; Harnett, J.; Huys, G.; Laulund, S.; Ouwehand, A.; et al. Food fermentations: Microorganisms with technological beneficial use. Int. J. Food Microbiol. 2012, 154, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Radecka, D.; Mukherjee, V.; Mateo, R.Q.; Stojiljkovic, M.; Foulquié-Moreno, M.R.; Thevelein, J.M. Looking beyond Saccharomyces: The potential of non-conventional yeast species for desirable traits in bioethanol fermentation. FEMS Yeast Res. 2015, 15, fov053. [Google Scholar] [CrossRef]
- Mukherjee, V.; Radecka, D.; Aerts, G.; Verstrepen, K.J.; Lievens, B.; Thevelein, J.M. Phenotypic landscape of non-conventional yeast species for different stress tolerance traits desirable in bioethanol fermentation. Biotechnol. Biofuels 2017, 10, 216. [Google Scholar] [CrossRef]
- Xiao, H.; Shao, Z.; Jiang, Y.; Dole, S.; Zhao, H. Exploiting Issatchenkia orientalis SD108 for succinic acid production. Microb. Cell Fact. 2014, 13, 121. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhuge, J.; Fang, H.; Prior, B.A. Glycerol production by microbial fermentation: A review. Biotechnol. Adv. 2001, 19, 201–223. [Google Scholar] [CrossRef]
- Nagarathnamma, T.; Chunchanur, S.K.; Rudramurthy, S.M.; Vineetha, K.R.; Ramamurthy, K.; Joseph, J.; Ambica, R. Outbreak of Pichia kudriavzevii fungaemia in a neonatal intensive care unit. J. Med. Microbiol. 2017, 66, 1759–1764. [Google Scholar] [CrossRef] [PubMed]
- Kumura, H.; Tanoue, Y.; Tsukahara, M.; Tanaka, T.; Shimazaki, K. Screening of Dairy Yeast Strains for Probiotic Applications. J. Dairy Sci. 2004, 87, 4050–4056. [Google Scholar] [CrossRef]
- Greppi, A.; Saubade, F.; Botta, C.; Humblot, C.; Guyot, J.P.; Cocolin, L. Potential probiotic Pichia kudriavzevii strains and their ability to enhance folate content of traditional cereal-based African fermented food. Food Microbiol. 2017, 62, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Passoth, V.; Fredlund, E.; Druvefors, U.Ä.; Schnürer, J. Biotechnology, physiology and genetics of the yeast Pichia anomala. FEMS Yeast Res. 2006, 6, 3–13. [Google Scholar] [CrossRef]
- Petruzzi, L.; Capozzi, V.; Berbegal, C.; Corbo, M.R.; Bevilacqua, A.; Spano, G.; Sinigaglia, M. Microbial resources and enological significance: Opportunities and benefits. Front. Microbiol. 2017, 8, 995. [Google Scholar] [CrossRef]
- Walker, G.M. Pichia anomala: Cell physiology and biotechnology relative to other yeasts. Antonie van Leeuwenhoek 2010, 99, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Sabel, A.; Martens, S.; Petri, A.; König, H.; Claus, H. Wickerhamomyces anomalus AS1: A new strain with potential to improve wine aroma. Ann. Microbiol. 2013, 64, 483–491. [Google Scholar] [CrossRef]
- Padilla, B.; Gil, J.V.; Manzanares, P. Challenges of the Non-Conventional Yeast Wickerhamomyces anomalus in Winemaking. Fermentation 2018, 4, 68. [Google Scholar] [CrossRef]
- Cappelli, A.; Ulissi, U.; Valzano, M.; Damiani, C.; Epis, S.; Gabrielli, M.G.; Conti, S.; Polonelli, L.; Bandi, C.; Favia, G.; et al. A Wickerhamomyces anomalus Killer Strain in the Malaria Vector Anopheles stephensi. PLoS ONE 2014, 9, e95988. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Wang, Y.; Dhanasekaran, S.; Guo, Z.; Chen, S.; Zhang, X.; Zhang, H. Efficacy of Wickerhamomyces anomalus yeast in the biocontrol of blue mold decay in apples and investigation of the mechanisms involved. BioControl 2021, 66, 547–558. [Google Scholar] [CrossRef]
- Raynaldo, F.A.; Dhanasekaran, S.; Ngea, G.L.N.; Yang, Q.; Zhang, X.; Zhang, H. Investigating the biocontrol potentiality of Wickerhamomyces anomalus against postharvest gray mold decay in cherry tomatoes. Sci. Hortic. (Amst.) 2021, 285, 110137. [Google Scholar] [CrossRef]
- Barchiesi, F.; Tortorano, A.M.; Di Francesco, L.F.; Rigoni, A.; Giacometti, A.; Spreghini, E.; Scalise, G.; Viviani, M.A. Genotypic variation and antifungal susceptibilities of Candida pelliculosa clinical isolates. J. Med. Microbiol. 2005, 54, 279–285. [Google Scholar] [CrossRef]
- Lin, H.C.; Lin, H.Y.; Su, B.H.; Ho, M.W.; Ho, C.M.; Lee, C.Y.; Lin, M.H.; Hsieh, H.Y.; Lin, H.C.; Li, T.C.; et al. Reporting an outbreak of Candida pelliculosa fungemia in a neonatal intensive care unit. J. Microbiol. Immunol. Infect. 2013, 46, 456–462. [Google Scholar] [CrossRef] [PubMed]
- Yılmaz-Semerci, S.; Demirel, G.; Taştekin, A. Wickerhamomyces anomalus blood stream infection in a term newborn with pneumonia. Turk. J. Pediatr. 2017, 59, 349–351. [Google Scholar] [CrossRef]
- Noni, M.; Stathi, A.; Velegraki, A.; Malamati, M.; Kalampaliki, A.; Zachariadou, L.; Michos, A. Rare Invasive Yeast Infections in Greek Neonates and Children, a Retrospective 12-Year Study. J. Fungi 2020, 6, 194. [Google Scholar] [CrossRef]
- Storgårds, E.; Tapani, K.; Hartwall, P.; Saleva, R.; Suihko, M.L. Microbial attachment and biofilm formation in brewery bottling plants. J. Am. Soc. Brew. Chem. 2006, 64, 8–15. [Google Scholar] [CrossRef]
- Ruiz, J.; Ortega, N.; Martín-Santamaría, M.; Acedo, A.; Marquina, D.; Pascual, O.; Rozès, N.; Zamora, F.; Santos, A.; Belda, I. Occurrence and enological properties of two new non-conventional yeasts (Nakazawaea ishiwadae and Lodderomyces elongisporus) in wine fermentations. Int. J. Food Microbiol. 2019, 305, 108255. [Google Scholar] [CrossRef]
- Ji, Z.-H.; Jia, J.H.; Bai, F.-Y. Four novel Candida species in the Candida albicans/Lodderomyces elongisporus clade isolated from the gut of flower beetles. Antonie van Leeuwenhoek 2008, 95, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Huh, W.K.; Falvo, J.V.; Gerke, L.C.; Carroll, A.S.; Howson, R.W.; Weissman, J.S.; O’Shea, E.K. Global analysis of protein localization in budding yeast. Nature 2003, 425, 686–691. [Google Scholar] [CrossRef] [PubMed]
- James, S.A.A.; Collins, M.D.D.; Roberts, I.N.N. The genetic relationship of Lodderomyces elongisporus to other ascomycete yeast species as revealed by small-subunit rRNA gene sequences. Lett. Appl. Microbiol. 1994, 19, 308–311. [Google Scholar] [CrossRef]
- Lockhart, S.R.; Messer, S.A.; Pfaller, M.A.; Diekema, D.J. Lodderomyces elongisporus masquerading as Candida parapsilosis as a cause of bloodstream infections. J. Clin. Microbiol. 2008, 46, 374–376. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ahmad, S.; Khan, Z.U.; Johny, M.; Ashour, N.M.; Al-Tourah, W.H.; Joseph, L.; Chandy, R. Isolation of lodderomyces elongisporus from the catheter tip of a fungemia patient in the Middle East. Case Rep. Med. 2013, 2013. [Google Scholar] [CrossRef] [PubMed]
- Hatanaka, S.; Nakamura, I.; Fukushima, S.; Ohkusu, K.; Matsumoto, T. Catheter-Related Bloodstream Infection Due to Lodderomyces elongisporus. Jpn. J. Infect. Dis. 2016, 69, 520–522. [Google Scholar] [CrossRef] [PubMed]
- Al-Obaid, K.; Ahmad, S.; Joseph, L.; Khan, Z. Lodderomyces elongisporus: A bloodstream pathogen of greater clinical significance. New Microbes New Infect. 2018, 26, 20–24. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.Y.; Kim, S.J.; Kim, D.; Jang, J.; Sung, H.; Kim, M.N.; Choi, C.M. Catheter-related Bloodstream Infection due to Lodderomyces elongisporus in a Patient with Lung Cancer. Ann. Lab. Med. 2018, 38, 182–184. [Google Scholar] [CrossRef]
- Koh, B.; Halliday, C.; Chan, R. Concurrent bloodstream infection with Lodderomyces elongisporus and Candida parapsilosis. Med. Mycol. Case Rep. 2020, 28, 23–25. [Google Scholar] [CrossRef] [PubMed]
- Thompson, C.M.; Warner, N.; Hurt, C.B.; Alby, K.; Miller, M.B. The brief case: A case of prosthetic valve endocarditis due to lodderomyces elongisporus. J. Clin. Microbiol. 2021, 59, 1225–1245. [Google Scholar] [CrossRef]
- Lee, A.C.; Fujio, Y. Microflora of banh men, a fermentation starter from Vietnam. World J. Microbiol. Biotechnol. 1999, 15, 51–55. [Google Scholar] [CrossRef]
- Simoncini, N.; Rotelli, D.; Virgili, R.; Quintavalla, S. Dynamics and characterization of yeasts during ripening of typical Italian dry-cured ham. Food Microbiol. 2007, 24, 577–584. [Google Scholar] [CrossRef]
- Simpson, V.R.; Borman, A.M.; Fox, R.I.; Mathews, F. Cutaneous mycosis in a Barbastelle bat (Barbastella barbastellus) caused by Hyphopichia burtonii. J. Vet. Diagn. Investig. 2013, 25, 551–554. [Google Scholar] [CrossRef]
- Chamroensakchai, T.; Kanjanabuch, T.; Saikong, W.; Panya, W.; Thaweekote, S.; Eiam-Ong, S.; Hurdeal, V.G.; Hyde, K.D. The first human report of Hyphopichia burtonii, initially misdiagnosed as sterile peritonitis in a patient on peritoneal dialysis. Med. Mycol. Case Rep. 2021, 33, 26–29. [Google Scholar] [CrossRef]
- Quirós, M.; Martorell, P.; Valderrama, M.J.; Querol, A.; Peinado, J.M.; de Silóniz, M.I. PCR-RFLP analysis of the IGS region of rDNA: A useful tool for the practical discrimination between species of the genus Debaryomyces. Antonie van Leeuwenhoek, Int. J. Gen. Mol. Microbiol. 2006, 90, 211–219. [Google Scholar] [CrossRef]
- Gupta, A.J.; Mi, H.; Wroe, C.; Jaques, B.; Talbot, D. Fatal Candida famata peritonitis complicating sclerosing peritonitis in a peritoneal dialysis patient. Nephrol. Dial. Transplant. 2006, 21, 2036–2037. [Google Scholar] [CrossRef][Green Version]
- St-Germain, G.; Laverdiere, M. Torulopsis candida, a new opportunistic pathogen. J. Clin. Microbiol. 1986, 24, 884–885. [Google Scholar] [CrossRef]
- Wagner, D.; Sander, A.; Bertz, H.; Finke, J.; Kern, W.V. Breakthrough invasive infection due to Debaryomyces hansenii (teleomorph Candida famata) and Scopulariopsis brevicaulis in a stem cell transplant patient receiving liposomal amphotericin B and caspofungin for suspected aspergillosis. Infection 2005, 33, 397–400. [Google Scholar] [CrossRef]
- Castanheira, M.; Woosley, L.N.; Diekema, D.J.; Jones, R.N.; Pfaller, M.A. Candida guilliermondii and other species of Candida misidentified as Candida famata: Assessment by Vitek 2, DNA sequencing analysis, and matrix-assisted laser desorption ionization-time of flight mass spectrometry in two global antifungal surveillance programs. J. Clin. Microbiol. 2013, 51, 117–124. [Google Scholar] [CrossRef]
- Kim, S.H.; Shin, J.H.; Mok, J.H.; Kim, S.Y.; Song, S.A.; Kim, H.R.; Kook, J.K.; Chang, Y.H.; Bae, I.K.; Lee, K. Misidentification of Candida guilliermondii as C. Famata among strains isolated from blood cultures by the VITEK 2 system. BioMed Res. Int. 2014, 2014, 250408. [Google Scholar] [CrossRef]
- Nishikawa, A.; Sugita, T.; Shinoda, T. Differentiation between Debaryomyces hansenii/Candida famata complex and Candida guilliermondii by polymerase chain reaction. FEMS Immunol. Med. Microbiol. 2006, 19, 125–129. [Google Scholar] [CrossRef]
- Nishikawa, A.; Tomomatsu, H.; Sugita, T.; Ikeda, R.; Shinoda, T. Taxonomic position of clinical isolates of Candida famata. J. Med. Vet. Mycol. 1996, 34, 411–419. [Google Scholar] [CrossRef]
- Gibson, B.; Liti, G. Saccharomyces pastorianus: Genomic insights inspiring innovation for industry. Yeast 2015, 32, 17–27. [Google Scholar] [CrossRef]
- Aucott, J.N.; Fayen, J.; Grossnicklas, H.; Morrissey, A.; Lederman, M.M.; Salata, R.A. Invasive infection with Saccharomyces cerevisiae: Report of three cases and review. Clin. Infect. Dis. 1990, 12, 406–411. [Google Scholar] [CrossRef]
- Posteraro, B.; Sanguinetti, M.; D’Amore, G.; Masucci, L.; Morace, G.; Fadda, G. Molecular and epidemiological characterization of vaginal Saccharomyces cerevisiae isolates. J. Clin. Microbiol. 1999, 37, 2230–2235. [Google Scholar] [CrossRef] [PubMed]
- Sobel, J.D.; Vazquez, J.; Lynch, M.; Meriwether, C.; Zervos, M.J. Vaginitis Due to Saccharomyces cerevisiae: Epidemiology, Clinical Aspects, and Therapy. Clin. Infect. Dis. 1993, 16, 93–99. [Google Scholar] [CrossRef]
- Pérez-Torrado, R.; Querol, A. Opportunistic strains of Saccharomyces cerevisiae: A potential risk sold in food products. Front. Microbiol. 2016, 6, 1522. [Google Scholar] [CrossRef] [PubMed]
- Enache-Angoulvant, A.; Hennequin, C. Invasive Saccharomyces Infection: A Comprehensive Review. Clin. Infect. Dis. 2005, 41, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Anoop, V.; Rotaru, S.; Shwed, P.S.; Tayabali, A.F.; Arvanitakis, G. Review of current methods for characterizing virulence and pathogenicity potential of industrial Saccharomyces cerevisiae strains towards humans. FEMS Yeast Res. 2015, 15, fov057. [Google Scholar] [CrossRef]
- Lherm, T.; Monet, C.; Nougière, B.; Soulier, M.; Larbi, D.; Le Gall, C.; Caen, D.; Malbrunot, C. Seven cases of fungemia with Saccharomyces boulardii in critically ill patients. Intensive Care Med. 2002, 28, 797–801. [Google Scholar] [CrossRef] [PubMed]
- Cassone, M.; Serra, P.; Mondello, F.; Girolamo, A.; Scafetti, S.; Pistella, E.; Venditti, M. Outbreak of Saccharomyces cerevisiae Subtype boulardii Fungemia in Patients Neighboring Those Treated with a Probiotic Preparation of the Organism. J. Clin. Microbiol. 2003, 41, 5340–5343. [Google Scholar] [CrossRef] [PubMed]
- Muñoz, P.; Bouza, E.; Cuenca-Estrella, M.; Eiros, J.M.; Pérez, M.J.; Sánchez-Somolinos, M.; Rincón, C.; Hortal, J.; Peláez, T. Saccharomyces cerevisiae Fungemia: An Emerging Infectious Disease. Clin. Infect. Dis. 2005, 40, 1–10. [Google Scholar] [CrossRef] [PubMed]
- de Llanos, R.; Querol, A.; Pemán, J.; Gobernado, M.; Fernández-Espinar, M.T. Food and probiotic strains from the Saccharomyces cerevisiae species as a possible origin of human systemic infections. Int. J. Food Microbiol. 2006, 110, 286–290. [Google Scholar] [CrossRef] [PubMed]
- Williams, J.S.; Mufti, G.J.; Powell, S.; Salisbury, J.R.; Higgins, E.M. Saccharomyces cerevisiae emboli in an immunocompromised patient with relapsed acute myeloid leukaemia. Clin. Exp. Dermatol. 2007, 32, 395–397. [Google Scholar] [CrossRef]
- Seng, P.; Cerlier, A.; Cassagne, C.; Coulange, M.; Legré, R.; Stein, A. Saccharomyces cerevisiae osteomyelitis in an immunocompetent baker. IDCases 2016, 5, 1–3. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pfaller, M.A.; Diekema, D.J. Epidemiology of invasive mycoses in North America. Crit. Rev. Microbiol. 2010, 36, 1–53. [Google Scholar] [CrossRef]
- Kliemann, D.A.; Antonello, V.S.; Severo, L.C.; Pasqualotto, A.C. Saccharomyces cerevisiae oesophagitis in a patient with oesophageal carcinoma. J. Infect. Dev. Ctries. 2011, 5, 493–495. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kelesidis, T.; Pothoulakis, C. Efficacy and safety of the probiotic Saccharomyces boulardii for the prevention and therapy of gastrointestinal disorders. Ther. Adv. Gastroenterol. 2012, 5, 111–125. [Google Scholar] [CrossRef]
- Landaburu, M.F.; López Daneri, G.A.; Relloso, S.; Zarlenga, L.J.; Vinante, M.A.; Mujica, M.T. Fungemia following Saccharomyces cerevisiae var. boulardii probiotic treatment in an elderly patient. Rev. Argent. Microbiol. 2020, 52, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Verstrepen, K.J.; Reynolds, T.B.; Fink, G.R. Origins of variation in the fungal cell surface. Nat. Rev. Microbiol. 2004, 2, 533–540. [Google Scholar] [CrossRef]
- Hennequin, C.; Kauffmann-Lacroix, C.; Jobert, A.; Viard, J.P.; Ricour, C.; Jacquemin, J.L.; Berche, P. Possible role of catheters in Saccharomyces boulardii fungemia. Eur. J. Clin. Microbiol. Infect. Dis. 2000, 19, 16–20. [Google Scholar] [CrossRef]
- Donlan, R.M.; Costerton, J.W. Biofilms: Survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 2002, 15, 167–193. [Google Scholar] [CrossRef] [PubMed]
- Jabra-Rizk, M.A.; Falkler, W.A.; Meiller, T.F. Fungal Biofilms and Drug Resistance. Emerg. Infect. Dis. 2004, 10, 14–19. [Google Scholar] [CrossRef] [PubMed]
- Sen, S.; Mansell, T.J. Yeasts as probiotics: Mechanisms, outcomes, and future potential. Fungal Genet. Biol. 2020, 137, 103333. [Google Scholar] [CrossRef] [PubMed]
- Pais, P.; Almeida, V.; Yılmaz, M.; Teixeira, M.C. Saccharomyces boulardii: What Makes It Tick as Successful Probiotic? J. Fungi 2020, 6, 78. [Google Scholar] [CrossRef] [PubMed]
- Czerucka, D.; Piche, T.; Rampal, P. Review article: Yeast as probiotics—Saccharomyces boulardii. Aliment. Pharmacol. Ther. 2007, 26, 767–778. [Google Scholar] [CrossRef] [PubMed]
- Thygesen, J.B.; Glerup, H.; Tarp, B. Saccharomyces boulardii fungemia caused by treatment with a probioticum. BMJ Case Rep. 2012, 2012, bcr0620114412. [Google Scholar] [CrossRef] [PubMed]
- Appel-da-Silva, M.C.; Narvaez, G.A.; Perez, L.R.R.; Drehmer, L.; Lewgoy, J. Saccharomyces cerevisiae var. boulardii fungemia following probiotic treatment. Med. Mycol. Case Rep. 2017, 18, 15–17. [Google Scholar] [CrossRef] [PubMed]
- Khatri, I.; Tomar, R.; Ganesan, K.; Prasad, G.S.; Subramanian, S. Complete genome sequence and comparative genomics of the probiotic yeast Saccharomyces boulardii. Sci. Rep. 2017, 7, 371. [Google Scholar] [CrossRef] [PubMed]
- Edwards-Ingram, L.; Gitsham, P.; Burton, N.; Warhurst, G.; Clarke, I.; Hoyle, D.; Oliver, S.G.; Stateva, L. Genotypic and physiological characterization of Saccharomyces boulardii, the probiotic strain of Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2007, 73, 2458–2467. [Google Scholar] [CrossRef] [PubMed]


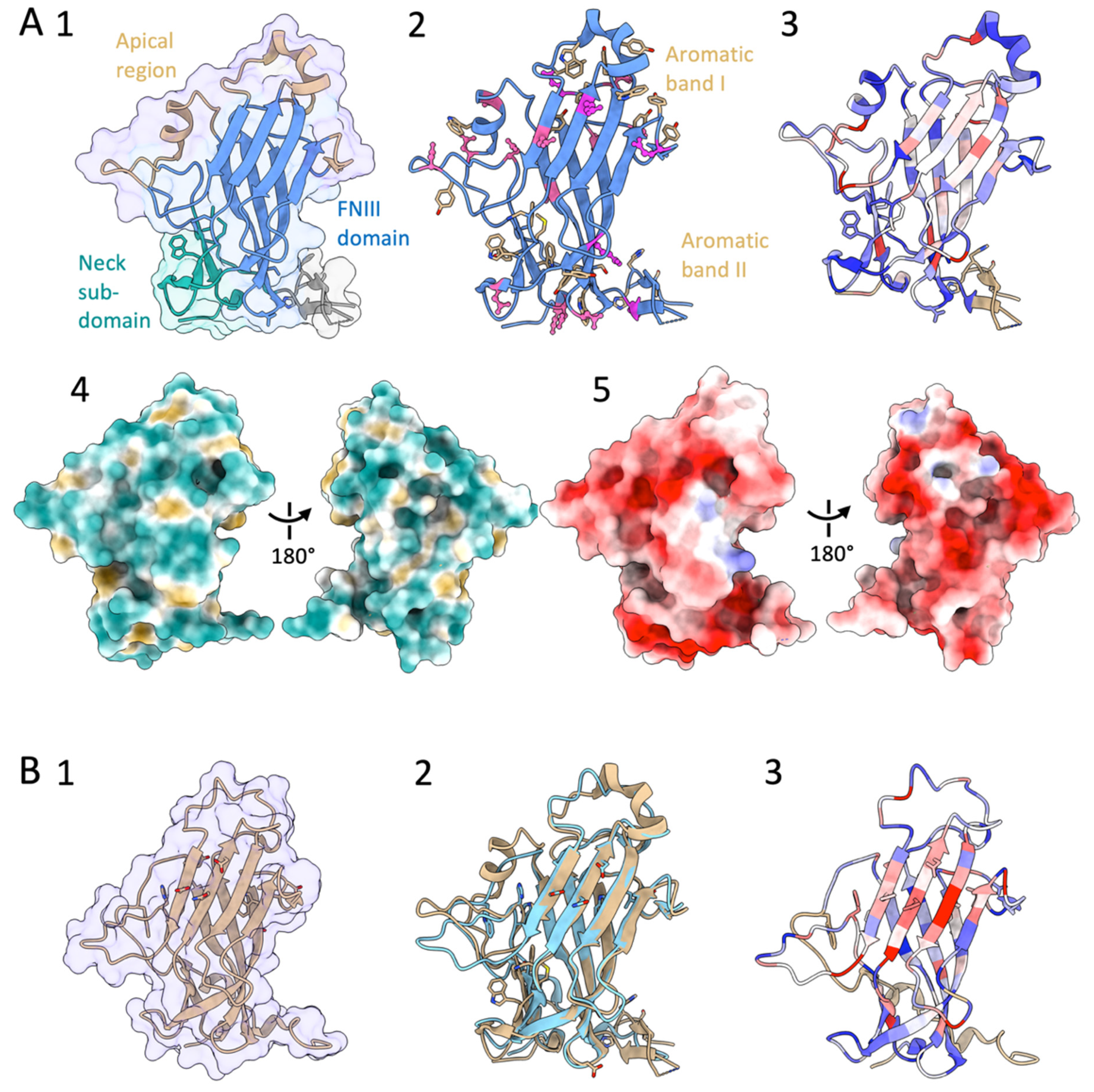
| Flo Adhesin Class/Subtype | Adhesin | Micro-Organism | Ligand in the Structure | Mutations | PDB Code | Interacting Substrate/Function Properties | Refs |
|---|---|---|---|---|---|---|---|
| Flo-type/PA14- | N-Flo1p | S. cerevisiae | Apo | - | 4LHL | Cell-cell interaction via cell surface mannans | [93] |
| Man | - | 4LHN | [93] | ||||
| Flo-type/PA14 | N-Lg-Flo1p | S. pastorianus | Apo | - | 4GQ7 | Cell-cell interaction via cell surface mannans and phospho-mannans | [96] |
| Manα-1,2-Man | - | 4LHK | [93] | ||||
| Flo-type/PA14 | N-Flo5p | S. cerevisiae | Apo | - | 2XJQ | Cell-cell interaction via cell surface mannans | [97] |
| Man | - | 2XJP | [97] | ||||
| Man3(D1) | - | 2XJT | [97] | ||||
| Man5(D2-3) | - | 2XJR | [97] | ||||
| Manα-1,2-Man | - | 2XJS | [97] | ||||
| Manα-1,2-Man | S277A | 2XJU | [97] | ||||
| Glc | - | 2XJV | [97] | ||||
| Flo-type/GLEYA | N-Epa1p | C. glabrata | Gal | - | 4A3X | Epithelial cells, fibronectin, mucin | [92] |
| Galβ-1,3-Glc | - | 4AF9 | [95] | ||||
| Galβ-1,3-Glc | E227D, Y228N | 4AFC | [95] | ||||
| Galβ-1,3-GalNAc (T antigen) | - | 4ASL | [95] | ||||
| Galβ-1,3-GalNAc (T antigen) | - | 4D3W | [98] | ||||
| Galβ-1,4-Glc (lactose) | - | 4COU | [98] | ||||
| Glycerol | E227D, Y228N, D229N | 4AFA | [95] | ||||
| Glycerol | R226I, E227G, Y228K | 4AFB | [95] | ||||
| N-Epa6p | C. glabrata | Galβ-1,4-Glc (lactose) | - | 4COU | Epithelial cells | [98] | |
| Galβ-1,3-GalNAc (T-antigen) | - | 4COW | [98] | ||||
| N-acetyl-D-lactosamine | - | 4COY | [98] | ||||
| Lacto-N-biose | - | 4COZ | [98] | ||||
| α1-3-galactobiose | - | 4COV | [98] | ||||
| N-Epa9p | C. glabrata | Galβ-1,4-Glc | - | 4CP0 | Epithelial cells | * | |
| Galβ-1,3-GlcNAc | - | 4CP1 | * | ||||
| Galβ-1,4-GlcNAc | - | 4CP2 | * | ||||
| Flo11-type | N-ScFlo11p | S. cerevisiae | - | - | 4UYR | Cell-cell and cell-hydrophobic plastic adhesion via hydrophobic interactions, biofilm formation, kin discrimination | [69] |
| - | - | 4UYS | [69] | ||||
| - | - | 4UYT | [69] | ||||
| Flo11-type | N-KpFlo11 | K. pastoris # | - | - | 5FV5 | Cell-cell adhesion interactions, biofilm formation, kin discrimination | [99] |
| Glycerol | - | 5FV6 |
| Flo Adhesin Subtype | Adhesin Uniprot Entry Pfam Protein Architecture | Description Uniprot | Adhesin Domain [Number of Repeats] | Flocculin [Number of Repeats] | Flocculin_t3 [Number of Repeats] | Organism NCBI Taxonomy ID |
|---|---|---|---|---|---|---|
| PA14 | FLO1_YEAST https://www.uniprot.org/uniprot/P32768 (accessed on 24 October 2021) | Flocculation protein Flo1p | 1 | 18 | 0 | Saccharomyces cerevisiae S288c 559292 https://www.uniprot.org/taxonomy/559292 (accessed on 24 October 2021) |
 | ||||||
| FLO10_YEAST https://www.uniprot.org/uniprot/P36170 (accessed on 24 October 2021) | Flocculation protein Flo10p | 1 | 0 | 2 | Saccharomyces cerevisiae S288c 559292 https://www.uniprot.org/taxonomy/559292 (accessed on 24 October 2021) | |
 | ||||||
| FLO5_YEAST https://www.uniprot.org/uniprot/P38894 (accessed on 24 October 2021) | Flocculation protein Flo5p | 1 | 8 | 3 | Saccharomyces cerevisiae S288c 559292 https://www.uniprot.org/taxonomy/559292 (accessed on 24 October 2021) | |
 | ||||||
| FLO9_YEAST https://www.uniprot.org/uniprot/P39712 (accessed on 24 October 2021) | Flocculation protein Flo9p | 1 | 13 | 3 | Saccharomyces cerevisiae S288c 559292 https://www.uniprot.org/taxonomy/559292 (accessed on 24 October 2021) | |
 | ||||||
| B3IUA8_SACPS https://www.uniprot.org/uniprot/B3IUA8 (accessed on 24 October 2021) | Flocculation protein Lg-Flo1p | 1 | 8 | 3 | Saccharomyces pastorianus 27292 https://www.uniprot.org/taxonomy/27292 (accessed on 24 October 2021) | |
 | ||||||
| A0A0L8VPM7_9SACH https://www.uniprot.org/uniprot/A0A0L8VPM7 (accessed on 24 October 2021) | YHR213Wp flocculin-like protein | 1 | 1 | 0 | S. cerevisiae var. boulardii | |
 | ||||||
| GLEYA | Q6FR82_CANGA https://www.uniprot.org/uniprot/Q6FR82 (accessed on 24 October 2021) | PA14 domain-containing protein, Epa23p | 1 | 5 | 0 | Candida glabrata 284593 https://www.uniprot.org/taxonomy/284593 (accessed on 24 October 2021) |
 | ||||||
| C4Y9G0_CLAL4 https://www.uniprot.org/uniprot/C4Y9G0 (accessed on 24 October 2021) | PA14 domain-containing protein | 1 | 0 | 1 | Clavispora lusitaniae 306902 https://www.uniprot.org/taxonomy/306902 (accessed on 24 October 2021) | |
 | ||||||
 | ||||||
| Adhesin Uniprot Entry Pfam Protein Architecture | Description Uniprot | Flo11 Domain [Number of Repeats] | Flocculin [Number of Repeats] | Flocculin_t3 [Number of Repeats] | Other Adhesin Domain [Number] [Pfam ID] | Organism NCBI Taxonomy ID |
|---|---|---|---|---|---|---|
| A0A1L0DFL1_9ASCOhttps://www.uniprot.org/uniprot/A0A1L0DFL1 (accessed on 24 October 2021) | CIC11C00000002180 | 1 | 0 | 0 | — | Candida intermedia 45354https://www.uniprot.org/taxonomy/45354 (accessed on 24 October 2021) |
 | ||||||
| A0A367Y9C2_9ASCOhttps://www.uniprot.org/uniprot/A0A367Y9C2 (accessed on 24 October 2021) | Cell wall protein RTB1 | 1 | 0 | 0 | — | Candida viswanathii 5486https://www.uniprot.org/taxonomy/5486 (accessed on 24 October 2021) |
 | ||||||
| C4YAK2_CLAL4https://www.uniprot.org/uniprot/C4YAK2 (accessed on 24 October 2021) | Flo11 domain-containing protein | 1 | 0 | 0 | — | Clavispora lusitaniae 306902https://www.uniprot.org/taxonomy/306902 (accessed on 24 October 2021) |
 | ||||||
| A0A1V2L8G6_CYBFAhttps://www.uniprot.org/uniprot/A0A1V2L8G6 (accessed on 24 October 2021) | Flocculation protein Flo11p | 2 | 0 | 0 | — | Cyberlindnera fabianii 36022https://www.uniprot.org/taxonomy/36022 (accessed on 24 October 2021) |
 | ||||||
| Q6BXK5_DEBHAhttps://www.uniprot.org/uniprot/Q6BXK5 (accessed on 24 October 2021) | DEHA2B02222p | 1 | 0 | 0 | — | Debaryomyces hansenii 284592https://www.uniprot.org/taxonomy/284592 (accessed on 24 October 2021) |
 | ||||||
| A5DNK5_PICGUhttps://www.uniprot.org/uniprot/A5DNK5 (accessed on 24 October 2021) | Flo11 domain-containing protein | 1 | 0 | 0 | — | Meyerozyma guilliermondii 294746https://www.uniprot.org/taxonomy/294746 (accessed on 24 October 2021) |
 | ||||||
| A0A2U9R2I2_PICKUhttps://www.uniprot.org/uniprot/A0A2U9R2I2 (accessed on 24 October 2021) | Flo11 domain-containing protein | 1 | 0 | 0 | — | Pichia kudriavzevii 4909https://www.uniprot.org/taxonomy/4909 (accessed on 24 October 2021) |
 | ||||||
| FLO11_YEASThttps://www.uniprot.org/uniprot/P08640 (accessed on 24 October 2021) | Flocculation protein Flo11p | 1 | 0 | 0 | — | Saccharomyces cerevisiae 559292https://www.uniprot.org/taxonomy/559292 (accessed on 24 October 2021) |
 | ||||||
| A0A1L0BS11_9ASCOhttps://www.uniprot.org/uniprot/A0A1L0BS11 (accessed on 24 October 2021) | CIC11C00000003144 | 6 | 0 | 5 | — | Candida intermedia 45354https://www.uniprot.org/taxonomy/45354 (accessed on 24 October 2021) |
 | ||||||
| G8B9T8_CANPChttps://www.uniprot.org/uniprot/G8B9T8 (accessed on 24 October 2021) | Flo11 domain-containing protein | 1 | 0 | 2 | — | Candida parapsilosis 578454https://www.uniprot.org/taxonomy/578454 (accessed on 24 October 2021) |
 | ||||||
| C5M338_CANTThttps://www.uniprot.org/uniprot/C5M338 (accessed on 24 October 2021) | Flo11 domain-containing protein | 1 | 0 | 2 | — | Candida tropicalis 294747https://www.uniprot.org/taxonomy/294747 (accessed on 24 October 2021) |
 | ||||||
| C4XZ24_CLAL4https://www.uniprot.org/uniprot/C4XZ24 (accessed on 24 October 2021) | Uncharacterized protein | 3 | 0 | 4 | — | Clavispora lusitaniae 306902https://www.uniprot.org/taxonomy/306902 (accessed on 24 October 2021) |
 | ||||||
| A0A1E4RJE3_9ASCOhttps://www.uniprot.org/uniprot/A0A1E4RJE3 (accessed on 24 October 2021) | Flo11 domain-containing protein | 1 | 0 | 1 | — | Hyphopichia burtonii 984485https://www.uniprot.org/taxonomy/984485 (accessed on 24 October 2021) |
 | ||||||
| A5E4F6_LODELhttps://www.uniprot.org/uniprot/A5E4F6 (accessed on 24 October 2021) | Flo11 domain-containing protein | 1 | 0 | 2 | — | Lodderomyces elongisporus 379508https://www.uniprot.org/taxonomy/379508(accessed on 24 October 2021) |
 | ||||||
| A5DGW9_PICGUhttps://www.uniprot.org/uniprot/A5DGW9 (accessed on 24 October 2021) | Flo11 domain-containing protein | 1 | 0 | 1 | — | Meyerozyma guilliermondii 294746https://www.uniprot.org/taxonomy/294746 (accessed on 24 October 2021) |
 | ||||||
| A0A2H1A319_CANARhttps://www.uniprot.org/uniprot/A0A2H1A319 (accessed on 24 October 2021) | Flo11 domain-containing protein | 1 | 0 | 0 | Collagen 1 1 PF01391 | Candida auris 498019https://www.uniprot.org/taxonomy/498019 (accessed on 24 October 2021) |
 | ||||||
| A0A2V1B0R1_9ASCOhttps://www.uniprot.org/uniprot/A0A2V1B0R1 (accessed on 24 October 2021) | Flo11 domain-containing protein | 1 | 0 | 0 | Collagen 1 1 PF01391 | Candida haemuloni 45357https://www.uniprot.org/taxonomy/45357 (accessed on 24 October 2021) |
 | ||||||
| A0A4T0 × 4H6_9ASCOhttps://www.uniprot.org/uniprot/A0A4T0X4H6 (accessed on 24 October 2021) | Uncharacterized protein | 1 | 0 | 0 | CMB_1 2 3 PF00734 | Candida inconspicua 52247https://www.uniprot.org/taxonomy/52247 (accessed on 24 October 2021) |
 | ||||||
| A5E3T4_LODELhttps://www.uniprot.org/uniprot/A5E3T4 (accessed on 24 October 2021) | Flo11 domain-containing protein | 1 | 0 | 0 | Candida_ALS 3 20 PF05792 | Lodderomyces elongisporus 379508https://www.uniprot.org/taxonomy/379508 (accessed on 24 October 2021) |
 | ||||||
| A0A099NZM2_PICKUhttps://www.uniprot.org/uniprot/A0A099NZM2 (accessed on 24 October 2021) | Flocculation protein Flo11p | 1 | 0 | 0 | CMB_1 2 2 PF00734 | Pichia kudriavzevii 4909https://www.uniprot.org/taxonomy/4909 (accessed on 24 October 2021) |
 | ||||||
| A0A1E3NVU9_WICAAhttps://www.uniprot.org/uniprot/A0A1E3NVU9 (accessed on 24 October 2021) | Uncharacterized protein | 1 | 0 | 0 | CMB_1 2 2 PF00734 | Wickerhamomyces anomalus 683960https://www.uniprot.org/taxonomy/683960 (accessed on 24 October 2021) |
 | ||||||
 | ||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Willaert, R.G.; Kayacan, Y.; Devreese, B. The Flo Adhesin Family. Pathogens 2021, 10, 1397. https://doi.org/10.3390/pathogens10111397
Willaert RG, Kayacan Y, Devreese B. The Flo Adhesin Family. Pathogens. 2021; 10(11):1397. https://doi.org/10.3390/pathogens10111397
Chicago/Turabian StyleWillaert, Ronnie G., Yeseren Kayacan, and Bart Devreese. 2021. "The Flo Adhesin Family" Pathogens 10, no. 11: 1397. https://doi.org/10.3390/pathogens10111397
APA StyleWillaert, R. G., Kayacan, Y., & Devreese, B. (2021). The Flo Adhesin Family. Pathogens, 10(11), 1397. https://doi.org/10.3390/pathogens10111397







