Colistin Resistance Onset Strategies and Genomic Mosaicism in Clinical Acinetobacter baumannii Lineages
Abstract
:1. Introduction
2. Results
2.1. COL MIC
2.2. COL Resistance Induction
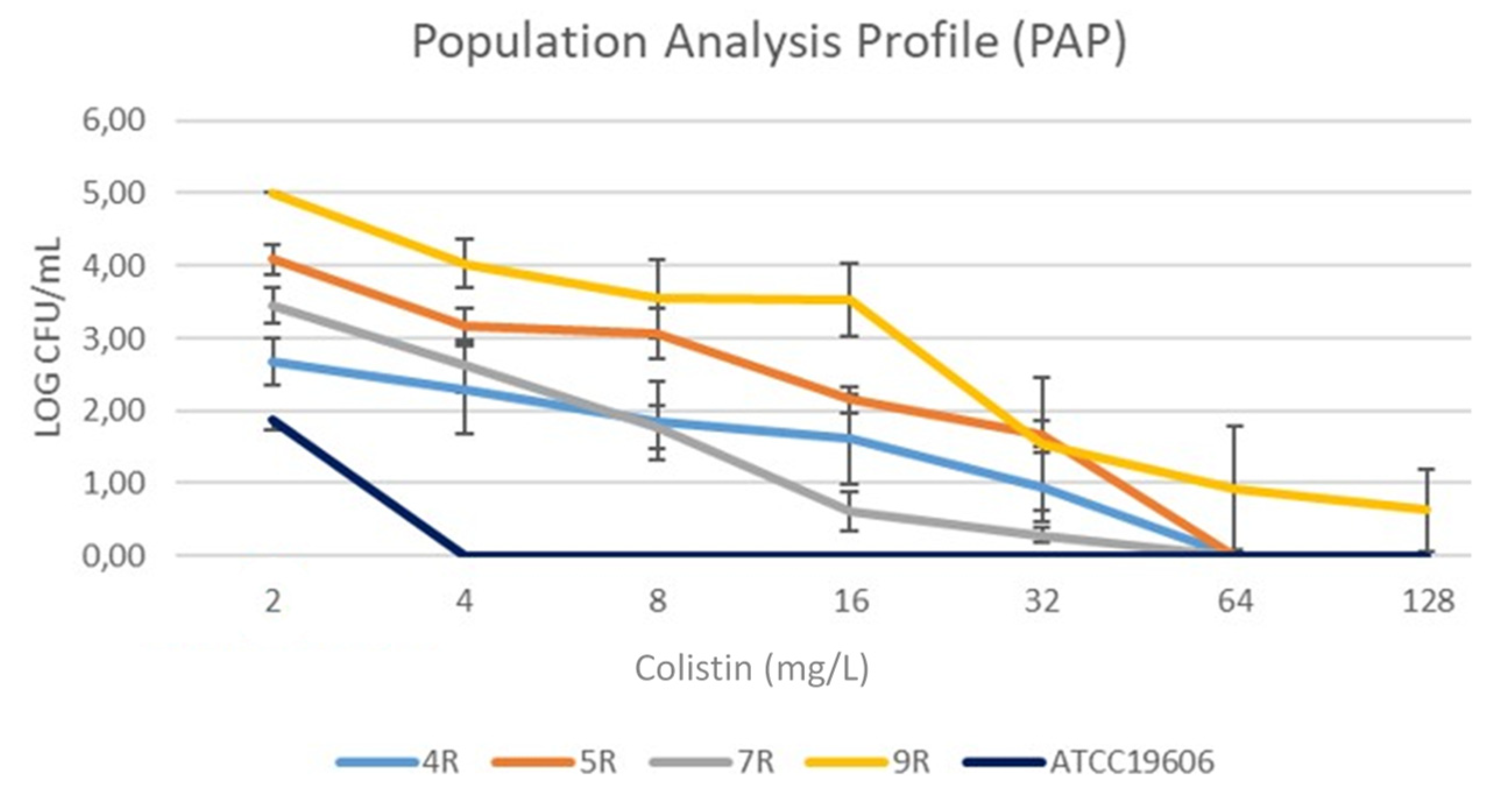
2.3. Hetero-Resistance
2.4. LPS Quantification and Cell-Envelope Charge
2.5. Phylogenetic Tree and Genomic Typing
2.6. Resistomes and nsSNPs in Genes Previously Associated to COL-R Mechanisms
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains
4.2. COL Minimum Inhibitory Concentrations (MICs)
4.3. COL Resistance Induction Assay
4.4. COL Hetero-Resistance Detection
4.5. Surface Charge Determination
4.6. Direct LPS Quantification
4.7. Genotyping
4.8. Whole Genome Sequencing (WGS)
4.9. Phylogeny and Genomic Epidemiology
4.10. Single Nucleotide Polymorphisms (SNPs)
4.11. Genomic Single-Nucleotide Polymorphism Effect Prediction
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sievert, D.M.; Ricks, P.; Edwards, J.R.; Schneider, A.; Patel, J.; Srinivasan, A.; Kallen, A.; Limbago, B.; Fridkin, S.; National Healthcare Safety Network (NHSN) Team and Participating NHSN Facilities. Antimicrobial-resistant pathogens associated with healthcare-associated infections: Summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2009–2010. Infect. Control Hosp. Epidemiol. 2013, 34, 1–14. [Google Scholar] [CrossRef]
- Cafiso, V.; Stracquadanio, S.; Lo Verde, F.; Gabriele, G.; Mezzatesta, M.L.; Caio, C.; Pigola, G.; Ferro, A.; Stefani, S. Colistin Resistant A. baumannii: Genomic and Transcriptomic Traits Acquired under Colistin Therapy. Front. Microbiol. 2019, 9, 3195. [Google Scholar] [CrossRef] [PubMed]
- Ilsan, N.A.; Lee, Y.-J.; Kuo, S.-C.; Lee, I.-H.; Huang, T.-W. Antimicrobial Resistance Mechanisms and Virulence of Colistin- and Carbapenem-Resistant Acinetobacter baumannii Isolated from a Teaching Hospital in Taiwan. Microorganisms 2021, 9, 1295. [Google Scholar] [CrossRef]
- Wand, M.E.; Bock, L.J.; Bonney, L.C.; Sutton, J.M. Retention of virulence following adaptation to colistin in Acinetobacter baumannii reflects the mechanism of resistance. J. Antimicrob. Chemother. 2015, 70, 2209–2216. [Google Scholar] [CrossRef] [Green Version]
- Geisinger, E.; Isberg, R.R. Antibiotic modulation of capsular exopolysaccharide and virulence in Acinetobacter baumannii. PLoS Pathog. 2015, 11, e1004691. [Google Scholar] [CrossRef] [Green Version]
- Martins-Sorenson, N.; Snesrud, E.; Xavier, D.E.; Cacci, L.C.; Iavarone, A.T.; McGann, P.; Riley, L.W.; Moreira, B.M. A novel plasmid-encoded mcr-4.3 gene in a colistin-resistant Acinetobacter baumannii clinical strain. J. Antimicrob. Chemother. 2020, 75, 60–64. [Google Scholar] [CrossRef]
- Sacco, F.; Visca, P.; Runci, F.; Antonelli, G.; Raponi, G. Susceptibility Testing of Colistin for Acinetobacter baumannii: How Far Are We from the Truth? Antibiotics 2021, 10, 48. [Google Scholar] [CrossRef] [PubMed]
- Karakonstantis, S.; Saridakis, I. Colistin heteroresistance in Acinetobacter spp.: Systematic review and meta-analysis of the prevalence and discussion of the mechanisms and potential therapeutic implications. Int. J. Antimicrob. Agents 2020, 56, 106065. [Google Scholar] [CrossRef]
- Li, J.; Rayner, C.R.; Nation, R.L.; Owen, R.J.; Spelman, D.; Tan, K.E.; Liolios, L. Heteroresistance to colistin in multidrug-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 2006, 50, 2946–2950. [Google Scholar] [CrossRef] [Green Version]
- Rasidin, R.S.M.; Suhaili, Z.; Mohamed, A.F.S.; Hod, R.; Neela, V.; Amin-Nordin, S. Time-kill and post-antibiotic effect of colistin at different static concentrations in in vitro Acinetobacter baumannii. Trop. Biomed. 2020, 37, 471–481. [Google Scholar] [PubMed]
- Andersson, D.I.; Nicoloff, H.; Hjort, K. Mechanisms and clinical relevance of bacterial heteroresistance. Nat. Rev. Microbiol. 2019, 17, 479–496. [Google Scholar] [CrossRef] [PubMed]
- Çağlan, E.; Nigiz, Ş.; Sancak, B.; Gür, D. Resistance and heteroresistance to colistin among clinical isolates of Acinetobacter baumannii. Acta Microbiol. Immunol. Hung. 2020, 67, 107–111. [Google Scholar] [CrossRef] [PubMed]
- Ezadi, F.; Jamali, A.; Heidari, A.; Javid, N.; Ardebili, A. Heteroresistance to colistin in oxacillinase-producing carbapenem-resistant Acinetobacter baumannii clinical isolates from Gorgan, Northern Iran. J. Glob. Antimicrob. Resist. 2020, 21, 380–385. [Google Scholar] [CrossRef] [PubMed]
- Thet, K.T.; Lunha, K.; Srisrattakarn, A.; Lulitanond, A.; Tavichakorntrakool, R.; Kuwatjanakul, W.; Charoensri, N.; Chanawong, A. Colistin heteroresistance in carbapenem-resistant Acinetobacter baumannii clinical isolates from a Thai university hospital. World J. Microbiol. Biotechnol. 2020, 36, 102. [Google Scholar] [CrossRef]
- Chen, L.; Lin, J.; Lu, H.; Zhang, X.; Wang, C.; Liu, H.; Zhang, X.; Li, J.; Cao, J.; Zhou, T. Deciphering colistin heteroresistance in Acinetobacter baumannii clinical isolates from Wenzhou, China. J. Antibiot. 2020, 73, 463–470. [Google Scholar] [CrossRef]
- Charretier, Y.; Diene, S.M.; Baud, D.; Chatellier, S.; Santiago-Allexant, E.; van Belkum, A.; Guigon, G.; Schrenzel, J. Colistin Heteroresistance and Involvement of the PmrAB Regulatory System in Acinetobacter baumannii. Antimicrob Agents Chemother. 2018, 62, e00788-18. [Google Scholar] [CrossRef] [Green Version]
- Machado, D.; Antunes, J.; Simões, A.; Perdigão, J.; Couto, I.; McCusker, M.; Martins, M.; Portugal, I.; Pacheco, T.; Batista, J.; et al. Contribution of efflux to colistin heteroresistance in a multidrug resistant Acinetobacter baumannii clinical isolate. J. Med. Microbiol. 2018, 67, 740–749. [Google Scholar] [CrossRef] [Green Version]
- El-Sayed Ahmed, M.A.E.; Zhong, L.L.; Shen, C.; Yang, Y.; Doi, Y.; Tian, G.B. Colistin and its role in the Era of antibiotic resistance: An extended review (2000–2019). Emerg. Microbes. Infect. 2020, 9, 868–885. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, Y.; Chai, D.; Wang, R.; Liang, B.; Bai, N. Colistin resistance of Acinetobacter baumannii: Clinical reports, mechanisms and antimicrobial strategies. J. Antimicrob. Chemother. 2012, 67, 1607–1615. [Google Scholar] [CrossRef] [PubMed]
- Herrera, M.E.; Mobilia, L.N.; Posse, G.R. Comparative evaluation of the sensitivity of Acinetobacter to colistin, using the prediffusion and minimum inhibitory concentration methods: Detection of heteroresistant isolates. Rev. Argent. Microbiol. 2011, 43, 115–119. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, C.H.; De Ambrosio, A.; Bajuk, M.; Spinozzi, M.; Nastro, M.; Bombicino, K.; Radice, M.; Gutkind, G.; Vay, C.; Famiglietti, A. In vitro antimicrobials activity against endemic Acinetobacter baumannii multiresistant clones. J. Infect. Dev. Ctries. 2010, 4, 164–167. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez, C.H.; Bombicino, K.; Granados, G.; Nastro, M.; Vay, C.; Famiglietti, A. Selection of colistin-resistant Acinetobacter baumannii isolates in postneurosurgical meningitis in an intensive care unit with high presence of heteroresistance to colistin. Diagn. Microbiol. Infect. Dis. 2009, 65, 188–191. [Google Scholar] [CrossRef]
- Yau, W.; Owen, R.J.; Poudyal, A.; Bell, J.M.; Turnidge, J.D.; Yu, H.H.; Nation, R.L.; Li, J. Colistin hetero-resistance in multidrug-resistant Acinetobacter baumannii clinical isolates from the Western Pacific region in the SENTRY antimicrobial surveillance programme. J. Infect. 2009, 58, 138–144. [Google Scholar] [CrossRef]
- Barin, J.; Martins, A.F.; Heineck, B.L.; Barth, A.L.; Zavascki, A.P. Hetero- and adaptive resistance to polymyxin B in OXA-23-producing carbapenem-resistant Acinetobacter baumannii isolates. Ann. Clin. Microbiol. Antimicrob. 2013, 12, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cafiso, V.; Stracquadanio, S.; Lo Verde, F.; Dovere, V.; Zega, A.; Pigola, G.; Aranda, J.; Stefani, S. COLR Acinetobacter baumannii sRNA Signatures: Computational Comparative Identification and Biological Targets. Front. Microbiol. 2020, 10, 3075. [Google Scholar] [CrossRef] [PubMed]
- Motta, S.S.; Cluzel, P.; Aldana, M. Adaptive resistance in bacteria requires epigenetic inheritance, genetic noise, and cost of efflux pumps. PLoS ONE 2015, 10, e0118464. [Google Scholar] [CrossRef] [Green Version]
- Falagas, M.E.; Makris, G.C.; Dimopoulos, G.; Matthaiou, D.K. Heteroresistance: A concern of increasing clinical significance? Clin. Microbiol. Infect. 2008, 14, 101–104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, S.S.; Lin, T.Y.; Wang, W.B.; Liu, M.C.; Hsueh, P.R.; Liaw, S.J. Characterization of UDP-glucose dehydrogenase and UDP-glucose pyrophosphorylase mutants of Proteus mirabilis: Defectiveness in polymyxin B resistance, swarming, and virulence. Antimicrob. Agents Chemother. 2010, 54, 2000–2009. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olaitan, A.O.; Morand, S.; Rolain, J.M. Mechanisms of polymyxin resistance: Acquired and intrinsic resistance in bacteria. Front. Microbiol. 2014, 5, 643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- French, S.; Farha, M.; Ellis, M.J.; Sameer, Z.; Côté, J.P.; Cotroneo, N.; Lister, T.; Rubio, A.; Brown, E.D. Potentiation of Antibiotics against Gram-Negative Bacteria by Polymyxin B Analogue SPR741 from Unique Perturbation of the Outer Membrane. ACS Infect. Dis. 2020, 6, 1405–1412. [Google Scholar] [CrossRef] [PubMed]
- Overview Microflex Series—Highest Performance Bench-Top MALDI-TOF, MS Bruker.com. Available online: https://www.bruker.com/products/mass-spectrometry-and-separations/maldi-toftof/microflex/overview.html (accessed on 16 September 2019).
- BD PhoenixTM M50 Instrument. Available online: https://www.bd.com/en-us/offerings/capabilities/microbiology-solutions/identification-and-susceptibility-testing/bd-phoenix-automated-identification-and-susceptibility-testing-system/bd-phoenix-m50-instrument (accessed on 16 September 2019).
- Sensititre Antimicrobial Susceptibility Testing System—IT. Available online: https://www.thermofisher.com/uk/en/home/clinical/clinical-microbiology/antimicrobial-susceptibility-testing/sensititre-antimicrobial-susceptibility-testing-system.html (accessed on 16 September 2019).
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing; 20-Third Informational Supplement; Document M100-S25; Springer: Wayne, PA, USA, 2015. [Google Scholar]
- EUCASTBreakpointV_9.0. 2019. Available online: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_9.0_Breakpoint_Tables.pdf (accessed on 11 November 2019).
- Acinetobacter baumannii: Methods and Protocols; Methods in Molecular Biology; Springer Protocol; Springer: New York, NY, USA, 2019.
- Yang, S.J.; Kreiswirth, B.N.; Sakoulas, G.; Yeaman, M.R.; Xiong, Y.Q.; Sawa, A.; Bayer, A.S. Enhanced expression of dltABCD is associated with the development of daptomycin nonsusceptibility in a clinical endocarditis isolate of Staphylococcus aureus. J. Infect. Dis. 2009, 200, 1916–1920. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, S.J.; Nast, C.C.; Mishra, N.N.; Yeaman, M.R.; Fey, P.D.; Bayer, A.S. Cell wall thickening is not a universal accompaniment of the daptomycin nonsusceptibility phenotype in Staphylococcus aureus: Evidence for multiple resistance mechanisms. Antimicrob. Agents Chemother. 2010, 54, 3079–3085, PMCID: PMC2916340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rezania, S.; Amirmozaffari, N.; Tabarraei, B.; Jeddi-Tehrani, M.; Zarei, O.; Alizadeh, R.; Masjedian, F.; Zarnani, A.H. Extraction, Purification and Characterization of Lipopolysaccharide from Escherichia coli and Salmonella typhi. Avicenna J. Med. Biotechnol. 2011, 3, 3–9. [Google Scholar]
- Mezzatesta, M.L.; D’Andrea, M.M.; Migliavacca, R.; Giani, T.; Gona, F.; Nucleo, E.; Fugazza, G.; Pagani, L.; Rossolini, G.M.; Stefani, S. Epidemiological characterization and distribution of carbapenem-resistant Acinetobacter baumannii clinical isolates in Italy. Clin. Microbiol. Infect. 2012, 18, 160–166. [Google Scholar] [CrossRef] [Green Version]
- Tenover, F.C.; Arbeit, R.D.; Goering, R.V.; Mickelsen, P.A.; Murray, B.E.; Persing, D.H.; Swaminathan, B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: Criteria for bacterial strain typing. J. Clin. Microbiol. 1995, 33, 2233–2239. [Google Scholar] [CrossRef] [Green Version]
- Towner, K.J.; Levi, K.; Vlassiadi, M.; ARPAC Steering Group. Genetic diversity of carbapenem-resistant isolates of Acinetobacter baumannii in Europe. Clin. Microbiol. Infect. 2008, 14, 161–167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCracken, M.; Mataseje, L.F.; Loo, V.; Walkty, A.; Adam, H.J.; Hoban, D.J.; Zhanel, G.G.; Mulvey, M.R.; Canadian Antimicrobial Resistance Alliance (CARA). Characterization of Acinetobacter baumannii and meropenem-resistant Pseudomonas aeruginosa in Canada: Results of the CANWARD 2007–2009 study. Diagn Microbiol. Infect. Dis. 2011, 69, 335–341. [Google Scholar] [CrossRef]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef] [PubMed]
- Larsen, M.V.; Cosentino, S.; Rasmussen, S.; Friis, C.; Hasman, H.; Marvig, R.L.; Jelsbak, L.; Sicheritz-Pontén, T.; Ussery, D.W.; Aarestrup, F.M.; et al. Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 2012, 50, 1355–1361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johansson, M.H.K.; Bortolaia, V.; Tansirichaiya, S.; Aarestrup, F.M.; Roberts, A.P.; Petersen, T.N. Detection of mobile genetic elements associated with antibiotic resistance in Salmonella enterica using a newly developed web tool: Mobile Element Finder. J. Antimicrob. Chemother. 2021, 76, 101–109. [Google Scholar] [CrossRef]
- Zhou, Y.; Liang, Y.; Lynch, K.H.; Dennis, J.J.; Wishart, D.S. PHAST: A Fast Phage Search Tool. Nucleic Acids Res. 2011, 39, W347–W352. [Google Scholar] [CrossRef] [PubMed]
- Pourcel, C.; Touchon, M.; Villeriot, N.; Vernadet, J.P.; Couvin, D.; Toffano-Nioche, C.; Vergnaud, G. CRISPRCasdb a successor of CRISPRdb containing CRISPR arrays and cas genes from complete genome sequences, and tools to download and query lists of repeats and spacers. Nucleic Acids Res. 2020, 48, D535–D544. [Google Scholar] [CrossRef] [PubMed]
- Wick, R.R.; Heinz, E.; Holt, K.E.; Wyres, K.L. Kaptive Web: User-friendly capsule and lipopolysaccharide serotype prediction for Klebsiella genomes. J. Clin. Microbiol. 2018, 56, e00197-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wyres, K.L.; Cahill, S.M.; Holt, K.E.; Hall, R.M.; Kenyon, J.J. Identification of Acinetobacter baumannii loci for capsular polysaccharide (KL) and lipooligosaccharide outer core (OCL) synthesis in genome assemblies using curated reference databases compatible with Kaptive. Microbial. Genom. 2020, 6, e000339. [Google Scholar] [CrossRef] [PubMed]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cafiso, V.; Stracquadanio, S.; Lo Verde, F.; De Guidi, I.; Zega, A.; Pigola, G.; Stefani, S. Genomic and Long-Term Transcriptomic Imprints Related to the Daptomycin Mechanism of Action Occurring in Daptomycin- and Methicillin-Resistant Staphylococcus aureus Under Daptomycin Exposure. Front. Microbiol. 2020, 11, 1893. [Google Scholar] [CrossRef] [PubMed]


| COL MIC | Most Frequent COL MIC | COL-R Induction | COL PAP VARIANT MICs | LPS Quantification | Cell Envelop Positive Charge | AA Substitution in Proteins already Associated to COL-R | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Strain | (mg/L) | (mg/L) | COL Induction | Post Induction COL MIC | Variant-1 | Variant-2 | (µg/mL) | COL Repulsion (%) | PmrB | LpxA | GalU | COL-R Phenotype |
| 1R | 128 | - | - | - | - | - | 1334.38 | 3.7 | L208F | - | Q140L I273V | Full Resistance |
| 2R | 128 | - | - | - | - | - | 1698.02 | 8.87 | R263H | - | Q140L I273V | Full Resistance |
| 3R | 128 | - | - | - | - | - | 2538.93 | 27.57 | P170L A138T | N5V | I273V | Full Resistance |
| 4R | 1–64 | 8 | NEG | - | 8 | 64 | 257.11 | 49.5 | - | - | I245T | Hetero Resistance |
| 5R | 2–16 | 16 | NEG | - | 2 | 128 | 1561.65 | 9.8 | - | - | I245T S199A | Hetero Resistance |
| 6R | 2–128 | 64 | POS | >256 | - | - | 139.93 | 6 | A226T | - | I245T S199A | Adaptive Resistance |
| 7R | 2–128 | 2 | NEG | - | 8 | 64 | 3288.93 | 34.11 | - | - | I245T S199A | Hetero Resistance |
| 8R | 2–32 | 32 | POS | >256 | - | - | 811.66 | 31.7 | S14P | - | I245T S199A | Adaptive Resistance |
| 9R | 1–128 | 4 | NEG | - | 64 | 128 | 2266.20 | 21.02 | S14P | - | I245T S199A | Hetero Resistance |
| Strain | gPhyl Cluster | A. baumannii MLST Pasteur Institute | A. baumannii MLST Oxford University | KL TYPE | OCL TYPE | MGEs | Prophage | PFGE Clone Profile | RESISTOME | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| β-Lactams | Aminoglycosides | Sulfonamides | Macrolides | Tetracycline | |||||||||
| 1R | gPhyl cluster-III | ST-187 | ST-1839; ST-281 | KL22 | OCL3 | IS26 (6 copies) IS6100 (1 copy) ISAba1 (16 copies) ISAba13 (5 copies) ISAba125 (7 copies) Tn6080 (1 copy) | vB_AbaS_TRS1 | A | blaADC-25 blaOXA-23 blaOXA-82 | aadA2 ant(2″)-Ia aph(3′)-VIa | sul1 | - | - |
| 2R | gPhyl cluster-III | ST-2 | ST 1839 | KL22 | OCL3 | IS26 (5 copies) IS6100 (1 copy) ISAba1 (14 copies) ISAba13 (13 copies) ISAba125 (3 copies) | vB_AbaS_TRS1 Bφ_B1251 | A | blaADC-25 blaOXA-23 blaOXA-82 | aac(3)-Ia aadA1 aadA2 ant(2″)-Ia aph(3′)-VIa | sul1 | - | - |
| 3R | gPhyl cluster-II | ST-2 | ST-1816; ST-195 | KL3 | OCL1 | IS26 (3 copies) ISAba1 (16 copies) ISAba24 (24 copies) IsAba26 (2 copies) IsVsa3 (1 copy) Tn2006 (1 copy) | vB_AbaS_TRS1 | A | blaADC-25 blaOXA-23 blaOXA-66 blaTEM-1D | aph(3″)-Ib aph(3′)-Ia aph(6)-Id armA | Sul2 | mphE msrE | tetB |
| 4R | gPhyl cluster-II | ST-2 | ST-218 | KL28 | OCL1 | ISVsa3 (1 copy) ISAba13 (1 copy) ISAba27 (1 copy) | Bφ_B1251 | A | blaADC-25 blaOXA-23 blaOXA-66 | aph(3″)-Ib aph(6)-Id | - | - | tetB |
| 5R | gPhyl cluster-I | ST-2 | ST-1808; ST-348 | KL9 | OCL1 | IS17 (1 copy) IS26 (1 copy) ISAba1 (1 copy) ISAba13 (1 copy) ISAba26 (1 copy) ISAba36 (1 copy) ISVsa3 (1 copy) ISEc29 (1 copy) | A | blaADC-25 blaOXA-66 blaOXA-72 | aac(6′)-Ip aph(3″)-Ib aph(6)-Id armA | sul1 sul2 | mphE msrE | tetB | |
| 6R | gPhyl cluster-I | ST-2 | ST-1808; ST-348 | KL9 | OCL1 | IS17 (1 copy) IS26 (1 copy) ISAba1 (1 copy) ISAba13 (1 copy) ISAba26 (1 copy) ISAba36 (1 copy) ISVsa3 (1 copy) ISEc29(1 copy) | A | blaADC-25 blaOXA-66 blaOXA-72 | aac(6′)-Ip aph(3″)-Ib aph(6)-Id armA | sul1 sul2 | mphE msrE | tetB | |
| 7R | gPhyl cluster-I | ST-2 | ST-1808; ST-348 | KL9 | OCL1 | IS17 (1 copy) IS26 (1 copy) ISAba1 (1 copy) ISAba13 (1 copy) ISAba26 (1 copy) ISAba36 (1 copy) ISVsa3 (1 copy) ISEc29(1 copy) | A | blaADC-25 blaOXA-66 blaOXA-72 | aph(3″)-Ib aph(6)-Id armA | sul1 sul2 | mphE msrE | tetB | |
| 8R | gPhyl cluster-I | ST-2 | ST-1808; ST-348 | KL9 | OCL1 | IS17 (1 copy) IS26 (1 copy) ISAba1 (1 copy) ISAba13 (1 copy) ISAba26 (1 copy) ISEc29 (1 copy) Tn6207 (1 copy) | A | blaADC-25 blaOXA-66 blaOXA-72 | aph(3″)-Ib aph(6)-Id armA | sul1 | mphE msrE | tetB | |
| 9R | gPhyl cluster-I | ST-2 | ST-1808; ST-348 | KL9 | OCL1 | IS17 (1 copy) IS26 (1 copy) ISAba1 (1 copy) ISAba13 (1 copy) ISAba26 (1 copy) ISEc29 (1 copy) Tn6207 (1 copy) | A | blaADC-25 blaOXA-66 blaOXA-72 | aph(3″)-Ib aph(6)-Id armA | sul1 | mphE msrE | tetB |
| Strain | Hospital Wards | Source |
|---|---|---|
| 1R | Anesthesia and Intensive care | Bronchial aspirate |
| 2R | Anesthesia and Intensive care | Bronchial aspirate |
| 3R | Anesthesia and Intensive care | Bronchial aspirate |
| 4R | Anesthesia and Intensive care | Bronchial aspirate |
| 5R | Anesthesia and Intensive care | Bronchial aspirate |
| 6R | Anesthesia and Intensive care | Bronchial aspirate |
| 7R | Burns Unit | Blood culture |
| 8R | Anesthesia and Intensive care | Blood culture |
| 9R | Orthopedics and Traumatology | Wound swab |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cafiso, V.; Stracquadanio, S.; Dovere, V.; Lo Verde, F.; Zega, A.; Pigola, G.; Barnini, S.; Ghelardi, E.; Stefani, S. Colistin Resistance Onset Strategies and Genomic Mosaicism in Clinical Acinetobacter baumannii Lineages. Pathogens 2021, 10, 1516. https://doi.org/10.3390/pathogens10111516
Cafiso V, Stracquadanio S, Dovere V, Lo Verde F, Zega A, Pigola G, Barnini S, Ghelardi E, Stefani S. Colistin Resistance Onset Strategies and Genomic Mosaicism in Clinical Acinetobacter baumannii Lineages. Pathogens. 2021; 10(11):1516. https://doi.org/10.3390/pathogens10111516
Chicago/Turabian StyleCafiso, Viviana, Stefano Stracquadanio, Veronica Dovere, Flavia Lo Verde, Alessandra Zega, Giuseppe Pigola, Simona Barnini, Emilia Ghelardi, and Stefania Stefani. 2021. "Colistin Resistance Onset Strategies and Genomic Mosaicism in Clinical Acinetobacter baumannii Lineages" Pathogens 10, no. 11: 1516. https://doi.org/10.3390/pathogens10111516
APA StyleCafiso, V., Stracquadanio, S., Dovere, V., Lo Verde, F., Zega, A., Pigola, G., Barnini, S., Ghelardi, E., & Stefani, S. (2021). Colistin Resistance Onset Strategies and Genomic Mosaicism in Clinical Acinetobacter baumannii Lineages. Pathogens, 10(11), 1516. https://doi.org/10.3390/pathogens10111516






