Development of a Multiplex Droplet Digital PCR Assay for the Detection of Babesia, Bartonella, and Borrelia Species
Abstract
:1. Introduction
2. Methods
2.1. Sample Reference Types and DNA Extraction
2.2. Mammalian Cell Line In Vitro Infection
2.3. Experimentally Infected Mouse, Rabbit and Hamster Tissue
2.4. Spiked Blood Samples
2.5. Previously Characterized Borrelia spp. DNA Samples
2.6. DNA Extraction
3. Molecular Methods
3.1. Housekeeping Gene DNA
3.2. Primers and Probes for Bartonella, Borrelia, and Babesia DNA Detection
3.3. DNA Amplification
4. Results
4.1. Naive Blood, Tissue and In Vitro Cultivated Samples
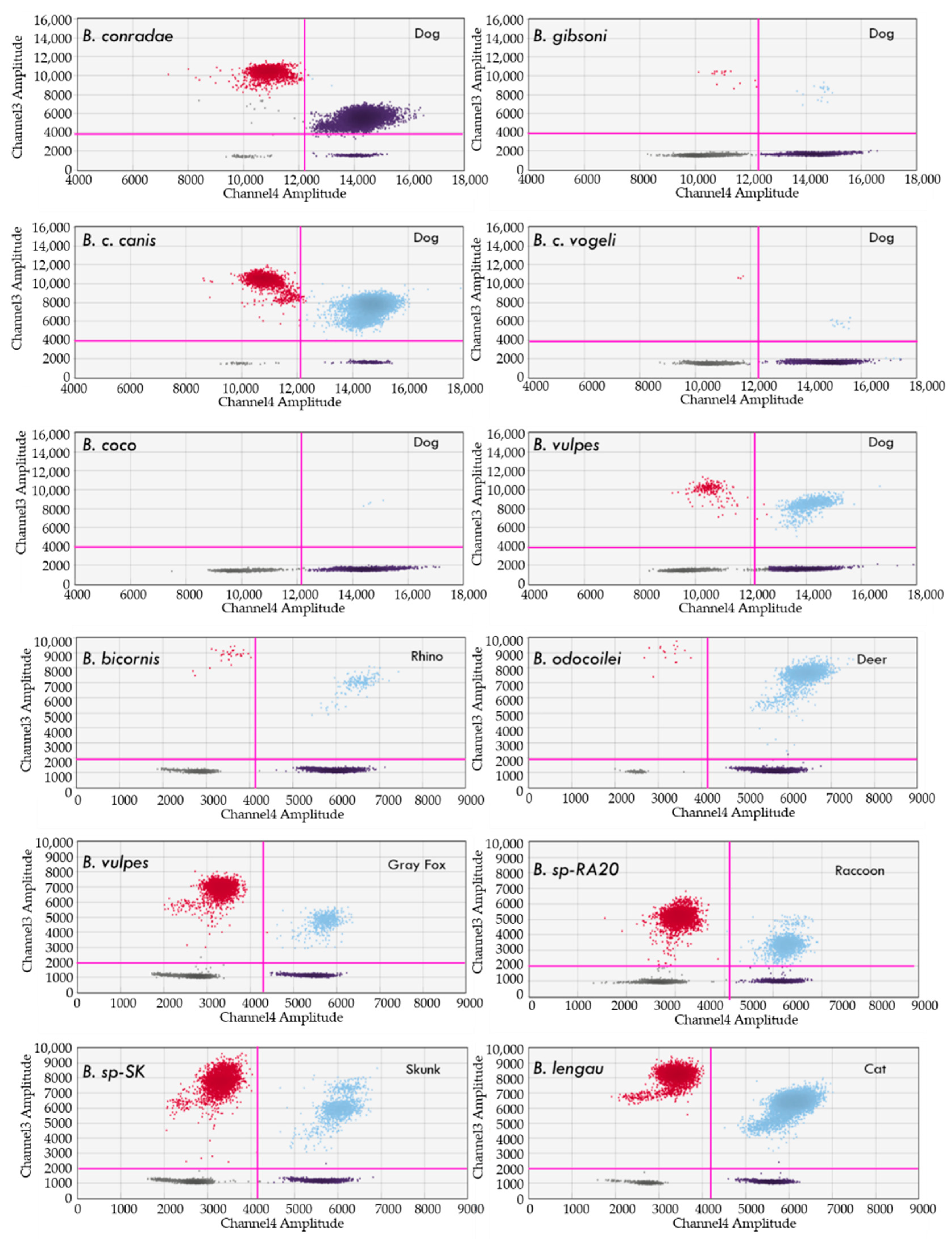
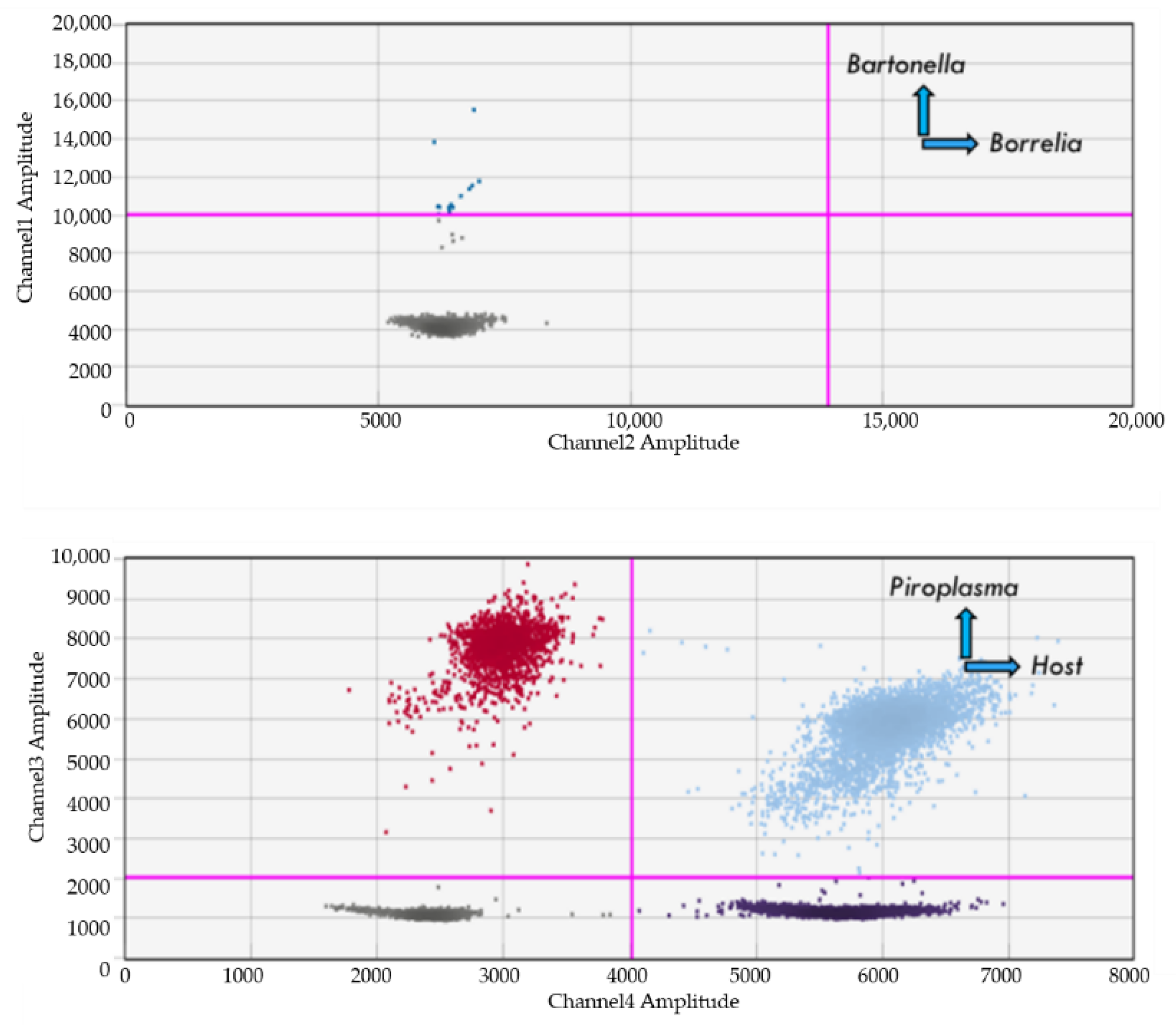
4.2. Detection of Babesia, Theileria and Cytauxzoon Species
Clinical Samples from Naturally and Experimentally Infected Animals
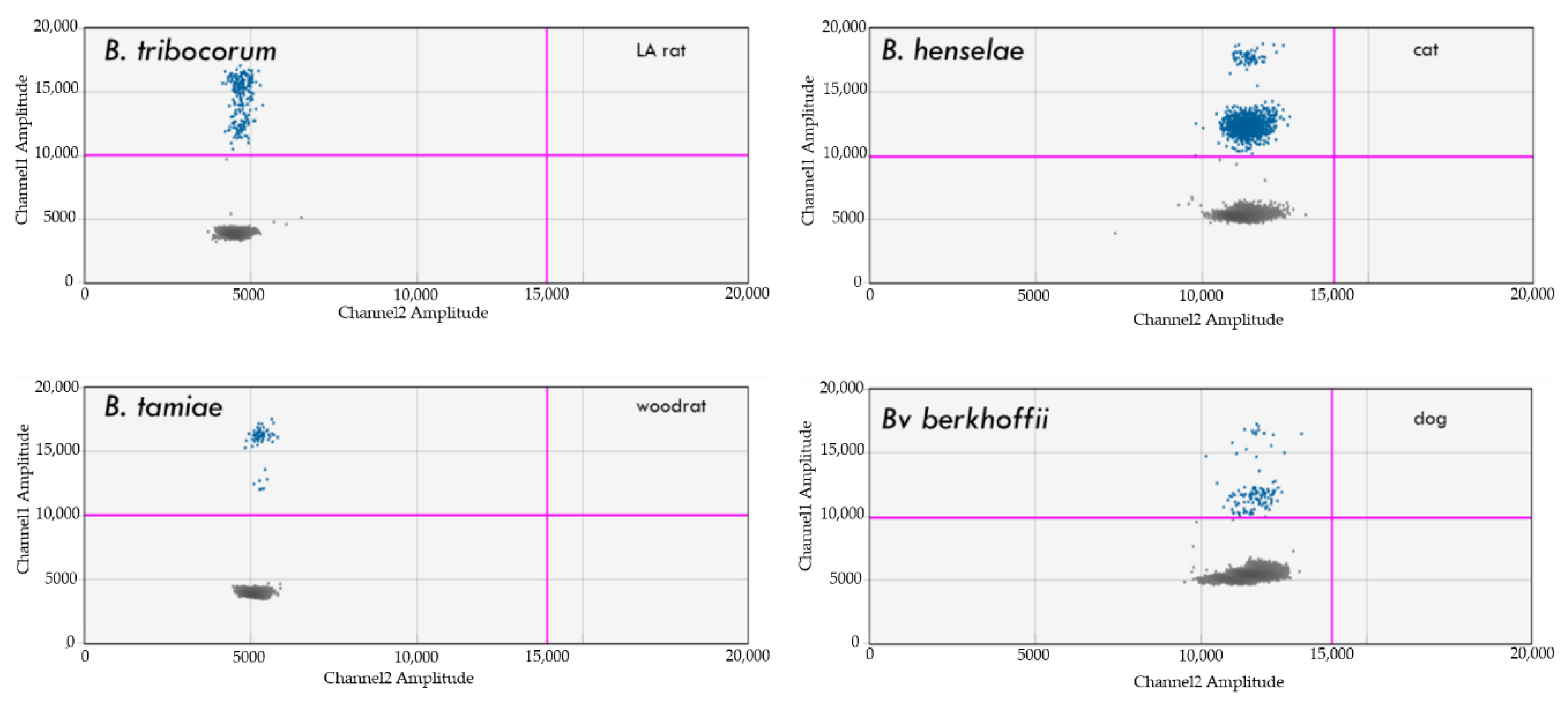
4.3. Detection of Bartonella spp.
4.4. Detection of Borrelia spp.
4.5. Spiked Naïve DNA Blood Samples
4.6. Previously Characterized Borrelia spp. DNA Samples
4.7. Tissues from Experimentally Infected Mice and Cell Lines and from Naturally Infected Vectors
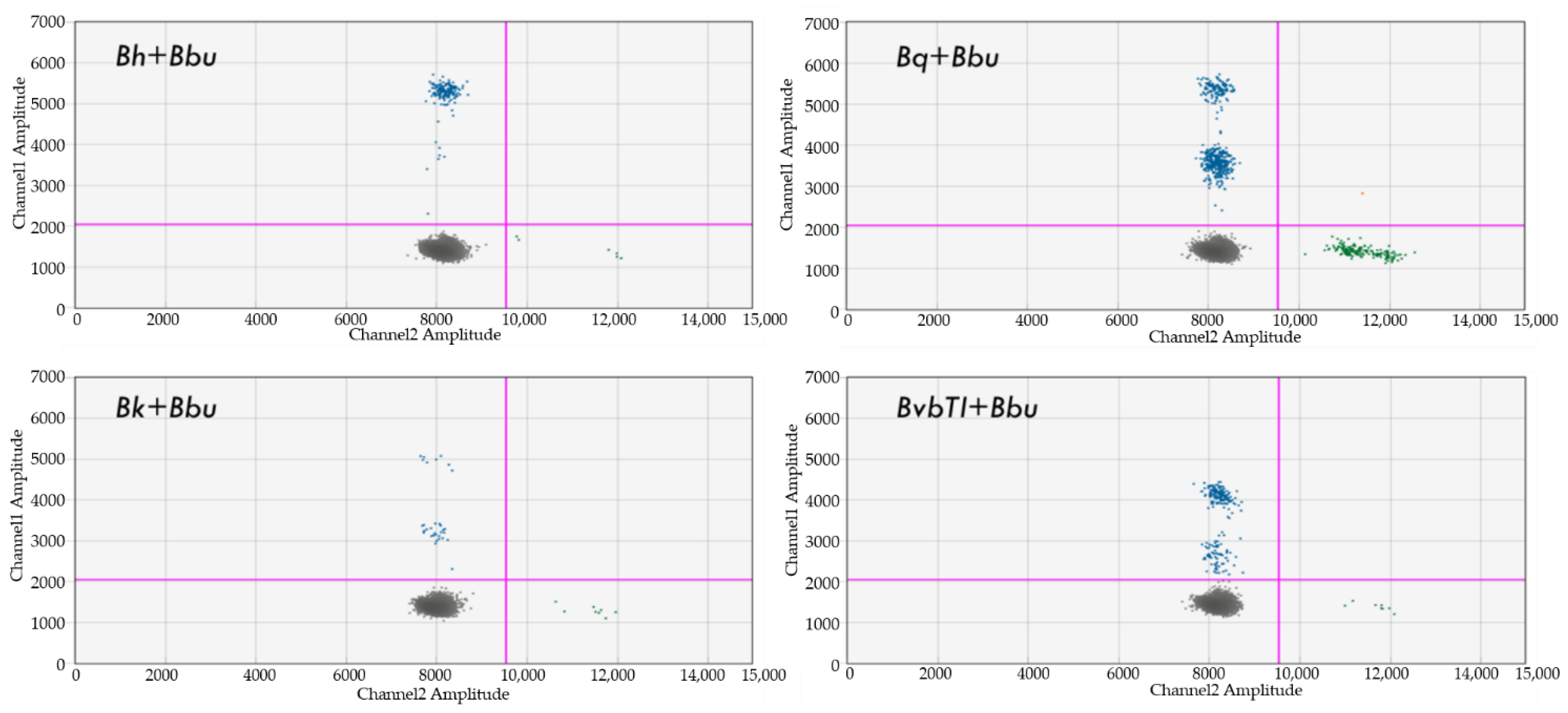
4.8. Dual Detection of Bartonella and Borrelia spp.
4.9. Dual Detection of Bartonella and Babesia
4.10. Babesia, Bartonella, and Borrelia Spiked Naïve Blood Samples
5. Discussion and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Disclosure
References
- Zhang, R.; Chen, B.; Tong, X.; Wang, Y.; Wang, C.; Jin, J.; Tian, P.; Li, W. Diagnostic accuracy of droplet digital PCR for detection of EGFR T790M mutation in circulating tumor DNA. Cancer Manag. Res. 2018, 10, 1209–1218. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.-S.; Choi, H.-J.; Kim, J.-S.; Kim, M.S.; Lee, I.-S.; Byun, B.; Jia, L.; Oh, M.R.; Moon, Y.; Park, S.; et al. Droplet digital PCR-based EGFR mutation detection with an internal quality control index to determine the quality of DNA. Sci. Rep. 2018, 8, 1–10. [Google Scholar] [CrossRef]
- Dong, L.; Wang, S.; Fu, B.; Wang, J. Evaluation of droplet digital PCR and next generation sequencing for characterizing DNA reference material for KRAS mutation detection. Sci. Rep. 2018, 8, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Xu, Y.; Zhong, W.; Zhao, J.; Chen, M.; Zhang, L.; Li, L.; Wang, M. Total DNA input is a crucial determinant of the sensitivity of plasma cell-free DNA EGFR mutation detection using droplet digital PCR. Oncotarget 2016, 8, 5861–5873. [Google Scholar] [CrossRef]
- Tong, Y.; Shen, S.; Jiang, H.; Chen, Z. Application of Digital PCR in Detecting Human Diseases Associated Gene Mutation. Cell. Physiol. Biochem. 2017, 43, 1718–1730. [Google Scholar] [CrossRef] [PubMed]
- Millier, M.J.; Stamp, L.K.; Hessian, P.A. Digital-PCR for gene expression: Impact from inherent tissue RNA degradation. Sci. Rep. 2017, 7, 17235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, R.; Ye, S.B.; He, Y.; Wang, X.; Wu, N.; Xia, Q.Y.; Shen, Q.; Shi, S.S. Detection of epidermal growth factor receptor gene mutations in different types of non-small cell lung cancer by droplet digital PCR and amplification refractory mutation system. Chin. J. Pathol. 2017, 46, 764–768. [Google Scholar]
- Taylor, S.C.; Carbonneau, J.; Shelton, D.N.; Boivin, G. Optimization of Droplet Digital PCR from RNA and DNA extracts with direct comparison to RT-qPCR: Clinical implications for quantification of Oseltamivir-resistant subpopulations. J. Virol. Methods 2015, 224, 58–66. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Bai, R.; Zhao, Z.; Tao, L.; Ma, M.; Ji, Z.; Jian, M.; Ding, Z.; Dai, X.; Bao, F.; et al. Application of droplet digital PCR to detect the pathogens of infectious diseases. Biosci. Rep. 2018, 38. [Google Scholar] [CrossRef] [Green Version]
- Srisutham, S.; Saralamba, N.; Malleret, B.; Rénia, L.; Dondorp, A.M.; Imwong, M. Four human Plasmodium species quantification using droplet digital PCR. PLoS ONE 2017, 12, e0175771. [Google Scholar] [CrossRef]
- Luo, J.; Li, J.; Yang, H.; Yu, J.; Wei, H. Accurate detection of methicillin-resistant Staphylococcus aureus in mixtures utilizing single bacterial duplex droplet digital PCR. J. Clin. Microbiol. 2017, 55, 2946–2955. [Google Scholar] [CrossRef] [Green Version]
- King, J.L.; Smith, A.D.; Mitchell, E.A.; Allen, M.S. Validation of droplet digital PCR for the detection and absolute quantification of Borrelia DNA in Ixodes scapularis ticks. Parasitology 2016, 144, 359–367. [Google Scholar] [CrossRef] [PubMed]
- Wilson, M.; Glaser, K.C.; Adams-Fish, D.; Boley, M.; Mayda, M.; Molestina, R.E. Development of droplet digital PCR for the detection of Babesia microti and Babesia duncani. Exp. Parasitol. 2014, 149, 24–31. [Google Scholar] [CrossRef] [Green Version]
- Gutiérrez-Aguirre, I.; Rački, N.; Dreo, T.; Ravnikar, M. Droplet Digital PCR for Absolute Quantification of Pathogens. Methods Mol. Biol. 2015, 1302, 331–347. [Google Scholar] [CrossRef]
- Bian, X.; Jing, F.; Li, G.; Fan, X.; Jia, C.; Zhou, H.; Jin, Q.; Zhao, J. A microfluidic droplet digital PCR for simultaneous detection of pathogenic Escherichia coli O157 and Listeria monocytogenes. Biosens. Bioelectron. 2015, 74, 770–777. [Google Scholar] [CrossRef]
- Ruelle, J.; Yfantis, V.; Duquenne, A.; Goubau, P. Validation of an ultrasensitive digital droplet PCR assay for HIV-2 plasma RNA quantification. J. Int. AIDS Soc. 2014, 17, 19675. [Google Scholar] [CrossRef] [PubMed]
- Lashnits, E.; Maggi, R.; Jarskog, F.; Bradley, J.; Breischwerdt, E.; Frohlic, F. Schizophrenia and Bartonella spp. Infection: A Pilot Case-Control Study. Vector Borne Zoonotic Dis. 2021, 21, 413–421. [Google Scholar] [CrossRef] [PubMed]
- Maggi, R.G.; Richardson, T.; Breitschwerdt, E.B.; Miller, J.C. Development and validation of a droplet digital PCR assay for the detection and quantification of Bartonella species within human clinical samples. J. Microbiol. Methods 2020, 176, 106022. [Google Scholar] [CrossRef]
- Birkenheuer, A.J.; Harms, C.A.; Neel, J.; Marr, H.S.; Tucker, M.D.; Acton, A.E.; Tuttle, A.D.; Stoskopf, M.K. The identification of a genetically unique piroplasma in North American river otters (Lontra canadensis). Parasitology 2006, 134, 631–635. [Google Scholar] [CrossRef]
- Westmoreland, L.S.H.; Stoskopf, M.K.; Sheppard, E.; Perno, C.F.; Gould, N.P.; Olfenbuttel, C.; Maggi, R.G. Detection and Prevalence of Babesia spp. in American Black Bears (Ursus americanus) from Eastern and Western North Carolina, USA. J. Wildl. Dis. 2019, 55, 678. [Google Scholar] [CrossRef] [PubMed]
- Birkenheuer, A.; Levy, M.G.; Savary, K.C.; Gager, R.B.; Breitschwerdt, E. Babesia gibsoni infections in dogs from North Carolina. J. Am. Anim. Hosp. Assoc. 1999, 35, 125–128. [Google Scholar] [CrossRef] [PubMed]
- Qurollo, B.A.; Archer, N.R.; Schreeg, M.E.; Marr, H.S.; Birkenheuer, A.J.; Haney, K.N.; Thomas, B.S.; Breitschwerdt, E.B. Improved molecular detection of Babesia infections in animals using a novel quantitative real-time PCR diagnostic assay targeting mitochondrial DNA. Parasites Vectors 2017, 10, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Hunfeld, K.-P.; Hildebrandt, A.; Gray, J.S. Babesiosis: Recent insights into an ancient disease. Int. J. Parasitol. 2008, 38, 1219–1237. [Google Scholar] [CrossRef]
- Joseph, J.T.; Purtill, K.; Wong, S.J.; Munoz, J.; Teal, A.; Madison-Antenucci, S.; Horowitz, H.W.; Aguero-Rosenfeld, M.E.; Moore, J.M.; Abramowsky, C.; et al. Vertical transmission of Babesia microti, United States. Emerg. Infect. Dis. 2012, 18, 1318–1321. [Google Scholar] [CrossRef]
- Lobo, C.A.; Cursino-Santos, J.R.; Alhassan, A.; Rodrigues, M. Babesia: An Emerging Infectious Threat in Transfusion Medicine. PLoS Pathog. 2013, 9, e1003387. [Google Scholar] [CrossRef] [Green Version]
- Johnson, S.T.; Cable, R.G.; Leiby, D.A. Lookback investigations of Babesia microti-seropositive blood donors: Seven-year experience in a Babesia-endemic area. Transfusion 2011, 52, 1509–1516. [Google Scholar] [CrossRef] [PubMed]
- Johnson, S.T.; Cable, R.G.; Tonnetti, L.; Spencer, B.; Rios, J.; Leiby, D.A. Seroprevalence of Babesia microti in blood donors from Babesia-endemic areas of the northeastern United States: 2000 through 2007. Transfusion 2009, 49, 2574–2582. [Google Scholar] [CrossRef] [PubMed]
- Johnson, S.T.; van Tassell, E.R.; Tonnetti, L.; Cable, R.G.; Berardi, V.P.; Leiby, D.A. Babesia microti real-time polymerase chain reaction testing of Connecticut blood donors: Potential implications for screening algorithms. Transfusion 2013, 53, 2644–2649. [Google Scholar] [CrossRef]
- Leiby, D.A.; Johnson, S.T.; Won, K.Y.; Nace, E.K.; Slemenda, S.B.; Pieniazek, N.J.; Cable, R.G.; Herwaldt, B.L. A longitudinal study of Babesia microti infection in seropositive blood donors. Transfusion 2014, 54, 2217–2225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moritz, E.D.; Winton, C.S.; Johnson, S.T.; Krysztof, D.E.; Townsend, R.L.; Foster, G.A.; Devine, P.; Molloy, P.; Brissette, E.; Berardi, V.P.; et al. Investigational screening for Babesia microti in a large repository of blood donor samples from nonendemic and endemic areas of the United States. Transfusion 2014, 54, 2226–2236. [Google Scholar] [CrossRef]
- Tonnetti, L.; Thorp, A.M.; Deisting, B.; Bachowski, G.; Johnson, S.T.; Wey, A.R.; Hodges, J.S.; Leiby, D.A.; Mair, D. Babesia microti seroprevalence in Minnesota blood donors. Transfusion 2013, 53, 1698–1705. [Google Scholar] [CrossRef]
- Knapp, K.L.; Rice, N.A. Human Coinfection with Borrelia burgdorferi and Babesia microti in the United States. J. Parasitol. Res. 2015, 2015, 587131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levin, A.E.; Williamson, P.C.; Bloch, E.M.; Clifford, J.; Cyrus, S.; Shaz, B.; Kessler, D.; Gorlin, J.; Erwin, J.L.; Krueger, N.X.; et al. Serologic screening of United States blood donors for Babesia microti using an investigational enzyme immunoassay. Transfusion 2016, 56, 1866–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ward, S.J.; Stramer, S.L.; Szczepiorkowski, Z.M. Assessing the risk of Babesia to the United States blood supply using a risk-based decision-making approach: Report of AABB’s Ad Hoc Babesia Policy Working Group (Original Report). Transfusion 2018, 58, 1916–1923. [Google Scholar] [CrossRef] [Green Version]
- Conrad, P.A.; Kjemtrup, A.M.; Carreno, R.A.; Thomford, J.; Wainwright, K.; Eberhard, M.; Quick, R.; Iii, S.R.T.; Herwaldt, B.L. Description of Babesia duncani n.sp. (Apicomplexa: Babesiidae) from humans and its differentiation from other piroplasms. Int. J. Parasitol. 2006, 36, 779–789. [Google Scholar] [CrossRef] [PubMed]
- Herwaldt, B.L.; Linden, J.V.; Bosserman, E.; Young, C.; Olkowska, D.; Wilson, M. Transfusion-Associated Babesiosis in the United States: A Description of Cases. Ann. Intern. Med. 2011, 155, 509–519. [Google Scholar] [CrossRef] [Green Version]
- Mayne, P. Emerging incidence of Lyme borreliosis, babesiosis, bartonellosis, and granulocytic ehrlichiosis in Australia. Int. J. Gen. Med. 2011, 4, 845–852. [Google Scholar] [CrossRef] [Green Version]
- Colson, P.; Lebrun, L.; Drancourt, M.; Boué, F.; Raoult, D.; Nordmann, P. Multiple recurrent bacillary angiomatosis due to Bartonella quintana in an HIV-infected patient. Eur. J. Clin. Microbiol. Infect. Dis. 1996, 15, 178–180. [Google Scholar] [CrossRef]
- Krause, P.J.; Gewurz, B.E.; Hill, D.; Marty, F.M.; Vannier, E.; Foppa, I.M.; Furman, R.R.; Neuhaus, E.; Skowron, G.; Gupta, S.; et al. Persistent and Relapsing Babesiosis in Immunocompromised Patients. Clin. Infect. Dis. 2008, 46, 370–376. [Google Scholar] [CrossRef] [Green Version]
- Chiang, E.; Haller, N. Babesiosis: An emerging infectious disease that can affect those who travel to the northeastern United States. Travel Med. Infect. Dis. 2011, 9, 238–242. [Google Scholar] [CrossRef]
- Genchi, C. Human babesiosis, an emerging zoonosis. Parassitologia 2007, 49, 29–31. [Google Scholar]
- Kjemtrup, A.; Conrad, P. Human babesiosis: An emerging tick-borne disease. Int. J. Parasitol. 2000, 30, 1323–1337. [Google Scholar] [CrossRef]
- Oz, H.S.; Westlund, K.H. “Human babesiosis”: An emerging transfusion dilemma. Int. J. Hepatol. 2012, 2012, 431761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vannier, E.; Krause, P.J. Babesiosis in China, an emerging threat. Lancet Infect. Dis. 2015, 15, 137–139. [Google Scholar] [CrossRef]
- Yang, Y.; Christie, J.; Köster, L.; Du, A.; Yao, C. Emerging Human Babesiosis with “Ground Zero” in North America. Microorganisms 2021, 9, 440. [Google Scholar] [CrossRef] [PubMed]
- Paddock, C.D.; Lane, R.S.; Staples, J.E.; Labruna, M.B.; Board on Global Health; Health and Medicine Division. Changing paradigms for tick-borne diseases in the Americas. In Global Health Impacts of Vector-Borne Diseases, In Proceedings of the Forum on Microbial Threats; National Academies Press: Washington, DC, USA, 2016. [Google Scholar]
- Lappin, M.R.; Elston, T.; Evans, L.; Glaser, C.; Jarboe, L.; Karczmar, P.; Lund, C.; Ray, M. 2019 AAFP Feline Zoonoses Guidelines. J. Feline Med. Surg. 2019, 21, 1008–1021. [Google Scholar] [CrossRef] [Green Version]
- Reynolds, C.A.; Lappin, M.R. “Candidatus Mycoplasma haemominutum” infections in 21 client-owned cats. J. Am. Anim. Hosp. Assoc. 2007, 43, 249–257. [Google Scholar] [CrossRef]
- Bouloy, R.P.; Lappin, M.R.; Holland, C.H.; Thrall, M.A.; Baker, D.; O’Neil, S. Clinical ehrlichiosis in a cat. J. Am. Vet. Med Assoc. 1994, 204, 1475–1478. [Google Scholar]
- Mullins, K.E.; Krueger, L.; Maina, A.N.; Cummings, R.; Williams, G.; Drusys, A.; Jiang, J.; Richards, A.L.; Dhillon, M. Rickettsial Infections among Cats and Cat Fleas in Riverside County, California. Am. J. Trop. Med. Hyg. 2018, 99, 291–296. [Google Scholar] [CrossRef]
- Ferreira, F.C.; Fonseca, D.M.; Hamilton, G.; Price, D. Metagenomic analysis of human-biting cat fleas in urban northeastern United States of America reveals an emerging zoonotic pathogen. Sci. Rep. 2020, 10, 1–8. [Google Scholar] [CrossRef]
- Horowitz, R.I.; Freeman, P.R. Precision medicine: Retrospective chart review and data analysis of 200 patients on dapsone combination therapy for chronic Lyme disease/post-treatment Lyme disease syndrome: Part 1. Int. J. Gen. Med. 2019, 12, 101–119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maia, C.; Cristóvão, J.M.; Pereira, A.; Parreira, R.; Campino, L. Detection of Rickettsia conorii israelensis DNA in the Blood of a Cat and a Dog from Southern Portugal. Top. Companion Anim. Med. 2019, 36, 12–15. [Google Scholar] [CrossRef] [PubMed]
- Mayne, P. Clinical determinants of Lyme borreliosis, babesiosis, bartonellosis, anaplasmosis, and ehrlichiosis in an Australian cohort. Int. J. Gen. Med. 2014, 8, 15–26. [Google Scholar] [CrossRef] [Green Version]
- Maggi, R.G.; Mozayeni, B.R.; Pultorak, E.L.; Hegarty, B.C.; Bradley, J.M.; Correa, M.; Breitscwerdt, E.B. Bartonella spp. bacteremia and rheumatic symptoms in patients from Lyme disease-endemic region. Emerg. Infect. Dis. 2012, 18, 783–791. [Google Scholar] [CrossRef]
- Tsachev, I.; Baymakova, M.; Zlateva, N.; Kundurzhiev, T.; Solano-Gallego, L. Seroprevalence Rates of Tick-Borne Pathogens in Cats from Southern Bulgaria. Vector-Borne Zoonotic Dis. 2020, 20, 864–867. [Google Scholar] [CrossRef]
- Register, K.B.; Sukumar, N.; Palavecino, E.L.; Rubin, B.K.; Deora, R. Bordetella bronchiseptica in a Paediatric Cystic Fibrosis Patient: Possible Transmission from a Household Cat. Zoonoses Public Health 2012, 59, 246–250. [Google Scholar] [CrossRef] [PubMed]
- Okaro, U.; Addisu, A.; Casanas, B.; Anderson, B. Bartonella Species, an Emerging Cause of Blood-Culture-Negative Endocarditis. Clin. Microbiol. Rev. 2017, 30, 709–746. [Google Scholar] [CrossRef] [Green Version]
- Breitschwerdt, E.B. Bartonellosis, One Health and all creatures great and small. Vet. Dermatol. 2017, 28, 96-e21. [Google Scholar] [CrossRef]
- Shapira, L.; Rasis, M.; Ehrenreich, I.B.; Maor, Y.; Katchman, E.A.; Treves, A.; Velan, A.; Halutz, O.; Graidy-Varon, M.; Leibovitch, C.; et al. Laboratory Diagnosis of 37 Cases of Bartonella Endocarditis Based on Enzyme Immunoassay and Real-Time PCR. J. Clin. Microbiol. 2021, 59. [Google Scholar] [CrossRef]
- Plantinga, N.L.; Vos, R.J.; Georgieva, L.; Roescher, N. Bartonella quintana as a cause for prosthetic valve endocarditis and post-sternotomy mediastinitis. Access Microbiol. 2021, 3, 000217. [Google Scholar] [CrossRef]
- Pecoraro, A.; Herbst, P.; Pienaar, C.; Taljaard, J.; Prozesky, H.; Janson, J.; Doubell, A. Bartonella species as a cause of culture-negative endocarditis in South Africa. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 40, 1873–1879. [Google Scholar] [CrossRef]
- Downey, R.D.; Russo, S.M.; Hauger, S.B.; Murphey, D.K.; Marx, G.; Huynh, T.; Denison, A.; Quirt, R.; Bailey, A.; Fernandez, M. Identification of an Emergent Pathogen, Bartonella vinsonii, Using Next-Generation Sequencing in a Patient with Culture-Negative Endocarditis. J. Pediatr. Infect. Dis. Soc. 2020, 10, 213–216. [Google Scholar] [CrossRef] [PubMed]
- Davis, A.Z.; Jaffe, D.A.; Honadel, T.E.; Lapsley, W.D.; Wilber-Raymond, J.L.; Kasten, R.W.; Chomel, B.B. Prevalence of Bartonella sp. in United States military working dogs with infectious endocarditis: A retrospective case-control study. J. Vet. Cardiol. 2020, 27, 1–9. [Google Scholar] [CrossRef]
- Boodman, C.; Wuerz, T.; Lagacé-Wiens, P. Endocarditis due to Bartonella quintana, the etiological agent of trench fever. Can. Med Assoc. J. 2020, 192, E1723–E1726. [Google Scholar] [CrossRef]
- Shtaya, A.A.; Perek, S.; Kibari, A.; Cohen, S. Bartonella henselae Endocarditis: An Usual Presentation of an Unusual Disease. Eur. J. Case Rep. Intern. Med. 2019, 6, 001038. [Google Scholar]
- Kuncová, K.; Žemličková, H.; Jäger, J.; Ryšková, L.; Plíšková, L.; Bolehovská, R.; Pojar, M. Diagnosis and treatment of Bartonella endocarditis. Epidemiol. Mikrobiol. Imunol. 2019, 68, 104–108. [Google Scholar] [PubMed]
- Pachirat, O.; Prathanee, S.; Watt, G. Echocardiographic Features in Bartonella Endocarditis: A Case Series. Cardiol. Res. 2018, 9, 116–119. [Google Scholar] [CrossRef] [Green Version]
- Shelnutt, L.; Balakrishnan, N.; de Vanna, J.; Batey, K.L.; Breitschwerdt, E. Death of Military Working Dogs Due to Bartonella vinsonii Subspecies berkhoffii Genotype III Endocarditis and Myocarditis. Mil. Med. 2017, 182, e1864–e1869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martín, L.; Vidal, L.; Campins, A.; Salvá, F.; Riera, M.; Carrillo, A.; De Ibarra, J.I.S. Bartonella as a Cause of Blood Culture-Negative Endocarditis. Description of 5 Cases. Rev. Esp. Cardiol. 2009, 62, 694–697. [Google Scholar] [CrossRef]
- Tsuneoka, H.; Yanagihara, M.; Tanimoto, A.; Tokuda, N.; Otsuyama, K.-I.; Nojima, J.; Ichihara, K. The utility of a country-specific Bartonella henselae antigen in an IgM-indirect fluorescent antibody assay for the improved diagnosis of cat scratch disease. Diagn. Microbiol. Infect. Dis. 2017, 87, 22–24. [Google Scholar] [CrossRef]
- Chi, S.L.; Stinnett, S.; Eggenberger, E.; Foroozan, R.; Golnik, K.; Lee, M.S.; Bhatti, M.T. Clinical Characteristics in 53 Patients with Cat Scratch Optic Neuropathy. Ophthalmology 2012, 119, 183–187. [Google Scholar] [CrossRef] [PubMed]
- Weinspach, S.; Tenenbaum, T.; Schönberger, S.; Schaper, J.; Engers, R.; Rueggeberg, J.; MacKenzie, C.R.; Wolf, A.; Mayatepek, E.; Schroten, H. Cat Scratch Disease—Heterogeneous in Clinical Presentation: Five Unusual Cases of an Infection Caused by Bartonella henselae. Klin. Pediatr. 2009, 222, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Scott, C.; Azwa, A.; Cohen, C.; McIntyre, M.; Desmond, N. Cat scratch disease: A diagnostic conundrum. Int. J. STD AIDS 2009, 20, 585–586. [Google Scholar] [CrossRef]
- Raoult, D. From Cat Scratch Disease to Bartonella henselae Infection. Clin. Infect. Dis. 2007, 45, 1541–1542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berkowitz, S.T.; Gannon, K.M.; Carberry, C.A.; Cortes, Y. Resolution of spontaneous hemoabdomen secondary to peliosis hepatis following surgery and azithromycin treatment in a Bartonella species infected dog. J. Vet. Emerg. Crit. Care 2016, 26, 851–857. [Google Scholar] [CrossRef]
- Buchmann, A.U.; Kempf, V.A.J.; Kershaw, O.; Gruber, A.D. Peliosis Hepatis in Cats Is Not Associated with Bartonella henselae Infections. Vet. Pathol. 2010, 47, 163–166. [Google Scholar] [CrossRef]
- Kitchell, B.E.; Fan, T.M.; Kordick, D.; Breitschwerdt, E.B.; Wollenberg, G.; Lichtensteiger, C.A. Peliosis hepatis in a dog infected with Bartonella henselae. J. Am. Vet. Med. Assoc. 2000, 216, 519–523. [Google Scholar] [CrossRef]
- Koehler, J.E.; Sanchez, M.A.; Garrido, C.S.; Whitfeld, M.; Chen, F.M.; Berger, T.G.; Rodriguez-Barradas, M.C.; LeBoit, P.E.; Tappero, J.W. Molecular Epidemiology of Bartonella Infections in Patients with Bacillary Angiomatosis–Peliosis. N. Engl. J. Med. 1997, 337, 1876–1883. [Google Scholar] [CrossRef]
- Barber, R.M.; Li, Q.; Diniz, P.P.V.P.; Forter, B.F.; Breitschwerdt, E.B.; Claiborne, M.K.; Birkenheuer, A.J.; Levine, J.M.; Levine, G.J.; Chandler, K.; et al. Evaluation of brain tissue or cerebrospinal fluid with broadly reactive polymerase chain reaction for Ehrlichia, Anaplasma, spotted fever group Rickettsia, Bartonella, and Borrelia species in canine neurological diseases (109 cases). J. Vet. Intern. Med. 2010, 24, 372–378. [Google Scholar] [CrossRef]
- Breitschwerdt, E.B.; Maggi, R.G.; Lantos, P.M.; Woods, C.W.; Hegarty, B.C.; Bradley, J.M. Bartonella vinsonii subsp. berkhoffii and Bartonella henselae bacteremia in a father and daughter with neurological disease. Parasites Vectors 2010, 3, 29. [Google Scholar] [CrossRef] [Green Version]
- Breitschwerdt, E.B.; Maggi, R.G.; Nicholson, W.L.; Cherry, N.A.; Woods, C.W. Bartonella sp. Bacteremia in Patients with Neurological and Neurocognitive Dysfunction. J. Clin. Microbiol. 2008, 46, 2856–2861. [Google Scholar] [CrossRef] [Green Version]
- Canneti, B.; Cabo-López, I.; Puy-Núñez, A.; García, J.C.G.; Cores, F.J.; Trigo, M.; Suárez-Gil, A.P.; Rodriguez-Regal, A. Neurological presentations of Bartonella henselae infection. Neurol. Sci. 2018, 40, 261–268. [Google Scholar] [CrossRef]
- Castel, A.; Olby, N.J.; Breitschwerdt, E.B.; Thomas, B.; Maggi, R.G.; Shelton, G.D. Co-infection with Bartonella henselae and Sarcocystis sp. in a 6-year-old male neutered domestic longhair cat with progressive multifocal neurological signs. Vet. Q. 2019, 39, 168–173. [Google Scholar] [CrossRef] [Green Version]
- Kaufman, D.L.; Kogelnik, A.M.; Mozayeni, R.B.; Cherry, N.A.; Breitschwerdt, E.B. Neurological and immunological dysfunction in two patients with Bartonella henselae bacteremia. Clin. Case Rep. 2017, 5, 931–935. [Google Scholar] [CrossRef]
- Mascarelli, P.E.; Maggi, R.G.; Hopkins, S.; Mozayeni, B.R.; Trull, C.L.; Bradley, J.M.; Hegarty, B.C.; Breitschwerdt, E.B. Bartonella henselae infection in a family experiencing neurological and neurocognitive abnormalities after woodlouse hunter spider bites. Parasites Vectors 2013, 6, 98. [Google Scholar] [CrossRef] [Green Version]
- Andersson, S.G.; Kempf, V.A. Host cell modulation by human, animal and plant pathogens. Int. J. Med Microbiol. 2004, 293, 463–470. [Google Scholar] [CrossRef] [PubMed]
- Franz, B.; Kempf, V.A.J. Adhesion and host cell modulation: Critical pathogenicity determinants of Bartonella henselae. Parasites Vectors 2011, 4, 54. [Google Scholar] [CrossRef] [Green Version]
- Arvand, M.; Ignatius, R.; Regnath, T.; Hahn, H.; Mielke, M.E.A. Bartonella henselae—Specific Cell-Mediated Immune Responses Display a Predominantly Th1 Phenotype in Experimentally Infected C57BL/6 Mice. Infect. Immun. 2001, 69, 6427–6433. [Google Scholar] [CrossRef] [Green Version]
- Il’Ina, T.S.; Bashkirov, V.N. Interaction of bacteria of the genus Bartonella with the host: Inhibition of apoptosis, induction of proliferation, and formation of tumors. Mol. Genet. Microbiol. Virol. 2008, 3, 3–11. [Google Scholar]
- Pulliainen, A.T.; Dehio, C. Bartonella henselae: Subversion of vascular endothelial cell functions by translocated bacterial effector proteins. Int. J. Biochem. Cell Biol. 2009, 41, 507–510. [Google Scholar] [CrossRef]
- Pulliainen, A.T.; Dehio, C. Persistence of Bartonella spp. stealth pathogens: From subclinical infections to vasoproliferative tumor formation. FEMS Microbiol. Rev. 2012, 36, 563–599. [Google Scholar] [CrossRef] [Green Version]
- Resto-Ruiz, S.; Burgess, A.; Anderson, B.E. The Role of the Host Immune Response in Pathogenesis of Bartonella henselae. DNA Cell Biol. 2003, 22, 431–440. [Google Scholar] [CrossRef]
- Chomel, B.B.; Boulouis, H.-J.; Breitschwerdt, E.B.; Kasten, R.W.; Vayssier-Taussat, M.; Birtles, R.J.; Koehler, J.E.; Dehio, C. Ecological fitness and strategies of adaptation of Bartonella species to their hosts and vectors. Vet. Res. 2009, 40, 29. [Google Scholar] [CrossRef] [Green Version]
- Maggi, R.G.; Duncan, A.W.; Breitschwerdt, E.B. Novel Chemically Modified Liquid Medium That Will Support the Growth of Seven Bartonella Species. J. Clin. Microbiol. 2005, 43, 2651–2655. [Google Scholar] [CrossRef] [Green Version]
- Hong, J.; Li, Y.; Hua, X.; Bai, Y.; Wang, C.; Zhu, C.; Du, Y.; Yang, Z.; Yuan, C. Lymphatic Circulation Disseminates Bartonella Infection into Bloodstream. J. Infect. Dis. 2017, 215, 303–311. [Google Scholar]
- Wolf, L.; Maggy, R.G.; Breitschwerdt, E.B.; Cherry, N.A. In Pursuit of a Stealth Pathogen: Laboratory Diagnosis of Bartonellosis. Clin. Microbiol. Newslett. 2014, 36, 33–38. [Google Scholar] [CrossRef]
- Harms, A.; Dehio, C. Intruders below the Radar: Molecular Pathogenesis of Bartonella spp. Clin. Microbiol. Rev. 2012, 25, 42–78. [Google Scholar] [CrossRef] [Green Version]
- Kaneko, K.; Osada, T.; Morse, M.A.; Gwin, W.R.; Ginzel, J.D.; Snyder, J.C.; Yang, X.-Y.; Liu, C.-X.; Diniz, M.A.; Bodoor, K.; et al. Heat shock protein 90-targeted photodynamic therapy enables treatment of subcutaneous and visceral tumors. Commun. Biol. 2020, 3, 226. [Google Scholar] [CrossRef]
- Billeter, S.A.; Breitschwerdt, E.B.; Levy, M.G. Invasion of canine erythrocytes by Bartonella vinsonii subsp. berkhoffii. Vet. Microbiol. 2012, 156, 213–216. [Google Scholar] [CrossRef]
- Sell, M.G.; Alcorta, D.A.; Padilla, A.E.; Nollner, D.W.; Hasenkampf, N.R.; Lambert, H.S.; Embers, M.E.; Spector, N.L. Visualizing Borrelia burgdorferi Infection Using a Small-Molecule Imaging Probe. J. Clin. Microbiol. 2021, 59. [Google Scholar] [CrossRef]
- Lager, M.; Faller, M.; Wilhelmsson, P.; Kjelland, V.; Andreassen, A.; Dargis, R.; Quarsten, H.; Dessau, R.; Fingerle, V.; Margos, G.; et al. Molecular detection of Borrelia burgdorferi sensu lato—An analytical comparison of real-time PCR protocols from five different Scandinavian laboratories. PLoS ONE 2017, 12, e0185434. [Google Scholar] [CrossRef] [Green Version]
- Portillo, A.; Maggi, R.; Oteo, J.A.; Bradley, J.; García-Álvarez, L.; San-Martín, M.; Roura, X.; Breitschwerdt, E. Bartonella spp. Prevalence (Serology, Culture, and PCR) in Sanitary Workers in La Rioja Spain. Pathogens 2020, 9, 189. [Google Scholar] [CrossRef] [Green Version]
- Lashnits, E.; Neupane, P.; Bradley, J.M.; Richardson, T.; Thomas, R.; Linder, K.E.; Breen, M.; Maggi, R.G.; Breitschwerdt, E.B. Molecular prevalence of Bartonella, Babesia, and hemotropic Mycoplasma species in dogs with hemangiosarcoma from across the United States. PLoS ONE 2020, 15, e0227234. [Google Scholar] [CrossRef]
- Breitschwerdt, E.B.; Maggi, R.G.; Quach, C.; Bradley, J.M. Bartonella spp. Bloodstream Infection in a Canadian Family. VectorBorne Zoonotic Dis. 2019, 19, 234–241. [Google Scholar] [CrossRef]
- Breitschwerdt, E.B.; Maggi, R.G. Bartonella quintana and Bartonella vinsonii subsp. vinsonii bloodstream co-infection in a girl from North Carolina, USA. Med. Microbiol. Immunol. 2018, 208, 101–107. [Google Scholar] [CrossRef]
- Carrasco, S.E.; Chomel, B.B.; Gill, V.A.; Kasten, R.W.; Maggi, R.; Breitschwerdt, E.; Byrne, B.A.; Burek-Huntington, K.A.; Miller, M.A.; Goldstein, T.; et al. Novel Bartonella infection in northern and southern sea otters (Enhydra lutris kenyoni and Enhydra lutris nereis). Vet. Microbiol. 2014, 170, 325–334. [Google Scholar] [CrossRef]
- Maggi, R.G.; Mascarelli, P.E.; Balakrishnan, N.; Rohde, C.M.; Kelly, C.M.; Ramaiah, L.; Leach, M.W.; Breitschwerdt, E.B. “Candidatus Mycoplasma haemomacaque” and Bartonella quintana Bacteremia in Cynomolgus Monkeys. J. Clin. Microbiol. 2013, 51, 1408–1411. [Google Scholar] [CrossRef] [Green Version]
- Chitwood, M.C.; Maggi, R.G.; Kennedy-Stoskopf, S.; Toliver, M.; de Perno, A.C.S. Bartonella vinsonii subsp. berkhoffii in Free-Ranging White-Tailed Deer (Odocoileus virginianus). J. Wildl. Dis. 2013, 49, 468–470. [Google Scholar] [CrossRef] [Green Version]
- Beerlage, C.; Varanat, M.; Linder, K.; Maggi, R.; Cooley, J.; Kempf, V.A.J.; Breitschwerdt, E.B. Bartonella vinsonii subsp. berkhoffii and Bartonella henselae as potential causes of proliferative vascular diseases in animals. Med. Microbiol. Immunol. 2012, 201, 319–326. [Google Scholar] [CrossRef]
- Chinnadurai, S.K.; Birkenheuer, A.J.; Blanton, H.L.; Maggi, R.G.; Belfiore, N.; Marr, H.S.; Breitschwerdt, E.B.; Stoskopf, M.K. Prevalence of Selected Vector-borne Organisms and Identification of Bartonella Species DNA in North American River Otters (Lontra canadensis). J. Wildl. Dis. 2010, 46, 947–950. [Google Scholar] [CrossRef] [Green Version]
- Breitschwerdt, E.B.; Maggi, R.G.; Mozayeni, B.R.; Hegarty, B.C.; Bradley, J.M.; Mascarelli, P.E. PCR amplification of Bartonella koehlerae from human blood and enrichment blood cultures. Parasites Vectors 2010, 3, 76. [Google Scholar] [CrossRef] [Green Version]
- Maggi, R.; Kosoy, M.; Mintzer, M.; Breitschwerdt, E.B. Isolation of Candidatus Bartonella melophagi from Human Blood1. Emerg. Infect. Dis. 2009, 15, 66–68. [Google Scholar] [CrossRef]
- Diniz, P.P.; Wood, M.; Maggi, R.G.; Sontakke, S.; Stepnik, M.; Breitschwerdt, E.B. Co-isolation of Bartonella henselae and Bartonella vinsonii subsp. berkhoffii from blood, joint and subcutaneous seroma fluids from two naturally infected dogs. Vet. Microbiol. 2009, 138, 368–372. [Google Scholar] [CrossRef]
- Cherry, N.A.; Maggi, R.G.; Cannedy, A.L.; Breitschwerdt, E.B. PCR detection of Bartonella bovis and Bartonella henselae in the blood of beef cattle. Vet. Microbiol. 2009, 135, 308–312. [Google Scholar] [CrossRef]
- Breitschwerdt, E.B.; Maggi, R.G.; Cadenas, M.B.; Diniz, P.P. A Groundhog, a Novel Bartonella Sequence, and My Father’s Death. Emerg. Infect. Dis. 2009, 15, 2080–2086. [Google Scholar] [CrossRef] [Green Version]
- Parker, A.M.; Sheehy, P.A.; Hazelton, M.S.; Bosward, K.L.; House, J. A review of mycoplasma diagnostics in cattle. J. Vet. Intern. Med. 2018, 32, 1241–1252. [Google Scholar] [CrossRef]
- Hazelton, M.; Morton, J.; Parker, A.; Sheehy, P.; Bosward, K.; Malmo, J.; House, J. Whole dairy herd sampling to detect subclinical intramammary Mycoplasma bovis infection after clinical mastitis outbreaks. Vet. Microbiol. 2020, 244, 108662. [Google Scholar] [CrossRef]
- Hazelton, M.; Sheehy, P.; Bosward, K.; Parker, A.; Morton, J.; Dwyer, C.; Niven, P.; House, J. Short communication: Shedding of Mycoplasma bovis and antibody responses in cows recently diagnosed with clinical infection. J. Dairy Sci. 2018, 101, 584–589. [Google Scholar] [CrossRef] [Green Version]
- Jordan, A.; Sadler, R.J.; Sawford, K.; van Andel, M.; Ward, M.P.; Cowled, B.D. Mycoplasma bovis outbreak in New Zealand cattle: An assessment of transmission trends using surveillance data. Transbound. Emerg. Dis. 2020. [Google Scholar] [CrossRef]
- Bürki, S.; Spergser, J.; Bodmer, M.; Pilo, P. A dominant lineage of Mycoplasma bovis is associated with an increased number of severe mastitis cases in cattle. Vet. Microbiol. 2016, 196, 63–66. [Google Scholar] [CrossRef] [Green Version]
- Schibrowski, M.; Gibson, J.; Hay, K.; Mahony, T.; Barnes, T. Mycoplasma bovis and bovine respiratory disease: A risk factor study in Australian feeder cattle. Prev. Vet. Med. 2018, 157, 152–161. [Google Scholar] [CrossRef] [PubMed]
- Binetruy, F.; Garnier, S.; Boulanger, N.; Talagrand-Reboul, E.; Loire, E.; Faivre, B.; Noël, V.; Buysse, M.; Duron, O. A novel Borrelia species, intermediate between Lyme disease and relapsing fever groups, in neotropical passerine-associated ticks. Sci. Rep. 2020, 10, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Maggi, R.G.; Reichelt, S.; Toliver, M.; Engber, B. Borrelia species in Ixodes affinis and Ixodes scapularis ticks collected from the coastal plain of North Carolina. Ticks Tick Borne Dis. 2010, 1, 168–171. [Google Scholar] [CrossRef]
- Beyer, H.L. Hawth’s Analysis Tools for ArcGIS. 2004. Available online: http://www.spatialecology.com/htools/tooldesc.php (accessed on 20 October 2021).
- Dietrich, F.; Schmidgen, T.; Maggi, R.G.; Richter, D.; Matuschka, F.-R.; Vonthein, R.; Breitschwerdt, E.B.; Kempf, V.A.J. Prevalence of Bartonella henselae and Borrelia burgdorferi Sensu Lato DNA in Ixodes ricinus Ticks in Europe. Appl. Environ. Microbiol. 2010, 76, 1395–1398. [Google Scholar] [CrossRef] [Green Version]
- Maggi, R.G.; Toliver, M.; Richardson, T.; Mather, T.; Breitschwerdt, E.B. Regional prevalences of Borrelia burgdorferi, Borrelia bissettiae, and Bartonella henselae in Ixodes affinis, Ixodes pacificus and Ixodes scapularis in the USA. Ticks Tick Borne Dis. 2018, 10, 360–364. [Google Scholar] [CrossRef] [PubMed]

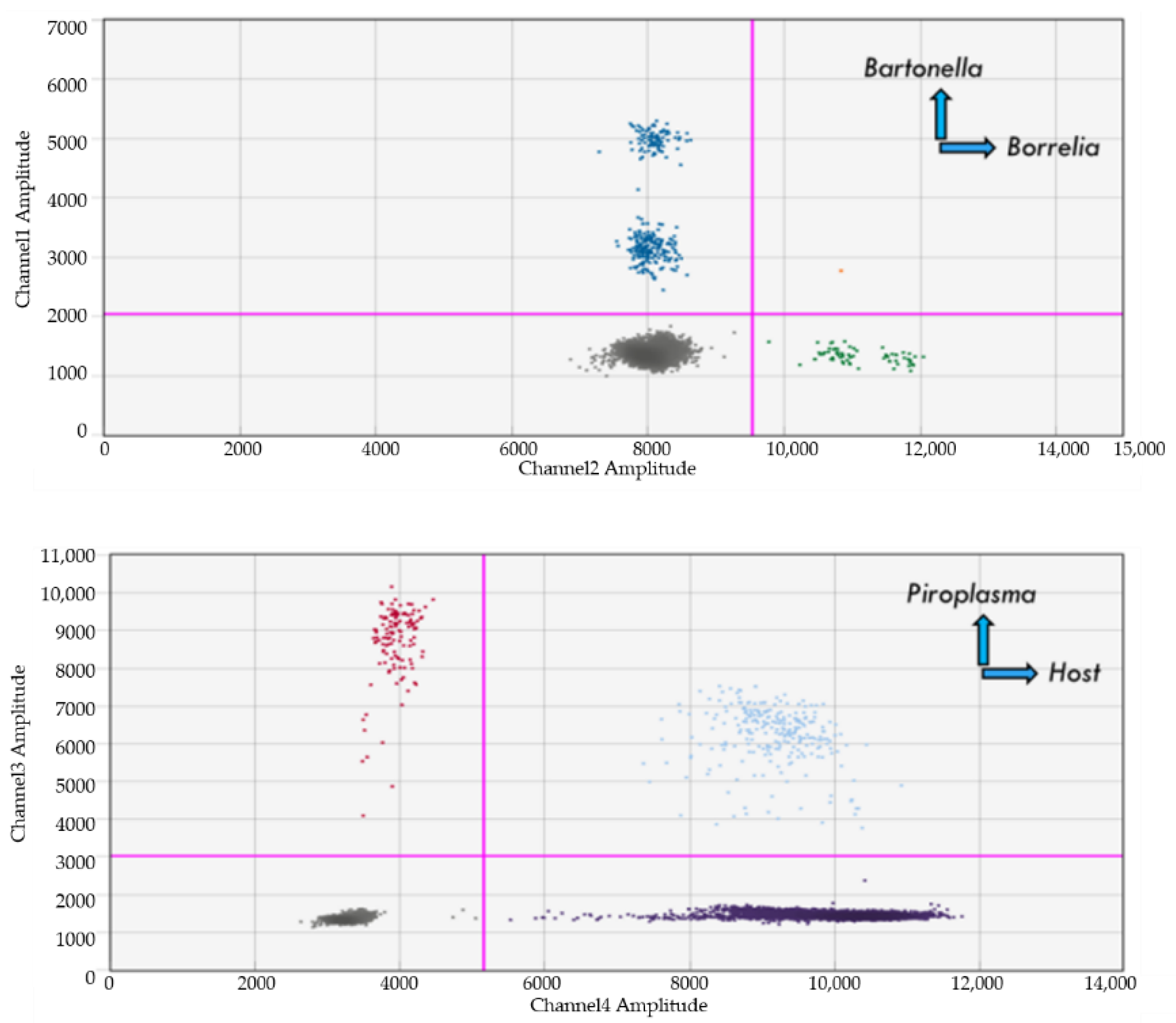
| Piroplasma | Sample Type | Host/s | Infection Type |
|---|---|---|---|
| B. bicornis | Blood | Rhinocerous | Naturally |
| B. canis canis | Blood | Dogs | Naturally |
| B. canis vogeli | Blood | Dogs | Naturally |
| B. coco | Blood | Dogs, Horse | Naturally |
| B. conrade | Blood | Dogs | Naturally |
| B. divergens | Blood | Rabbit | Experimentally |
| B. duncani | Blood | Hamster | Experimentally |
| B. felis | Blood | Cats | Naturally |
| B. gibsoni | Blood | Dogs | Naturally |
| B. gibsoni + B. vulpes | Blood | Dog | Naturally |
| B. lengau | Blood | Cats | Naturally |
| B. microti | Blood | Hamster | Experimentally |
| B. odoncoilei | Blood | Deer, Reindeer | Naturally |
| B. vulpes | Blood | Dogs, Foxes | Naturally |
| B. vulpes + Babesia spp. | Blood | Raccoon | Naturally |
| Babesia spp. | Blood | Dog | Naturally |
| Babesia spp. AJB 2006 | Blood | Bear | Naturally |
| Babesia spp. NC08 | Blood | Bear | Naturally |
| Babesia spp. NC13 | Blood | Bear | Naturally |
| Babesia spp. R-214 | Blood | Racoon | Naturally |
| Babesia spp. R431 | Blood | Racoon | Naturally |
| Babesia spp. R-9879 | Blood | Racoon | Naturally |
| Babesia spp. R-CO8 | Blood | Racoon | Naturally |
| Babesia spp. SK04 | Blood | Skunk | Naturally |
| Babesia spp. SK08 | Blood | Skunk | Naturally |
| Babesia spp. WR3 | Blood | Woodrat | Naturally |
| C. felis | Blood | Cats | Naturally |
| Maned wolf Babesia | Blood | Dogs, Racoons | Naturally |
| T. cervi | Blood | Deer, Bears | Naturally |
| T. equi | Blood | Horses | Naturally |
| Bartonella | Sample Type | Host/s | Infection Type |
|---|---|---|---|
| B. alsatica | Blood | Rabbits | Naturally |
| B. bovis | Blood | Cows | Naturally |
| B. chomelii | Blood | Elk | Naturally |
| B. clarridgeiae | Blood | Cats, Dogs | Naturally |
| B. henselae | Blood | Humans, Cats, Dogs | Naturally |
| B. henselae | Tissue (Brain) | Dolphins | Naturally |
| B. henselae | Tissue (Body) | Fleas, Sanflies | Naturally |
| B. henselae | Tissue (Skin) | Mice | Experimentally |
| B. henselae | Tissue (Body) | Tick (I. scapularis) | Naturally |
| B. henselae H1 | Blood | Human, Cat, Dog | Spiked |
| B. henselae SA2 | Blood | Human, Dogs, Birds | Naturally |
| B. henselae SA2 | Blood | Human, Dog | Spiked |
| B. henselae SA2 | DH82 cell-line | Dog | Experimentally |
| B. henselae SA2 | MCF10A cell-line | Human | Experimentally |
| B. koehlerae | Blood | Human, Dog | Spiked |
| B. melophagi | Blood | Human, Dog | Spiked |
| B. melophagi | Blood | Sheep | Naturally |
| B. quintana | Blood | Human, Dog | Spiked |
| B. quintana | Blood | Human, Monkey | Naturally |
| B. rochalimae | Blood | Dogs | Naturally |
| B. tamiae | Blood | Human | Spiked |
| B. tamiae | Blood | Woodrats | Naturally |
| B. tribocorum | Blood | Rats | Naturally |
| B. v. berkhoffii TI | Blood | Human, Dog | Spiked |
| B. v. berkhoffii TII | Blood | Human, Dog | Spiked |
| B. v. berkhoffii TII | Blood | Human, Coyotes, Foxes | Naturally |
| B. v. berkhoffii TIII | Blood | Human, Dog | Spiked |
| B. v. berkhoffii TIII | Blood | Skunk | Naturally |
| B. volans | Tissue (Heart) | River otter | Naturally |
| Bartonella spp. | Blood | Mongooses | Naturally |
| Bartonella spp. Rc | Blood | Racoons | Naturally |
| Borrelia | Sample Type | Host | Infection Type |
|---|---|---|---|
| B. afzelii | Blood | Dog | Spiked |
| B. bissetii | Tissue (Body) | Tick (I. affinis) | Naturally |
| B. burgdorferi | Tissue (Body) | Tick (I. scapularis) | Naturally |
| B. burgdorferi B31 | Blood | Human, Dog | Spiked |
| B. burgdorferi B31 | DH82 cell line | Dog | Experimentally |
| B. burgdorferi B31 | MCF10A cell line | Human | Experimentally |
| B. burgdorferi B31 | Tissue (Skin) | Mice | Experimentally |
| B. coreaceae | Blood | Dog | Spiked |
| B. garinii | Blood | Dog | Spiked |
| B. hermsii | Blood | Dog | Spiked |
| B. lusitanae | Blood | Dog | Spiked |
| B. turicatae | Blood | Dog | Spiked |
| Co-Infection | Sample Type | Host | Infection Type |
|---|---|---|---|
| B. burgdorferi + B. gibsonii | Blood | Dog | Naturally + Spiked |
| B. burgdorferi + B. gibsonii | Blood | Dog | Naturally + Spiked |
| B. burgdorferi + B. microti | Blood | Human | Spiked |
| B. henselae + B. burgdorferi | Blood | Dog | Spiked |
| B. henselae + B. burgdorferi | Blood | Human | Spiked |
| B. henselae + B. burgdorferi | Tissue (Brain) | Human | Experimentally |
| B. henselae + B. burgdorferi | Tissue (Body) | Tick (I. affinis) | Naturally |
| B. henselae + B. burgdorferi + B. gibsonii | Blood | Dog | Naturally + Spiked |
| B. henselae + B. burgdorferi + B. microti | Blood | Human | Spiked |
| B. henselae + B. burgdorferi B31 | Tissue (Skin) | Mice | Experimentally |
| B. henselae + B. microti | Blood | Human | Spiked |
| B. odoncoilei + B. henselae | Tissue (Body) | Tick (I. affinis) | Naturally |
| B. quintana + B. burgdorferi | Blood | Dog | Spiked |
| B. quintana + B. burgdorferi | Blood | Human | Spiked |
| B. v. berkhoffii TII + B. burgdorferi | Blood | Dog | Spiked |
| B. v. berkhoffii TII + B. burgdorferi | Blood | Human | Spiked |
| B. vinsonii + B. vulpes | Blood | Gray fox | Naturally |
| Borrelia sp. | Strain | Conc. Range (Genome Copies per 5 µL Sample) |
|---|---|---|
| B. afzelii | Pko | 0.1–10,000 |
| B. bavariensis | VS116 | 0.1–10,000 |
| B. bissettii | Pgeb | 10,000 |
| B. burgdorferi | B31, Pbre | 0.1–10,000 |
| B. garinii | PBr, Phei, Pla, Pref, PWudII | 0.1–10,000 |
| B. hermsii | 0.1–10,000 | |
| B. lusitanae | PotiB2 | 0.1–10,000 |
| B. miyamotoi | 10,000 | |
| B. spielmanii | PSigHH | 0.1–10,000 |
| B. turcica | 10,000 | |
| B. valaisiana | VS116 | 0.1–10,000 |
| Primer/Probe | Name | Channel | Sequence |
|---|---|---|---|
| Babesia sense | Piro18S-238s | 3 | 5′-TCGGTGATTCATAATAAACTRGCGAATCGC-3′ |
| Babesia antisense | Piro18S-380as | 5′-GAATCGAACCCCAATTCCCCGTTACCCG-3′ | |
| Babesia probe | Piro18S-340 | 5′-Cy5- GACGGTAGGGTATTGGCCTACCG-BHQ2-3′ | |
| Bartonella sense | BsppITS325s | 1 | 5’-CCTCAGATGATGATCCCAAGCCTTCTGGCG-3’ |
| Bartonella antisense | BsppITS543as | 5′-TAAAYTGGTGGGCCTGGGAGGACTTG-3′ | |
| Bartonella probe | BsppITS500 | 5′-FAM-GTTAGAGCGCGCGCTTGATAAG -IABkFQ-3′ | |
| Borrelia sense | BobuITS120s | 2 | 5′-AGGTCATTTTGGGGGTTTAGCTCAGTTGGCT-3′ |
| Borrelia antisense | BoLymeITS200as | 5′-AATGGAGGTTAAGGGACTCGAACCCT-3′ | |
| Borrelia probe | BobuITS160 | 5′-HEX-CGGCTTTGCAAGCCGAGGGTCAAG-BHQ-2-3′ |
| Babesia Species | Theileria-Cytauxzoon | |
|---|---|---|
| B. bicornis | B. microti | T. cervi |
| B. bigemina | B. negevi-like | T. equi |
| B. c. canis | B. vulpes (former B. annae) | C. felis |
| B. c. vogeli | maned Wolf Babesia | |
| B. coco | Bsp AJB 2006 * | |
| B. conradae | Bsp woodrat * | |
| B. divergens | Bspp skunks (SK04 *, SK08 *) | |
| B. duncani | Bspp raccoons (R214 *, 9879 *, CO8 *) | |
| B. gibsoni | Bspp bears (NC08 *, NC13 *) | |
| B. lengau | ||
| Bartonella Species | |||
|---|---|---|---|
| B. henselae | B. alsatica | B. doshiae | B. rochalimae |
| B. koehlerae | B. melophagi | B. schoembuchensis | B. tamiae |
| B. quintana | B. volans | B. rattimassiliensis | Ca. B. k. boulousii |
| B. v. berkhoffii TI | B. elizabethae | B. tribocorum | Ca. B. k. bothierii |
| B. v. berkhoffii TII | B. monaki | B. chomelii | Bsp deer (D2 *, D4 *, D5 *, D6 *) |
| B. v. berkhoffii TIII | B.v. baker | B. washoensis | Bsp river otter * |
| B. clarridgeiae | B. bovis | B. vinsonii | Bsp mongoose * |
| Borrelia Species (Strains) | |
|---|---|
| B. afzelii (Pko) | B. lusitanae (PotB2) |
| B. bavariensis (PBi) | B. miyamotoi |
| B. bissettii (PGeb) | B. spielmanii (PSigII) |
| B. burgdorferi (B31, PBre) | B. turcica |
| B. coriaceae | B. turicatae |
| B. garinii (PBr, PHei, Pla, PRef, PWudII) | B. valaisiana (VS116) |
| B. hermsii | |
| Borrelia sp. | Strain | Detection Limit (Genome Copies per 5 µL Sample) |
|---|---|---|
| B. afzelii | Pko | 1–10 |
| B. bavariensis | VS116 | 0.1–10 |
| B. bissettii | Pgeb | 1–10 |
| B. burgdorferi | B31, Pbre | 1–10 |
| B. garinii | PBr, Phei, Pla, Pref, PWudII | 1–10 |
| B. hermsii | NA | |
| B. lusitanae | PotiB2 | 1–10 |
| B. miyamotoi | NA | |
| B. spielmanii | PSigHH | 1–10 |
| B. turcica | 1–10 | |
| B. valaisiana | VS116 | 10 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maggi, R.; Breitschwerdt, E.B.; Qurollo, B.; Miller, J.C. Development of a Multiplex Droplet Digital PCR Assay for the Detection of Babesia, Bartonella, and Borrelia Species. Pathogens 2021, 10, 1462. https://doi.org/10.3390/pathogens10111462
Maggi R, Breitschwerdt EB, Qurollo B, Miller JC. Development of a Multiplex Droplet Digital PCR Assay for the Detection of Babesia, Bartonella, and Borrelia Species. Pathogens. 2021; 10(11):1462. https://doi.org/10.3390/pathogens10111462
Chicago/Turabian StyleMaggi, Ricardo, Edward B. Breitschwerdt, Barbara Qurollo, and Jennifer C. Miller. 2021. "Development of a Multiplex Droplet Digital PCR Assay for the Detection of Babesia, Bartonella, and Borrelia Species" Pathogens 10, no. 11: 1462. https://doi.org/10.3390/pathogens10111462
APA StyleMaggi, R., Breitschwerdt, E. B., Qurollo, B., & Miller, J. C. (2021). Development of a Multiplex Droplet Digital PCR Assay for the Detection of Babesia, Bartonella, and Borrelia Species. Pathogens, 10(11), 1462. https://doi.org/10.3390/pathogens10111462







