Stringent Response in Mycobacteria: From Biology to Therapeutic Potential
Abstract
:1. Introduction
2. Metabolism of (p)ppGpp in Mycobacteria
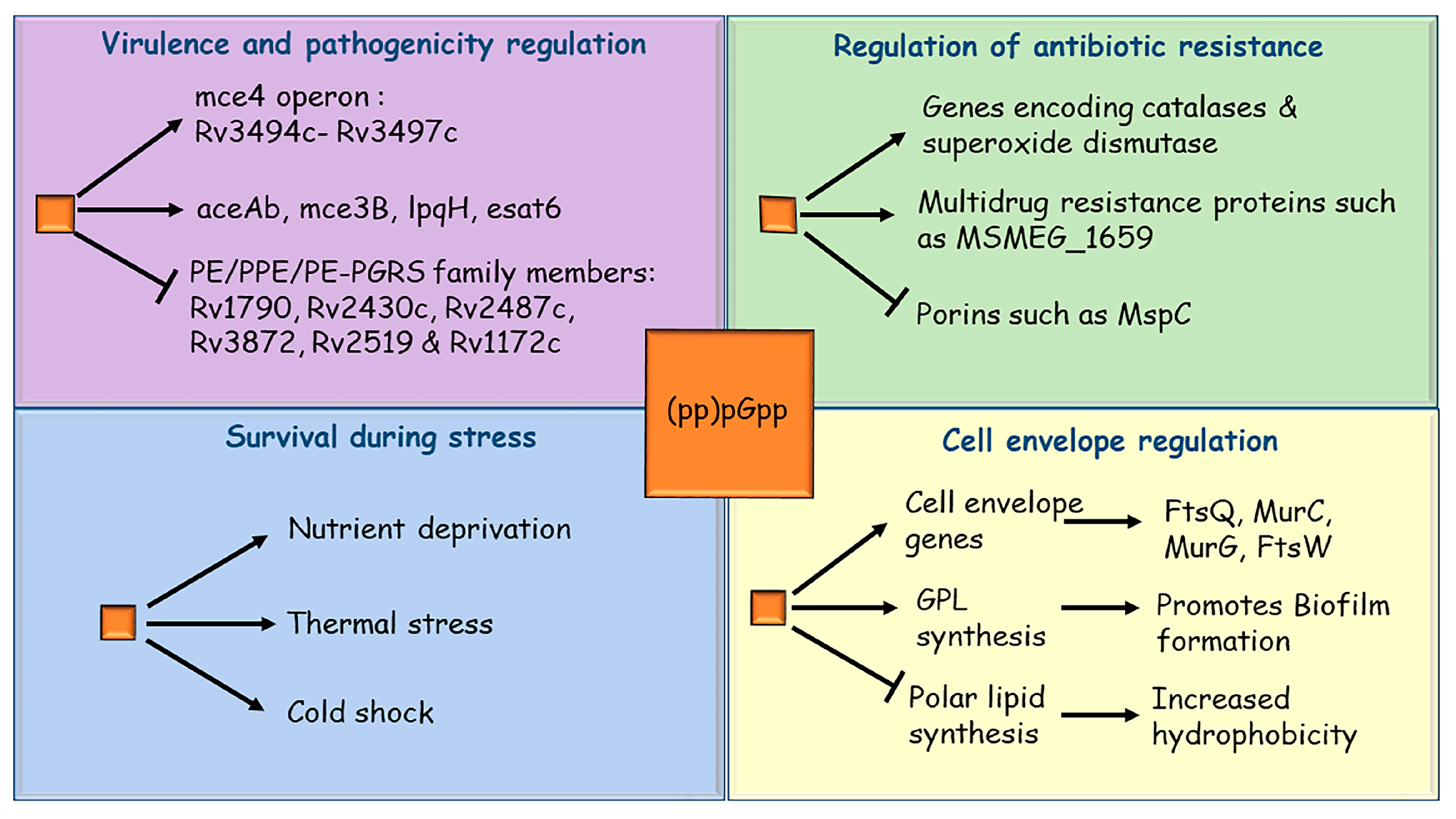
3. Mycobacterial Stringent Response and Its Role in Survival during Stress
4. Stringent Response Regulates Mycobacterial Virulence
5. Role of Stringent Response in Mycobacterial Drug Resistance
6. Stringent Response and Biofilm Formation
7. Chemical Inhibition of Stringent Response as a Therapeutic Tool
8. Outlook
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Arora, G.; Misra, R.; Sajid, A. Model Systems for Pulmonary Infectious Diseases: Paradigms of Anthrax and Tuberculosis. Curr. Top. Med. Chem. 2017, 17, 2077–2099. [Google Scholar] [CrossRef]
- Konstantinidis, K.T.; Viver, T.; Conrad, E.R.; Venter, S.N.; Rossello-Mora, R. Solar salterns as model systems to study the units of bacterial diversity that matter for ecosystem functioning. Curr. Opin. Biotechnol. 2021, 73, 151–157. [Google Scholar] [CrossRef]
- Liu, H.; Deutschbauer, A.M. Rapidly moving new bacteria to model-organism status. Curr. Opin. Biotechnol. 2018, 51, 116–122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mehta, P.; Ray, A.; Mazumder, S. TLRs in Mycobacterial Pathogenesis: Black and White or Shades of Gray. Curr. Microbiol. 2021, 78, 2183–2193. [Google Scholar] [CrossRef] [PubMed]
- Bellinzoni, M.; Wehenkel, A.M.; Durán, R.; Alzari, P.M. Novel mechanistic insights into physiological signaling pathways mediated by mycobacterial Ser/Thr protein kinases. Genes Immun. 2019, 20, 383–393. [Google Scholar] [CrossRef]
- Virmani, R.; Sajid, A.; Singhal, A.; Gaur, M.; Joshi, J.; Bothra, A.; Garg, R.; Misra, R.; Singh, V.P.; Molle, V.; et al. The Ser/Thr protein kinase PrkC imprints phenotypic memory in Bacillus anthracis spores by phosphorylating the glycolytic enzyme enolase. J. Biol. Chem. 2019, 294, 8930–8941. [Google Scholar] [CrossRef] [PubMed]
- Arora, G.; Sajid, A.; Singhal, A.; Joshi, J.; Virmani, R.; Gupta, M.; Verma, N.; Maji, A.; Misra, R.; Baronian, G.; et al. Identification of Ser/Thr kinase and Forkhead Associated Domains in Mycobacterium ulcerans: Characterization of Novel Association between Protein Kinase Q and MupFHA. PLoS Negl. Trop. Dis. 2014, 8, e3315. [Google Scholar] [CrossRef] [Green Version]
- Arora, G.; Bothra, A.; Prosser, G.; Arora, K.; Sajid, A. Role of post-translational modifications in the acquisition of drug resistance in Mycobacterium tuberculosis. FEBS J. 2020, 288, 3375–3393. [Google Scholar] [CrossRef] [PubMed]
- Romaní-Pérez, M.; Bullich-Vilarrubias, C.; López-Almela, I.; Liébana-García, R.; Olivares, M.; Sanz, Y. The Microbiota and the Gut–Brain Axis in Controlling Food Intake and Energy Homeostasis. Int. J. Mol. Sci. 2021, 22, 5830. [Google Scholar] [CrossRef]
- Papon, N.; Stock, A.M. What do archaeal and eukaryotic histidine kinases sense? F1000Research 2019, 8, 2145. [Google Scholar] [CrossRef]
- Arora, G.; Sajid, A.; Gupta, M.; Bhaduri, A.; Kumar, P.; Basu-Modak, S.; Singh, Y. Understanding the Role of PknJ in Mycobacterium tuberculosis: Biochemical Characterization and Identification of Novel Substrate Pyruvate Kinase A. PLoS ONE 2010, 5, e10772. [Google Scholar] [CrossRef]
- Sajid, A.; Arora, G.; Gupta, M.; Singhal, A.; Chakraborty, K.; Nandicoori, V.K.; Singh, Y. Interaction of Mycobacterium tuberculosis Elongation Factor Tu with GTP Is Regulated by Phosphorylation. J. Bacteriol. 2011, 193, 5347–5358. [Google Scholar] [CrossRef] [Green Version]
- Hauryliuk, V.; Atkinson, G.; Murakami, K.S.; Tenson, T.; Gerdes, K. Recent functional insights into the role of (p) ppGpp in bacterial physiology. Nat. Rev. Genet. 2015, 13, 298–309. [Google Scholar] [CrossRef] [Green Version]
- Potrykus, K.; Cashel, M. (p)ppGpp: Still Magical? Annu. Rev. Microbiol. 2008, 62, 35–51. [Google Scholar] [CrossRef] [Green Version]
- Srivatsan, A.; Wang, J.D. Control of bacterial transcription, translation and replication by (p)ppGpp. Curr. Opin. Microbiol. 2008, 11, 100–105. [Google Scholar] [CrossRef]
- Dalebroux, Z.D.; Svensson, S.L.; Gaynor, E.C.; Swanson, M.S. ppGpp Conjures Bacterial Virulence. Microbiol. Mol. Biol. Rev. 2010, 74, 171–199. [Google Scholar] [CrossRef] [Green Version]
- Pacios, O.; Blasco, L.; Bleriot, I.; Fernandez-Garcia, L.; Ambroa, A.; López, M.; Bou, G.; Cantón, R.; Garcia-Contreras, R.; Wood, T.K.; et al. (p)ppGpp and Its Role in Bacterial Persistence: New Challenges. Antimicrob. Agents Chemother. 2020, 64, e01283-20. [Google Scholar] [CrossRef]
- Cashel, M.; Gallant, J. Two Compounds implicated in the Function of the RC Gene of Escherichia coli. Nature 1969, 221, 838–841. [Google Scholar] [CrossRef]
- Stent, G.S.; Brenner, S. A Genetic Locus for the Regulation of Ribonucleic Acid Synthesis. Proc. Natl. Acad. Sci. USA 1961, 47, 2005–2014. [Google Scholar] [CrossRef] [Green Version]
- Wagner, R. Regulation of ribosomal RNA synthesis in E. coli: Effects of the global regulator guanosine tetraphosphate (ppGpp). J. Mol. Microbiol. Biotechnol. 2002, 4, 331–340. [Google Scholar]
- Paul, B.J.; Ross, W.; Gaal, T.; Gourse, R.L. rRNA Transcription in Escherichia coli. Annu. Rev. Genet. 2004, 38, 749–770. [Google Scholar] [CrossRef]
- Lamond, A.; Travers, A.A. Stringent control of bacterial transcription. Cell 1985, 41, 6–8. [Google Scholar] [CrossRef]
- Chatterji, D.; Ojha, A.K. Revisiting the stringent response, ppGpp and starvation signaling. Curr. Opin. Microbiol. 2001, 4, 160–165. [Google Scholar] [CrossRef]
- Primm, T.P.; Andersen, S.J.; Mizrahi, V.; Avarbock, D.; Rubin, H.; Barry, C.E. The Stringent Response of Mycobacterium tuberculosis Is Required for Long-Term Survival. J. Bacteriol. 2000, 182, 4797–4802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ojha, A.K.; Mukherjee, T.K.; Chatterji, D. High Intracellular Level of Guanosine Tetraphosphate in Mycobacterium smegmatis Changes the Morphology of the Bacterium. Infect. Immun. 2000, 68, 4084–4091. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, K.; Bittner, A.N.; Wang, J.D. Diversity in (p) ppGpp metabolism and effectors. Curr. Opin. Microbiol. 2015, 24, 72–79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jain, V.; Saleem-Batcha, R.; China, A.; Chatterji, D. Molecular dissection of the mycobacterial stringent response protein Rel. Protein Sci. 2006, 15, 1449–1464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boutte, C.; Crosson, S. Bacterial lifestyle shapes stringent response activation. Trends Microbiol. 2013, 21, 174–180. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Anderson, B.W.; Turdiev, A.; Turdiev, H.; Stevenson, D.M.; Amador-Noguez, D.; Lee, V.T.; Wang, J.D. The nucleotide pGpp acts as a third alarmone in Bacillus, with functions distinct from those of (p) ppGpp. Nat. Commun. 2020, 11, 5388. [Google Scholar] [CrossRef]
- Petchiappan, A.; Naik, S.Y.; Chatterji, D. RelZ-Mediated Stress Response in Mycobacterium smegmatis: pGpp Synthesis and Its Regulation. J. Bacteriol. 2020, 202, e00444-19. [Google Scholar] [CrossRef]
- Irving, S.E.; Choudhury, N.R.; Corrigan, R.M. The stringent response and physiological roles of (pp) pGpp in bacteria. Nat. Rev. Genet. 2020, 19, 256–271. [Google Scholar] [CrossRef]
- Ooga, T.; Ohashi, Y.; Kuramitsu, S.; Koyama, Y.; Tomita, M.; Soga, T.; Masui, R. Degradation of ppGpp by Nudix Pyrophosphatase Modulates the Transition of Growth Phase in the Bacterium Thermus thermophilus. J. Biol. Chem. 2009, 284, 15549–15556. [Google Scholar] [CrossRef] [Green Version]
- Krishnan, S.; Chatterji, D. Pleiotropic Effects of Bacterial Small Alarmone Synthetases: Underscoring the Dual-Domain Small Alarmone Synthetases in Mycobacterium smegmatis. Front. Microbiol. 2020, 11, 594024. [Google Scholar] [CrossRef]
- Gaca, A.O.; Kudrin, P.; Winter, C.C.; Beljantseva, J.; Liu, K.; Anderson, B.; Wang, J.; Rejman, D.; Potrykus, K.; Cashel, M.; et al. From (p)ppGpp to (pp)pGpp: Characterization of Regulatory Effects of pGpp Synthesized by the Small Alarmone Synthetase of Enterococcus faecalis. J. Bacteriol. 2015, 197, 2908–2919. [Google Scholar] [CrossRef] [Green Version]
- Forbes, B.A. Mycobacterial Taxonomy. J. Clin. Microbiol. 2017, 55, 380–383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Furin, J.; Cox, H.; Pai, M. Tuberculosis. Lancet 2019, 393, 1642–1656. [Google Scholar] [CrossRef]
- Gupta, V.K.; Kumar, M.M.; Singh, D.; Bisht, D.; Sharma, S. Drug targets in dormant Mycobacterium tuberculosis: Can the conquest against tuberculosis become a reality? Infect. Dis. 2017, 50, 81–94. [Google Scholar] [CrossRef]
- Pai, M.; Behr, M.A.; Dowdy, D.; Dheda, K.; Divangahi, M.; Boehme, C.C.; Ginsberg, A.; Swaminathan, S.; Spigelman, M.; Getahun, H.; et al. Tuberculosis. Nat. Rev. Dis. Prim. 2016, 2, 16076. [Google Scholar] [CrossRef]
- Lange, C.; Dheda, K.; Chesov, D.; Mandalakas, A.M.; Udwadia, Z.; Horsburgh, C.R. Management of drug-resistant tuberculosis. Lancet 2019, 394, 953–966. [Google Scholar] [CrossRef]
- Lange, C.; Chesov, D.; Heyckendorf, J.; Leung, C.C.; Udwadia, Z.; Dheda, K. Drug-resistant tuberculosis: An update on disease burden, diagnosis and treatment. Respirology 2018, 23, 656–673. [Google Scholar] [CrossRef]
- Gupta, K.R.; Kasetty, S.; Chatterji, D. Novel Functions of (p)ppGpp and Cyclic di-GMP in Mycobacterial Physiology Revealed by Phenotype Microarray Analysis of Wild-Type and Isogenic Strains of Mycobacterium smegmatis. Appl. Environ. Microbiol. 2015, 81, 2571–2578. [Google Scholar] [CrossRef] [Green Version]
- Gupta, K.R.; Baloni, P.; Indi, S.S.; Chatterji, D. Regulation of Growth, Cell Shape, Cell Division, and Gene Expression by Second Messengers (p)ppGpp and Cyclic Di-GMP in Mycobacterium smegmatis. J. Bacteriol. 2016, 198, 1414–1422. [Google Scholar] [CrossRef] [Green Version]
- Atkinson, G.C.; Tenson, T.; Hauryliuk, V. The RelA/SpoT Homolog (RSH) Superfamily: Distribution and Functional Evolution of ppGpp Synthetases and Hydrolases across the Tree of Life. PLoS ONE 2011, 6, e23479. [Google Scholar] [CrossRef] [Green Version]
- Prusa, J.; Zhu, D.; Stallings, C.L. The stringent response and Mycobacterium tuberculosis pathogenesis. Pathog. Dis. 2018, 76, fty054. [Google Scholar] [CrossRef]
- Sajish, M.; Kalayil, S.; Verma, S.K.; Nandicoori, V.K.; Prakash, B. The significance of EXDD and RXKD motif conservation in Rel proteins. J. Biol. Chem. 2009, 14, 9115–9123. [Google Scholar] [CrossRef] [Green Version]
- Avarbock, D.; Salem, J.; Li, L.-S.; Wang, Z.-M.; Rubin, H. Cloning and characterization of a bifunctional RelA/SpoT homologue from Mycobacterium tuberculosis. Gene 1999, 233, 261–269. [Google Scholar] [CrossRef]
- Murdeshwar, M.S.; Chatterji, D. MS_RHII-RSD, a Dual-Function RNase HII-(p) ppGpp Synthetase from Mycobacterium smegmatis. J. Bacteriol. 2012, 194, 4003–4014. [Google Scholar] [CrossRef] [Green Version]
- Lemos, J.A.; Lin, V.K.; Nascimento, M.M.; Abranches, J.; Burne, R.A. Three gene products govern (p)ppGpp production by Streptococcus mutans. Mol. Microbiol. 2007, 65, 1568–1581. [Google Scholar] [CrossRef]
- Das, B.; Pal, R.R.; Bag, S.; Bhadra, R.K. Stringent response in Vibrio cholerae: Genetic analysis of spoT gene function and identification of a novel (p) ppGpp synthetase gene. Mol. Microbiol. 2009, 72, 380–398. [Google Scholar] [CrossRef]
- Abranches, J.; Martinez, A.R.; Kajfasz, J.K.; Chaávez, V.; Garsin, D.A.; Lemos, J.A. The Molecular Alarmone (p)ppGpp Mediates Stress Responses, Vancomycin Tolerance, and Virulence in Enterococcus faecalis. J. Bacteriol. 2009, 191, 2248–2256. [Google Scholar] [CrossRef] [Green Version]
- Weiss, L.A.; Stallings, C.L. Essential Roles for Mycobacterium tuberculosis Rel beyond the Production of (p) ppGpp. J. Bacteriol. 2013, 195, 5629–5638. [Google Scholar] [CrossRef] [Green Version]
- Krishnan, S.; Petchiappan, A.; Singh, A.; Bhatt, A.; Chatterji, D. R-loop induced stress response by second (p)ppGpp synthetase in Mycobacterium smegmatis: Functional and domain interdependence. Mol. Microbiol. 2016, 102, 168–182. [Google Scholar] [CrossRef] [PubMed]
- Laurie, A.D.; Bernardo, L.M.D.; Sze, C.C.; Skärfstad, E.; Szalewska-Palasz, A.; Nyström, T.; Shingler, V. The Role of the Alarmone (p)ppGpp in ςN Competition for Core RNA Polymerase. J. Biol. Chem. 2003, 278, 1494–1503. [Google Scholar] [CrossRef] [Green Version]
- Jishage, M.; Kvint, K.; Shingler, V.; Nyström, T. Regulation of sigma factor competition by the alarmone ppGpp. Genes Dev. 2002, 16, 1260–1270. [Google Scholar] [CrossRef] [Green Version]
- Ramnaresh Gupta, K.; Chatterji, D. Sigma Factor Competition in Escherichia Coli: Kinetic and Thermodynamic Perspectives. In Stress and Environmental Regulation of Gene Expression and Adaptation in Bacteria; de Bruijn, F.J., Ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2016; pp. 196–202. [Google Scholar] [CrossRef]
- Rasouly, A.; Pani, B.; Nudler, E. A Magic Spot in Genome Maintenance. Trends Genet. 2016, 33, 58–67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szalewska-Palasz, A.; Wegrzyn, G.; Wegrzyn, A. Mechanisms of physiological regulation of RNA synthesis in bacteria: New discoveries breaking old schemes. J. Appl. Genet. 2007, 3, 281–294. [Google Scholar] [CrossRef]
- Anderson, B.W.; Fung, D.K.; Wang, J.D. Regulatory Themes and Variations by the Stress-Signaling Nucleotide Alarmones (p) ppGpp in Bacteria. Annu. Rev. Genet. 2021, 55. [Google Scholar] [CrossRef]
- Milon, P.; Tischenko, E.; Tomšic, J.; Caserta, E.; Folkers, G.; La Teana, A.; Rodnina, M.; Pon, C.L.; Boelens, R.; Gualerzi, C.O. The nucleotide-binding site of bacterial translation initiation factor 2 (IF2) as a metabolic sensor. Proc. Natl. Acad. Sci. USA 2006, 103, 13962–13967. [Google Scholar] [CrossRef] [Green Version]
- Stallings, C.L.; Glickman, M.S. Is Mycobacterium tuberculosis stressed out? A critical assessment of the genetic evidence. Microbes Infect. 2010, 12, 1091–1101. [Google Scholar] [CrossRef] [Green Version]
- Misra, R.; Menon, D.; Arora, G.; Virmani, R.; Gaur, M.; Naz, S.; Jaisinghani, N.; Bhaduri, A.; Bothra, A.; Maji, A.; et al. Tuning the Mycobacterium tuberculosis Alternative Sigma Factor SigF through the Multidomain Regulator Rv1364c and Osmosensory Kinase Protein Kinase D. J. Bacteriol. 2019, 201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barry, E.C.; Boshoff, H.I.; Dartois, V.; Dick, T.; Ehrt, S.; Flynn, J.; Schnappinger, D.; Wilkinson, R.; Young, D. The spectrum of latent tuberculosis: Rethinking the biology and intervention strategies. Nat. Rev. Genet. 2009, 7, 845–855. [Google Scholar] [CrossRef]
- Traxler, M.F.; Summers, S.M.; Nguyen, H.-T.; Zacharia, V.M.; Hightower, G.A.; Smith, J.T.; Conway, T. The global, ppGpp-mediated stringent response to amino acid starvation in Escherichia coli. Mol. Microbiol. 2008, 68, 1128–1148. [Google Scholar] [CrossRef] [Green Version]
- Eymann, C.; Homuth, G.; Scharf, C.; Hecker, M. Bacillus subtilis functional genomics: Global characterization of the stringent response by proteome and transcriptome analysis. J. Bacteriol. 2002, 184, 2500–2520. [Google Scholar] [CrossRef] [Green Version]
- Dahl, J.L.; Kraus, C.N.; Boshoff, H.I.M.; Doan, B.; Foley, K.; Avarbock, D.; Kaplan, G.; Mizrahi, V.; Rubin, H.; Barry, C.E. The role of RelMtb-mediated adaptation to stationary phase in long-term persistence of Mycobacterium tuberculosis in mice. Proc. Natl. Acad. Sci. USA 2003, 100, 10026–10031. [Google Scholar] [CrossRef] [Green Version]
- Stallings, C.L.; Stephanou, N.C.; Chu, L.; Hochschild, A.; Nickels, B.E.; Glickman, M.S. CarD Is an Essential Regulator of rRNA Transcription Required for Mycobacterium tuberculosis Persistence. Cell 2009, 138, 146–159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sureka, K.; Ghosh, B.; Dasgupta, A.; Basu, J.; Kundu, M.; Bose, I. Positive Feedback and Noise Activate the Stringent Response Regulator Rel in Mycobacteria. PLoS ONE 2008, 3, e1771. [Google Scholar] [CrossRef]
- Sureka, K.; Dey, S.; Datta, P.; Singh, A.K.; Dasgupta, A.; Rodrigue, S.; Basu, J.; Kundu, M. Polyphosphate kinase is involved in stress-induced mprAB-sigE-rel signalling in mycobacteria. Mol. Microbiol. 2007, 65, 261–276. [Google Scholar] [CrossRef]
- Rifat, D.; Bishai, W.R.; Karakousis, P.C. Phosphate Depletion: A Novel Trigger for Mycobacterium tuberculosis Persistence. J. Infect. Dis. 2009, 200, 1126–1135. [Google Scholar] [CrossRef] [Green Version]
- Singh, R.; Singh, M.; Arora, G.; Kumar, S.; Tiwari, P.; Kidwai, S. Polyphosphate Deficiency in Mycobacterium tuberculosis Is Associated with Enhanced Drug Susceptibility and Impaired Growth in Guinea Pigs. J. Bacteriol. 2013, 195, 2839–2851. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chuang, Y.-M.; Dutta, N.K.; Hung, C.-F.; Wu, T.-C.; Rubin, H.; Karakousis, P.C. Stringent Response Factors PPX1 and PPK2 Play an Important Role in Mycobacterium tuberculosis Metabolism, Biofilm Formation, and Sensitivity to Isoniazid In Vivo. Antimicrob. Agents Chemother. 2016, 60, 6460–6470. [Google Scholar] [CrossRef] [Green Version]
- Chuang, Y.-M.; Belchis, D.A.; Karakousis, P.C. The Polyphosphate Kinase Gene ppk2 is Required for Mycobacterium tuberculosis Inorganic Polyphosphate Regulation and Virulence. mBio 2013, 4, e00039-13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chuang, Y.-M.; Bandyopadhyay, N.; Rifat, D.; Rubin, H.; Bader, J.S.; Karakousis, P.C. Deficiency of the Novel Exopolyphosphatase Rv1026/PPX2 Leads to Metabolic Downshift and Altered Cell Wall Permeability in Mycobacterium tuberculosis. mBio 2015, 6, e02428. [Google Scholar] [CrossRef] [Green Version]
- Avarbock, A.; Avarbock, D.; Teh, J.-S.; Buckstein, M.; Wang, A.Z.-M.; Rubin, H. Functional Regulation of the Opposing (p)ppGpp Synthetase/Hydrolase Activities of RelMtb from Mycobacterium tuberculosis. Biochemistry 2005, 44, 9913–9923. [Google Scholar] [CrossRef]
- Mathew, R.; Ojha, A.K.; Karande, A.A.; Chatterji, D. Deletion of the rel gene in Mycobacterium smegmatis reduces its stationary phase survival without altering the cell-surface associated properties. Curr. Sci. 2004, 86, 149–153. [Google Scholar]
- Dahl, J.L.; Arora, K.; Boshoff, H.I.; Whiteford, D.C.; Pacheco, S.A.; Walsh, O.J.; Lau-Bonilla, D.; Davis, W.B.; Garza, A.G. The relA Homolog of Mycobacterium smegmatis Affects Cell Appearance, Viability, and Gene Expression. J. Bacteriol. 2005, 187, 2439–2447. [Google Scholar] [CrossRef] [Green Version]
- Klinkenberg, L.G.; Lee, J.-H.; Bishai, W.R.; Karakousis, P.C. The Stringent Response Is Required for Full Virulence of Mycobacterium tuberculosis in Guinea Pigs. J. Infect. Dis. 2010, 202, 1397–1404. [Google Scholar] [CrossRef] [Green Version]
- Post, F.A.; Manca, C.; Neyrolles, O.; Ryffel, B.; Young, D.B.; Kaplan, G. Mycobacterium tuberculosis 19-Kilodalton Lipoprotein Inhibits Mycobacterium smegmatis—Induced Cytokine Production by Human Macrophages In Vitro. Infect. Immun. 2001, 69, 1433–1439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cole, S.T.; Brosch, R.; Parkhill, J.; Garnier, T.; Churcher, C.; Harris, D.; Gordon, S.; Eiglmeier, K.; Gas, S.; Barry, E.C.; et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 1998, 393, 537–544. [Google Scholar] [CrossRef] [PubMed]
- Banu, S.; Honoré, N.; Saint-Joanis, B.; Philpott, D.; Prévost, M.-C.; Cole, S.T. Are the PE-PGRS proteins of Mycobacterium tuberculosis variable surface antigens? Mol. Microbiol. 2002, 44, 9–19. [Google Scholar] [CrossRef]
- Ramakrishnan, L.; Federspiel, N.A.; Falkow, S. Granuloma-Specific Expression of Mycobacterium Virulence Proteins from the Glycine-Rich PE-PGRS Family. Science 2000, 288, 1436–1439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Maio, F.; Battah, B.; Palmieri, V.; Petrone, L.; Corrente, F.; Salustri, A.; Palucci, I.; Bellesi, S.; Papi, M.; Rubino, S.; et al. PE_PGRS3 of Mycobacterium tuberculosis is specifically expressed at low phosphate concentration, and its arginine-rich C-terminal domain mediates adhesion and persistence in host tissues when expressed in Mycobacterium smegmatis. Cell. Microbiol. 2018, 20, e12952. [Google Scholar] [CrossRef]
- De Maio, F.; Salustri, A.; Battah, B.; Palucci, I.; Marchionni, F.; Bellesi, S.; Palmieri, V.; Papi, M.; Kramarska, E.; Sanguinetti, M.; et al. PE_PGRS3 ensures provision of the vital phospholipids cardiolipin and phosphatidylinositols by promoting the interaction between M. tuberculosis and host cells. Virulence 2021, 12, 868–884. [Google Scholar] [CrossRef]
- Singh, V.; Chibale, K. Strategies to Combat Multi-Drug Resistance in Tuberculosis. Acc. Chem. Res. 2021, 54, 2361–2376. [Google Scholar] [CrossRef]
- Seki, M.; Choi, H.J.; Kim, K.; Whang, J.; Sung, J.; Mitarai, S. Tuberculosis: A persistent unpleasant neighbour of humans. J. Infect. Public Health 2021, 14, 508–513. [Google Scholar] [CrossRef]
- Allué-Guardia, A.; García, J.I.; Torrelles, J.B. Evolution of Drug-Resistant Mycobacterium tuberculosis Strains and Their Adaptation to the Human Lung Environment. Front. Microbiol. 2021, 12. [Google Scholar] [CrossRef] [PubMed]
- Vinella, D.; D’Ari, R.; Jaffé, A.; Bouloc, P. Penicillin binding protein 2 is dispensable in Escherichia coli when ppGpp synthesis is induced. EMBO J. 1992, 11, 1493–1501. [Google Scholar] [CrossRef] [PubMed]
- Rodionov, D.G.; Ishiguro, E.E. Direct correlation between overproduction of guanosine 3′,5′-bispyrophosphate (ppGpp) and penicillin tolerance in Escherichia coli. J. Bacteriol. 1995, 177, 4224–4229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joseleau-Petit, D.; Thévenet, D.; D’Arl, R. ppGpp concentration, growth without PBP2 activity, and growth-rate control in Escherichia coli. Mol. Microbiol. 1994, 13, 911–917. [Google Scholar] [CrossRef]
- Greenway, D.L.A.; England, R.R. The intrinsic resistance of Escherichia coli to various antimicrobial agents requires ppGpp and sigmas. Lett. Appl. Microbiol. 1999, 29, 323–326. [Google Scholar] [CrossRef]
- Nguyen, D.; Joshi-Datar, A.; Lepine, F.; Bauerle, E.; Olakanmi, O.; Beer, K.; McKay, G.; Siehnel, R.; Schafhauser, J.; Wang, Y.; et al. Active Starvation Responses Mediate Antibiotic Tolerance in Biofilms and Nutrient-Limited Bacteria. Science 2011, 334, 982–986. [Google Scholar] [CrossRef] [Green Version]
- Dutta, N.K.; Klinkenberg, L.G.; Vazquez, M.-J.; Segura-Carro, D.; Colmenarejo, G.; Ramon, F.; Rodriguez-Miquel, B.; Mata-Cantero, L.; Francisco, E.P.-D.; Chuang, Y.-M.; et al. Inhibiting the stringent response blocks Mycobacterium tuberculosis entry into quiescence and reduces persistence. Sci. Adv. 2019, 5, eaav2104. [Google Scholar] [CrossRef] [Green Version]
- Baloni, P.; Padiadpu, J.; Singh, A.; Gupta, K.R.; Chandra, N. Identifying feasible metabolic routes in Mycobacterium smegmatis and possible alterations under diverse nutrient conditions. BMC Microbiol. 2014, 14, 276. [Google Scholar] [CrossRef] [Green Version]
- Bhaskar, A.; De Piano, C.; Gelman, E.; McKinney, J.D.; Dhar, N. Elucidating the role of (p) ppGpp in mycobacterial persistence against antibiotics. IUBMB Life 2018, 70, 836–844. [Google Scholar] [CrossRef] [Green Version]
- Costerton, J.W.; Stewart, P.S.; Greenberg, E.P. Bacterial Biofilms: A Common Cause of Persistent Infections. Science 1999, 284, 1318–1322. [Google Scholar] [CrossRef] [Green Version]
- Arora, G.; Sajid, A.; Virmani, R.; Singhal, A.; Kumar, C.M.S.; Dhasmana, N.; Khanna, T.; Maji, A.; Misra, R.; Molle, V.; et al. Ser/Thr protein kinase PrkC-mediated regulation of GroEL is critical for biofilm formation in Bacillus anthracis. NPJ Biofilms Microbiomes 2017, 3, 7. [Google Scholar] [CrossRef]
- Siddam, A.D.; Zaslow, S.J.; Wang, Y.; Phillips, K.S.; Silverman, M.D.; Regan, P.M.; Amarasinghe, J.J. Characterization of Biofilm Formation by Mycobacterium chimaera on Medical Device Materials. Front. Microbiol. 2021, 11, 199. [Google Scholar] [CrossRef]
- Vega-Dominguez, P.; Peterson, E.; Pan, M.; Di Maio, A.; Singh, S.; Umapathy, S.; Saini, D.K.; Baliga, N.; Bhatt, A. Biofilms of the non-tuberculous Mycobacterium chelonae form an extracellular matrix and display distinct expression patterns. Cell Surf. 2020, 6, 100043. [Google Scholar] [CrossRef] [PubMed]
- Katoch, P.; Gupta, K.; Yennamalli, R.M.; Vashistt, J.; Bisht, G.S.; Shrivastava, R. Random insertion transposon mutagenesis of Mycobacterium fortuitum identified mutant defective in biofilm formation. Biochem. Biophys. Res. Commun. 2019, 521, 991–996. [Google Scholar] [CrossRef]
- Falkinham, J.O. Mycobacterium avium complex: Adherence as a way of life. AIMS Microbiol. 2018, 4, 428–438. [Google Scholar] [CrossRef] [PubMed]
- Esteban, J.; García-Coca, M. Mycobacterium Biofilms. Front. Microbiol. 2018, 8, 2651. [Google Scholar] [CrossRef] [Green Version]
- Dokic, A.; Peterson, E.; Arrieta-Ortiz, M.L.; Pan, M.; Di Maio, A.; Baliga, N.; Bhatt, A. Mycobacterium abscessus biofilms produce an extracellular matrix and have a distinct mycolic acid profile. Cell Surf. 2021, 7, 100051. [Google Scholar] [CrossRef]
- Sharma, S.K.; Upadhyay, V. Epidemiology, diagnosis & treatment of non-tuberculous mycobacterial diseases. Indian J. Med. Res. 2020, 152, 185–226. [Google Scholar] [CrossRef]
- Ratnatunga, C.; Lutzky, V.P.; Kupz, A.; Doolan, D.L.; Reid, D.W.; Field, M.; Bell, S.; Thomson, R.M.; Miles, J.J. The Rise of Non-Tuberculosis Mycobacterial Lung Disease. Front. Immunol. 2020, 11, 303. [Google Scholar] [CrossRef] [Green Version]
- Faria, S.; Joao, I.; Jordao, L. General Overview on Nontuberculous Mycobacteria, Biofilms, and Human Infection. J. Pathog. 2015, 2015, 809014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sousa, S.; Bandeira, M.; Carvalho, P.A.; Duarte, A.; Jordao, L. Nontuberculous mycobacteria pathogenesis and biofilm assembly. Int. J. Mycobacteriol. 2015, 4, 36–43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Victoria, L.; Gupta, A.; Gómez, J.L.; Robledo, J. Mycobacterium abscessus complex: A Review of Recent Developments in an Emerging Pathogen. Front. Cell. Infect. Microbiol. 2021, 11, 338. [Google Scholar] [CrossRef] [PubMed]
- Degiacomi, G.; Sammartino, J.C.; Chiarelli, L.R.; Riabova, O.; Makarov, V.; Pasca, M.R. Mycobacterium abscessus, an Emerging and Worrisome Pathogen among Cystic Fibrosis Patients. Int. J. Mol. Sci. 2019, 20, 5868. [Google Scholar] [CrossRef] [Green Version]
- Ojha, A.; Hatfull, G.F. The role of iron in Mycobacterium smegmatis biofilm formation: The exochelin siderophore is essential in limiting iron conditions for biofilm formation but not for planktonic growth. Mol. Microbiol. 2007, 66, 468–483. [Google Scholar] [CrossRef] [Green Version]
- Ojha, A.; Anand, M.; Bhatt, A.; Kremer, L.; Jacobs, W.; Hatfull, G.F. GroEL1: A Dedicated Chaperone Involved in Mycolic Acid Biosynthesis during Biofilm Formation in Mycobacteria. Cell 2005, 123, 861–873. [Google Scholar] [CrossRef] [Green Version]
- Chakraborty, P.; Bajeli, S.; Kaushal, D.; Radotra, B.D.; Kumar, A. Biofilm formation in the lung contributes to virulence and drug tolerance of Mycobacterium tuberculosis. Nat. Commun. 2021, 12, 1606. [Google Scholar] [CrossRef]
- Chakraborty, P.; Kumar, A. The extracellular matrix of mycobacterial biofilms: Could we shorten the treatment of mycobacterial infections? Microb. Cell 2019, 6, 105–122. [Google Scholar] [CrossRef]
- Basaraba, R.J.; Ojha, A.K. Mycobacterial Biofilms: Revisiting Tuberculosis Bacilli in Extracellular Necrotizing Lesions. Microbiol. Spectr. 2017, 5, 15. [Google Scholar] [CrossRef]
- Taylor, C.M.; Beresford, M.; Epton, H.A.S.; Sigee, D.C.; Shama, G.; Andrew, P.W.; Roberts, I.S. Listeria monocytogenes relA and hpt Mutants Are Impaired in Surface-Attached Growth and Virulence. J. Bacteriol. 2002, 184, 621–628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lemos, J.; Brown, T.A.; Burne, R.A. Effects of RelA on Key Virulence Properties of Planktonic and Biofilm Populations of Streptococcus mutans. Infect. Immun. 2004, 72, 1431–1440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Paz, L.E.C.; Lemos, J.; Wickstrom, C.; Sedgley, C.M. Role of (p) ppGpp in Biofilm Formation by Enterococcus faecalis. Appl. Environ. Microbiol. 2012, 78, 1627–1630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, H.; Cooper, J.N.; Mishra, A.; Raskin, D. Stringent Response Regulation of Biofilm Formation in Vibrio cholerae. J. Bacteriol. 2012, 194, 2962–2972. [Google Scholar] [CrossRef] [Green Version]
- Schorey, J.S.; Sweet, L. The mycobacterial glycopeptidolipids: Structure, function, and their role in pathogenesis. Glycobiology 2008, 18, 832–841. [Google Scholar] [CrossRef] [Green Version]
- Arora, K.; Whiteford, D.C.; Lau-Bonilla, D.; Davitt, C.M.; Dahl, J.L. Inactivation of lsr2 results in a hypermotile phenotype in Mycobacterium smegmatis. J. Bacteriol. 2008, 12, 4291–4300. [Google Scholar] [CrossRef] [Green Version]
- Pereira, A.C.; Ramos, B.; Reis, A.C.; Cunha, M.V. Non-Tuberculous Mycobacteria: Molecular and Physiological Bases of Virulence and Adaptation to Ecological Niches. Microorganisms 2020, 8, 1380. [Google Scholar] [CrossRef]
- Falkinham, I.J. Surrounded by mycobacteria: Nontuberculous mycobacteria in the human environment. J. Appl. Microbiol. 2009, 107, 356–367. [Google Scholar] [CrossRef]
- Wexselblatt, E.; Oppenheimer-Shaanan, Y.; Kaspy, I.; London, N.; Schueler-Furman, O.; Yavin, E.; Glaser, G.; Katzhendler, J.; Ben-Yehuda, S. Relacin, a Novel Antibacterial Agent Targeting the Stringent Response. PLoS Pathog. 2012, 8, e1002925. [Google Scholar] [CrossRef] [Green Version]
- De La Fuente-Núñez, C.; Reffuveille, F.; Haney, E.F.; Straus, S.; Hancock, R. Broad-Spectrum Anti-biofilm Peptide That Targets a Cellular Stress Response. PLoS Pathog. 2014, 10, e1004152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tkachenko, A.G.; Kashevarova, N.M.; Sidorov, R.Y.; Nesterova, L.Y.; Akhova, A.V.; Tsyganov, I.V.; Vaganov, V.Y.; Shipilovskikh, S.A.; Rubtsov, A.E.; Malkov, A.V. A synthetic diterpene analogue inhibits mycobacterial persistence and biofilm formation by targeting (p) ppGpp synthetases. Cell Chem. Biol. 2021, 28, 1420–1432.e9. [Google Scholar] [CrossRef]
- Syal, K.; Flentie, K.; Bhardwaj, N.; Maiti, K.; Jayaraman, N.; Stallings, C.L.; Chatterji, D. Synthetic (p)ppGpp Analogue Is an Inhibitor of Stringent Response in Mycobacteria. Antimicrob. Agents Chemother. 2017, 61, e00443-17. [Google Scholar] [CrossRef] [Green Version]
- Syal, K.; Bhardwaj, N.; Chatterji, D. Vitamin C targets (p)ppGpp synthesis leading to stalling of long-term survival and biofilm formation in Mycobacterium smegmatis. FEMS Microbiol. Lett. 2016, 364, fnw282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, B.; Dai, P.; Ding, D.; Del Rosario, A.; Grant, R.A.; Pentelute, B.L.; Laub, M.T. Affinity-based capture and identification of protein effectors of the growth regulator ppGpp. Nat. Chem. Biol. 2018, 15, 141–150. [Google Scholar] [CrossRef]
- Sun, Z.; Wu, R.; Zhao, B.; Zeinert, R.; Chien, P.; You, M. Live-Cell Imaging of (p) ppGpp with RNA-based Fluorescent Sensors. bioRxiv 2021. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gupta, K.R.; Arora, G.; Mattoo, A.; Sajid, A. Stringent Response in Mycobacteria: From Biology to Therapeutic Potential. Pathogens 2021, 10, 1417. https://doi.org/10.3390/pathogens10111417
Gupta KR, Arora G, Mattoo A, Sajid A. Stringent Response in Mycobacteria: From Biology to Therapeutic Potential. Pathogens. 2021; 10(11):1417. https://doi.org/10.3390/pathogens10111417
Chicago/Turabian StyleGupta, Kuldeepkumar Ramnaresh, Gunjan Arora, Abid Mattoo, and Andaleeb Sajid. 2021. "Stringent Response in Mycobacteria: From Biology to Therapeutic Potential" Pathogens 10, no. 11: 1417. https://doi.org/10.3390/pathogens10111417
APA StyleGupta, K. R., Arora, G., Mattoo, A., & Sajid, A. (2021). Stringent Response in Mycobacteria: From Biology to Therapeutic Potential. Pathogens, 10(11), 1417. https://doi.org/10.3390/pathogens10111417







