Host Range, Biology, and Species Specificity of Seven-Segmented Influenza Viruses—A Comparative Review on Influenza C and D
Abstract
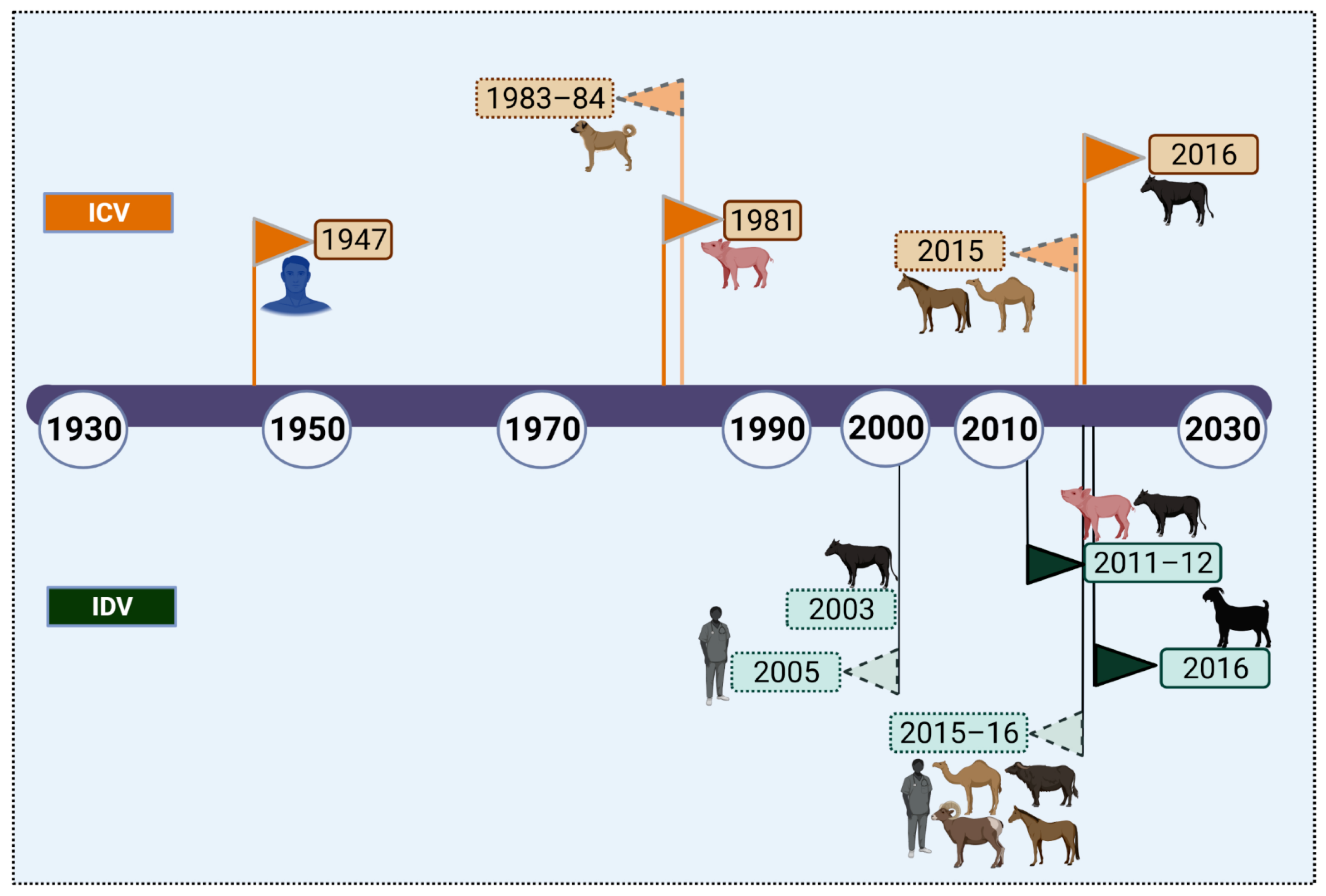
:1. Introduction
2. Cross-Species Transmission of Influenza C and D Viruses
3. Ecology of Seven-Segmented Influenza Viruses
3.1. Natural Hosts of Influenza C and D Viruses
3.1.1. Human
3.1.2. Bovines
Influenza D Virus
Influenza D Linked to Bovine Respiratory Disease
Influenza C Virus
3.1.3. Small Ruminants
3.1.4. Pigs
3.1.5. Dogs
3.1.6. Horses
3.1.7. Camels
3.1.8. Poultry
3.2. Experimental Hosts
3.2.1. Influenza C
3.2.2. Influenza D
4. Clinical Manifestations/Pathobiology of ICV and IDV in Principal Host Species
5. Bottlenecks for Viral Adaptation and Interspecies Transmission
6. Phylodynamics of Seven-Segmented Viruses
7. Reassortment and Species Specificity
7.1. Reassortment between IDV Lineages
7.2. Reassortment between ICV and IDV
8. Broad Cell and Tissue Tropism of IDV over ICV
9. Origin, Classification, and Morphology
10. Genome Structure
10.1. Influenza C Virus
10.2. Influenza D Virus
11. Viral and Host-Determinants of Influenza C and D Replication
11.1. Glycosylation of the HEF as Determinants of Virulence and Transmission
11.2. Receptor Preferences
11.3. 9-O-Acetyl Sialic Acids in Mammalian Tissues and Virus Tropism
11.4. Species-Specific Host Restriction Factors of ICV and IDV
11.5. Antiviral Innate Immunity to ICV and IDV
12. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Potter, C.W. A history of influenza. J. Appl. Microbiol. 2001, 91, 572–579. [Google Scholar] [CrossRef]
- Fields Virology: Emerging Viruses; Wolters Kluwer: Philadelphia, PA, USA, 2021; Volume 1.
- Callan, R.J.; Early, G.; Kida, H.; Hinshaw, V.S. The appearance of H3 influenza viruses in seals. J. Gen. Virol. 1995, 76, 199–203. [Google Scholar] [CrossRef]
- Kawano, J.; Onta, T.; Kida, H.; Yanagawa, R. Distribution of antibodies in animals against influenza B and C viruses. Jpn. J. Vet. Res. 1978, 26, 74–80. [Google Scholar] [PubMed]
- Chang, C.P.; New, A.E.; Taylor, J.F.; Chiang, H.S. Influenza virus isolations from dogs during a human epidemic in Taiwan. Int. J. Zoonoses 1976, 3, 61–64. [Google Scholar] [PubMed]
- Ditchfield, J.; Macpherson, L.W. Zbitnew A: Upper Respiratory Disease in Thouroughbred Horses: Studies of Its Viral Etiology in the Toronto Area, 1960 to 1963. Can. J. Comp. Med. Vet. Sci. 1965, 29, 18–22. [Google Scholar]
- Ohishi, K.; Ninomiya, A.; Kida, H.; Park, C.H.; Maruyama, T.; Arai, T.; Katsumata, E.; Tobayama, T.; Boltunov, A.N.; Khuraskin, L.S.; et al. Serological evidence of transmission of human influenza A and B viruses to Caspian seals (Phoca caspica). Microbiol. Immunol. 2002, 46, 639–644. [Google Scholar] [CrossRef] [PubMed]
- Onta, T.; Kida, H.; Kawano, J.; Matsuoka, Y.; Yanagawa, R. Distribution of antibodies against various influenza A viruses in animals. Nihon Juigaku Zasshi 1978, 40, 451–454. [Google Scholar] [CrossRef]
- Ran, Z.; Shen, H.; Lang, Y.; Kolb, E.A.; Turan, N.; Zhu, L.; Ma, J.; Bawa, B.; Liu, Q.; Liu, H.; et al. Domestic pigs are susceptible to infection with influenza B viruses. J. Virol. 2015, 89, 4818–4826. [Google Scholar] [CrossRef] [Green Version]
- Saito, K. An outbreak of cattle influenza in Japan in the fall of 1949. J. Am. Vet. Med. Assoc. 1951, 118, 316–319. [Google Scholar]
- Sreenivasan, C.C.; Thomas, M.; Kaushik, R.S.; Wang, D.; Li, F. Influenza A in Bovine Species: A Narrative Literature Review. Viruses 2019, 11, 561. [Google Scholar] [CrossRef] [Green Version]
- Lopez, J.W.; Woods, G.T. Influenza virus in ruminants: A review. Res. Commun. Chem. Pathol. Pharmacol. 1984, 45, 445–462. [Google Scholar] [PubMed]
- Taylor, R.M. Studies on survival of influenza virus between epidemics and antigenic variants of the virus. Am. J. Public Health Nations Health 1949, 39, 171–178. [Google Scholar] [CrossRef]
- Hilleman, M.R.; Werner, J.H.; Gauld, R.L. Influenza antibodies in the population of the USA; an epidemiological investigation. Bull. World Health Organ. 1953, 8, 613–631. [Google Scholar]
- Andrews, B.E.; McDonald, J.C. Influenza virus C infection in England. Br. Med. J. 1955, 2, 992–994. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Darke, C.S.; Watkins, P.H.; Whitehead, J.E. Fulminating staphylococcal pneumonia associated with influenza virus C: Report of a fatal case. Br. Med. J. 1957, 2, 606–609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grist, N.R. Influenza A and C in Glasgow, 1954. Br. Med. J. 1955, 2, 994–997. [Google Scholar] [CrossRef]
- Styk, B. Epidemic outbreak of influenza caused by virus C. Zh. Mikrobiol. Epidemiol. Immunobiol. 1955, 6, 68–75. [Google Scholar]
- Webster, R.G. Influenza Viruses (Orthomyxoviridae) | General Features. In Encyclopedia of Virology, 2nd ed.; Granoff, A., Webster, R.G., Eds.; Elsevier: Oxford, UK, 1999; pp. 824–829. [Google Scholar]
- Manuguerra, J.C.; Hannoun, C. Natural infection of dogs by influenza C virus. Res. Virol. 1992, 143, 199–204. [Google Scholar] [CrossRef]
- Ohwada, K.; Kitame, F.; Sugawara, K.; Nishimura, H.; Homma, M.; Nakamura, K. Distribution of the antibody to influenza C virus in dogs and pigs in Yamagata Prefecture, Japan. Microbiol. Immunol. 1987, 31, 1173–1180. [Google Scholar] [CrossRef]
- Zhang, H.; Porter, E.; Lohman, M.; Lu, N.; Peddireddi, L.; Hanzlicek, G.; Marthaler, D.; Liu, X.; Bai, J. Influenza C Virus in Cattle with Respiratory Disease, United States, 2016-2018. Emerg. Infect. Dis. 2018, 24, 1926–1929. [Google Scholar] [CrossRef]
- Trombetta, C.M.; Marchi, S.; Manini, I.; Kistner, O.; Li, F.; Piu, P.; Manenti, A.; Biuso, F.; Sreenivasan, C.; Druce, J.; et al. Influenza D Virus: Serological Evidence in the Italian Population from 2005 to 2017. Viruses 2019, 12, 30. [Google Scholar] [CrossRef] [Green Version]
- Borkenhagen, L.K.; Mallinson, K.A.; Tsao, R.W.; Ha, S.J.; Lim, W.H.; Toh, T.H.; Anderson, B.D.; Fieldhouse, J.K.; Philo, S.E.; Chong, K.S.; et al. Surveillance for respiratory and diarrheal pathogens at the human-pig interface in Sarawak, Malaysia. PLoS ONE 2018, 13, e0201295. [Google Scholar] [CrossRef]
- Bailey, E.S.; Choi, J.Y.; Zemke, J.; Yondon, M.; Gray, G.C. Molecular surveillance of respiratory viruses with bioaerosol sampling in an airport. Trop. Dis. Travel Med. Vaccines 2018, 4, 11. [Google Scholar] [CrossRef] [PubMed]
- White, S.K.; Ma, W.; McDaniel, C.J.; Gray, G.C.; Lednicky, J.A. Serologic evidence of exposure to influenza D virus among persons with occupational contact with cattle. J. Clin. Virol. Off. Publ. Pan Am. Soc. Clin. Virol. 2016, 81, 31–33. [Google Scholar] [CrossRef] [PubMed]
- Quast, M.; Sreenivasan, C.; Sexton, G.; Nedland, H.; Singrey, A.; Fawcett, L.; Miller, G.; Lauer, D.; Voss, S.; Pollock, S.; et al. Serological evidence for the presence of influenza D virus in small ruminants. Vet. Microbiol. 2015, 180, 281–285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bailey, E.S.; Fieldhouse, J.K.; Alarja, N.A.; Chen, D.D.; Kovalik, M.E.; Zemke, J.N.; Choi, J.Y.; Borkenhagen, L.K.; Toh, T.-H.; Lee, J.S.Y.; et al. First sequence of influenza D virus identified in poultry farm bioaerosols in Sarawak, Malaysia. Trop. Dis. Travel Med. Vaccines 2020, 6, 5. [Google Scholar] [CrossRef] [PubMed]
- Eda, S.; Suzuki, Y.; Kase, T.; Kawai, T.; Ohtani, K.; Sakamoto, T.; Kurimura, T.; Wakamiya, N. Recombinant bovine conglutinin, lacking the N-terminal and collagenous domains, has less conglutination activity but is able to inhibit haemagglutination by influenza A virus. Biochem J. 1996, 316 Pt 1, 43–48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Superti, F.; Agamennone, M.; Pietrantoni, A.; Ammendolia, M.G. Bovine Lactoferrin Prevents Influenza A Virus Infection by Interfering with the Fusogenic Function of Viral Hemagglutinin. Viruses 2019, 11, 51. [Google Scholar] [CrossRef] [Green Version]
- Wakamiya, N.; Okuno, Y.; Sasao, F.; Ueda, S.; Yoshimatsu, K.; Naiki, M.; Kurimura, T. Isolation and characterization of conglutinin as an influenza A virus inhibitor. Biochem. Biophys. Res. Commun. 1992, 187, 1270–1278. [Google Scholar] [CrossRef]
- Salem, E.; Cook, E.A.J.; Lbacha, H.A.; Oliva, J.; Awoume, F.; Aplogan, G.L.; Hymann, E.C.; Muloi, D.; Deem, S.L.; Alali, S.; et al. Serologic Evidence for Influenza C and D Virus among Ruminants and Camelids, Africa, 1991–2015. Emerg. Infect. Dis. 2017, 23, 1556–1559. [Google Scholar] [CrossRef] [Green Version]
- Helten, A.; Marschall, M.; Reininger, A.J.; Meier-Ewert, H. Experimental infection with a persistent influenza C virus variant leads to prolonged genome detection in the chicken lung. Acta. Virol. 1996, 40, 223–226. [Google Scholar]
- Yu, J.; Hika, B.; Liu, R.; Sheng, Z.; Hause, B.M.; Li, F.; Wang, D. Complete Genome Sequence of an Influenza D Virus Strain Identified in a Pig with Subclinical Infection in the United States. mSphere 2017, 2. [Google Scholar] [CrossRef] [Green Version]
- Guo, Y.J.; Jin, F.G.; Wang, P.; Wang, M.; Zhu, J.M. Isolation of influenza C virus from pigs and experimental infection of pigs with influenza C virus. J. Gen. Virol. 1983, 64, 177–182. [Google Scholar] [CrossRef]
- Yamaoka, M.; Hotta, H.; Itoh, M.; Homma, M. Prevalence of antibody to influenza C virus among pigs in Hyogo Prefecture, Japan. J. Gen. Virol. 1991, 72, 711–714. [Google Scholar] [CrossRef]
- Yuanji, G.; Desselberger, U. Genome analysis of influenza C viruses isolated in 1981/82 from pigs in China. J. Gen. Virol. 1984, 65, 1857–1872. [Google Scholar] [CrossRef] [PubMed]
- Nissly, R.H.; Zaman, N.; Ibrahim, P.A.S.; McDaniel, K.; Lim, L.; Kiser, J.N.; Bird, I.; Chothe, S.K.; Bhushan, G.L.; Vandegrift, K.; et al. Influenza C and D viral load in cattle correlates with bovine respiratory disease (BRD): Emerging role of orthomyxoviruses in the pathogenesis of BRD. Virology 2020, 551, 10–15. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, I.J.; Fort, M.; Pasucci, J.; Moreno, F.; Gimenez, H.; Näslund, K.; Hägglund, S.; Zohari, S.; Valarcher, J.F. Seroprevalence of influenza D virus in bulls in Argentina. J. Vet. Diagn. Investig. 2020, 32, 585–588. [Google Scholar] [CrossRef] [PubMed]
- Flynn, O.; Gallagher, C.; Mooney, J.; Irvine, C.; Ducatez, M.; Hause, B.; McGrath, G.; Ryan, E. Influenza D Virus in Cattle, Ireland. Emerg. Infect. Dis. 2018, 24, 389–391. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horimoto, T.; Hiono, T.; Mekata, H.; Odagiri, T.; Lei, Z.; Kobayashi, T.; Norimine, J.; Inoshima, Y.; Hikono, H.; Murakami, K.; et al. Nationwide Distribution of Bovine Influenza D Virus Infection in Japan. PLoS ONE 2016, 11, e0163828. [Google Scholar] [CrossRef] [Green Version]
- Murakami, S.; Odagiri, T.; Melaku, S.K.; Bazartseren, B.; Ishida, H.; Takenaka-Uema, A.; Muraki, Y.; Sentsui, H.; Horimoto, T. Influenza D Virus Infection in Dromedary Camels, Ethiopia. Emerg. Infect. Dis. 2019, 25, 1224–1226. [Google Scholar] [CrossRef] [PubMed]
- Snoeck, C.J.; Oliva, J.; Pauly, M.; Losch, S.; Wildschutz, F.; Muller, C.P.; Hubschen, J.M.; Ducatez, M.F. Influenza D Virus Circulation in Cattle and Swine, Luxembourg, 2012–2016. Emerg. Infect. Dis. 2018, 24, 1388–1389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thielen, P.; Nolting, J.M.; Nelson, S.W.; Mehoke, T.S.; Howser, C.; Bowman, A.S. Complete Genome Sequence of an Influenza D Virus Strain Identified in a Pig with Subclinical Infection in the United States. Microbiol. Resour. Announc. 2019, 8, e01462-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhai, S.L.; Zhang, H.; Chen, S.N.; Zhou, X.; Lin, T.; Liu, R.; Lv, D.H.; Wen, X.H.; Wei, W.K.; Wang, D.; et al. Influenza D Virus in Animal Species in Guangdong Province, Southern China. Emerg. Infect. Dis. 2017, 23, 1392–1396. [Google Scholar] [CrossRef]
- Nedland, H.; Wollman, J.; Sreenivasan, C.; Quast, M.; Singrey, A.; Fawcett, L.; Christopher-Hennings, J.; Nelson, E.; Kaushik, R.S.; Wang, D.; et al. Serological evidence for the co-circulation of two lineages of influenza D viruses in equine populations of the Midwest United States. Zoonoses Public Health 2018, 65, e148–e154. [Google Scholar] [CrossRef]
- Taylor, R.M. A further note on 1233 influenza C virus. Arch Gesamte Virusforsch 1951, 4, 485–500. [Google Scholar] [CrossRef]
- Francis, T.; Quilligan, J.J.; Minuse, E. Identification of Another Epidemic Respiratory Disease. Science 1950, 112, 495. [Google Scholar] [CrossRef] [PubMed]
- Cabezas, J.A.; Villar, E.; García-Sastre, A.; Manuguerra, J.C.; Hannoun, C. New data on influenza virus type C confirm its peculiarities as a new genus. Intervirology 1991, 32, 325–326. [Google Scholar] [CrossRef]
- Calvo, C.; Garcia-Garcia, M.L.; Borrell, B.; Pozo, F.; Casas, I. Prospective study of influenza C in hospitalized children. Pediatric Infect. Dis. J. 2013, 32, 916–919. [Google Scholar] [CrossRef]
- Principi, N.; Scala, A.; Daleno, C.; Esposito, S. Influenza C virus-associated community-acquired pneumonia in children. Influenza Other Respir. Viruses 2013, 7, 999–1003. [Google Scholar] [CrossRef] [Green Version]
- Moriuchi, H.; Katsushima, N.; Nishimura, H.; Nakamura, K.; Numazaki, Y. Community-acquired influenza C virus infection in children. J. Pediatr. 1991, 118, 235–238. [Google Scholar] [CrossRef]
- Takagi, Y.; Imamura, T.; Endo, S.; Hayashi, K.; Akiyama, S.; Ikuta, Y.; Kawaguchi, T.; Sumita, T.; Katori, T.; Hashino, M.; et al. Neurogenic pulmonary edema following febrile status epilepticus in a 22-month-old infant with multiple respiratory virus co-detection: A case report. BMC Infect. Dis. 2020, 20, 388. [Google Scholar] [CrossRef]
- Minuse, E.; Davenport, F.M. Simultaneous recovery of type A' and type C influenza viruses from a patient. J. Lab. Clin. Med. 1951, 38, 747–750. [Google Scholar]
- Atsumi, A.; Ono, Y.; Ishida, N.; Konno, J.; Tada, K. Isolation of influenza C virus. Nihon Saikingaku Zasshi 1966, 147–150. [Google Scholar] [CrossRef]
- Chakraverty, P.; Adhami, Z.; Wise, R.; Mathews, R.S. Influenza C virus in the United Kingdom. J. Infect. 1984, 8, 177–178. [Google Scholar] [CrossRef]
- Farkas, E.; Domok, I. Isolation of influenza C virus in Hungary. Acta. Microbiol. Acad. Sci. Hung. 1954, 1, 85–97. [Google Scholar] [PubMed]
- Iatel, T.P. Biological and antigenic properties of influenza virus C isolated in Kiev in 1956. Mikrobiol. Zh. 1958, 20, 56–64. [Google Scholar]
- Lee, H.S.; Lim, S.; Noh, J.Y.; Song, J.Y.; Cheong, H.J.; Lee, J.H.; Woo, S.I.; Kim, W.J. Identification of influenza C virus in young South Korean children, from October 2013 to September 2016. J. Clin. Virol. Off. Publ. Pan Am. Soc. Clin. Virol. 2019, 115, 47–52. [Google Scholar] [CrossRef]
- Wilczyński, J.; Jankowski, M.; Torbicka, E.; Tranda, I.; Roszkowska-Sliz, L. Respiratory viral infections in young children 1988–1990. Przegl. Epidemiol. 1990, 44, 293–297. [Google Scholar]
- El-Rai, F.M.; Shaheen, Y.A.; El-Diwani, K.M.; Abdel-Al, M.H.; Imam, I.Z.; Hosny, A.H.; El-Senousy, A.A. The incidence of antibodies to influenza C virus in Egyptian sera. J. Egypt. Public Health Assoc. 1977, 52, 6–11. [Google Scholar]
- Dykes, A.C.; Cherry, J.D.; Nolan, C.E. A clinical, epidemiologic, serologic, and virologic study of influenza C virus infection. Arch. Intern. Med. 1980, 140, 1295–1298. [Google Scholar] [CrossRef]
- O'Callaghan, R.J.; Gohd, R.S.; Labat, D.D. Human antibody to influenza C virus: Its age-related distribution and distinction from receptor analogs. Infect. Immun. 1980, 30, 500–505. [Google Scholar] [CrossRef]
- Liao, F.; Nishimura, H.; Ito, H.; Zhang, Y.; Matsuzaki, Y. Longitudinal course of influenza C virus antibody titers of healthy adults in Sendai, Japan. J. Clin. Virol. Off. Publ. Pan Am. Soc. Clin. Virol. 2020, 133, 104662. [Google Scholar] [CrossRef]
- Daniels, R.S.; Tse, H.; Ermetal, B.; Xiang, Z.; Jackson, D.J.; Guntoro, J.; Nicod, J.; Stewart, A.; Cross, K.J.; Hussain, S.; et al. Molecular Characterization of Influenza C Viruses from Outbreaks in Hong Kong SAR, China. J. Virol. 2020, 94, e01051-20. [Google Scholar] [CrossRef]
- Kaji, M.; Hiromatsu, Y.; Kashiwagi, S.; Hayashi, J.; Oyama, S.; Katagiri, S.; Homma, M. Distribution of antibodies to influenza C virus. Kurume. Med. J. 1983, 30, 121–123. [Google Scholar] [CrossRef]
- Govorkova, E.A.; Zakstel'skaia, L.; Demidova, S.A. Detection of influenza virus C antibodies in the gamma globulins and native sera of children. Vopr. Virusol. 1984, 29, 420–423. [Google Scholar]
- Vasil'eva, V.I.; Zakstel'skaia, L.; Govorkova, E.A.; Rusakova, E.V.; Alekseenkova, L.I. Immunostructure of the population to the influenza C virus. Vopr. Virusol. 1985, 30, 661–664. [Google Scholar]
- Manuguerra, J.C.; Hannoun, C.; Aymard, M. Influenza C virus infection in France. J. Infect. 1992, 24, 91–99. [Google Scholar] [CrossRef]
- Manuguerra, J.C.; Hannoun, C.; Simón, F.; Villar, E.; Cabezas, J.A. Natural infection of dogs by influenza C virus: A serological survey in Spain. New Microbiol. 1993, 16, 367–371. [Google Scholar]
- Youzbashi, E.; Marschall, M.; Chaloupka, I.; Meier-Ewert, H. Distribution of influenza C virus infection in dogs and pigs in Bavaria. Tierarztl. Prax. 1996, 24, 337–342. [Google Scholar]
- Motta, F.C.; Luiz, M.O.; Couceiro, J.N. Serological analysis reveals circulation of influenza C viruses, Brazil. Rev. Saude. Publica. 2000, 34, 204–205. [Google Scholar] [CrossRef] [Green Version]
- Smith, D.B.; Gaunt, E.R.; Digard, P.; Templeton, K.; Simmonds, P. Detection of influenza C virus but not influenza D virus in Scottish respiratory samples. J. Clin. Virol. Off. Publ. Pan Am. Soc. Clin. Virol. 2016, 74, 50–53. [Google Scholar] [CrossRef] [Green Version]
- Holwerda, M.; Kelly, J.; Laloli, L.; Stürmer, I.; Portmann, J.; Stalder, H.; Dijkman, R. Determining the Replication Kinetics and Cellular Tropism of Influenza D Virus on Primary Well-Differentiated Human Airway Epithelial Cells. Viruses 2019, 11, 377. [Google Scholar] [CrossRef] [Green Version]
- Song, H.; Qi, J.; Khedri, Z.; Diaz, S.; Yu, H.; Chen, X.; Varki, A.; Shi, Y.; Gao, G.F. An Open Receptor-Binding Cavity of Hemagglutinin-Esterase-Fusion Glycoprotein from Newly-Identified Influenza D Virus: Basis for Its Broad Cell Tropism. PLoS Pathog. 2016, 12, e1005411. [Google Scholar] [CrossRef] [Green Version]
- Murakami, S.; Sato, R.; Ishida, H.; Katayama, M.; Takenaka-Uema, A.; Horimoto, T. Influenza D Virus of New Phylogenetic Lineage, Japan. Emerg. Infect. Dis. 2020, 26, 168–171. [Google Scholar] [CrossRef] [Green Version]
- Chiapponi, C.; Faccini, S.; De Mattia, A.; Baioni, L.; Barbieri, I.; Rosignoli, C.; Nigrelli, A.; Foni, E. Detection of Influenza D Virus among Swine and Cattle, Italy. Emerg. Infect. Dis. 2016, 22, 352–354. [Google Scholar] [CrossRef] [Green Version]
- Foni, E.; Chiapponi, C.; Baioni, L.; Zanni, I.; Merenda, M.; Rosignoli, C.; Kyriakis, C.S.; Luini, M.V.; Mandola, M.L.; Bolzoni, L.; et al. Influenza D in Italy: Towards a better understanding of an emerging viral infection in swine. Sci. Rep. 2017, 7, 11660. [Google Scholar] [CrossRef] [Green Version]
- Ducatez, M.F.; Pelletier, C.; Meyer, G. Influenza D virus in cattle, France, 2011–2014. Emerg. Infect. Dis. 2015, 21, 368–371. [Google Scholar] [CrossRef]
- Oliva, J.; Eichenbaum, A.; Belin, J.; Gaudino, M.; Guillotin, J.; Alzieu, J.P.; Nicollet, P.; Brugidou, R.; Gueneau, E.; Michel, E.; et al. Serological Evidence of Influenza D Virus Circulation Among Cattle and Small Ruminants in France. Viruses 2019, 11, 516. [Google Scholar] [CrossRef] [Green Version]
- Sanogo, I.N.; Kouakou, C.; Batawui, K.; Djegui, F.; Byarugaba, D.K.; Adjin, R.; Adjabli, K.; Wabwire-Mangen, F.; Erima, B.; Atim, G.; et al. Serological Surveillance of Influenza D Virus in Ruminants and Swine in West and East Africa, 2017–2020. Viruses 2021, 13, 1749. [Google Scholar] [CrossRef]
- Hause, B.M.; Ducatez, M.; Collin, E.A.; Ran, Z.; Liu, R.; Sheng, Z.; Armien, A.; Kaplan, B.; Chakravarty, S.; Hoppe, A.D.; et al. Isolation of a novel swine influenza virus from Oklahoma in 2011 which is distantly related to human influenza C viruses. PLoS Pathog. 2013, 9, e1003176. [Google Scholar] [CrossRef] [Green Version]
- Luo, J.; Ferguson, L.; Smith, D.R.; Woolums, A.R.; Epperson, W.B.; Wan, X.F. Serological evidence for high prevalence of Influenza D Viruses in Cattle, Nebraska, United States, 2003–2004. Virology 2017, 501, 88–91. [Google Scholar] [CrossRef]
- Ferguson, L.; Eckard, L.; Epperson, W.B.; Long, L.P.; Smith, D.; Huston, C.; Genova, S.; Webby, R.; Wan, X.F. Influenza D virus infection in Mississippi beef cattle. Virology 2015, 486, 28–34. [Google Scholar] [CrossRef] [Green Version]
- USDA. Feedlot 2011. Part IV: Health and Health Management on U.S. Feedlots with a Capacity of 1000 or More Head; USDA: Washington, DC, USA, 2011.
- Collin, E.A.; Sheng, Z.; Lang, Y.; Ma, W.; Hause, B.M.; Li, F. Cocirculation of two distinct genetic and antigenic lineages of proposed influenza D virus in cattle. J. Virol. 2015, 89, 1036–1042. [Google Scholar] [CrossRef] [Green Version]
- Mitra, N.; Cernicchiaro, N.; Torres, S.; Li, F.; Hause, B.M. Metagenomic characterization of the virome associated with bovine respiratory disease in feedlot cattle identified novel viruses and suggests an etiologic role for influenza D virus. J. Gen. Virol. 2016, 97, 1771–1784. [Google Scholar] [CrossRef]
- Studer, E.; Schönecker, L.; Meylan, M.; Stucki, D.; Dijkman, R.; Holwerda, M.; Glaus, A.; Becker, J. Prevalence of BRD-Related Viral Pathogens in the Upper Respiratory Tract of Swiss Veal Calves. Animals 2021, 11, 1940. [Google Scholar] [CrossRef]
- Zhang, M.; Hill, J.E.; Alexander, T.W.; Huang, Y. The nasal viromes of cattle on arrival at western Canadian feedlots and their relationship to development of bovine respiratory disease. Transbound. Emerg. Dis. 2021, 68, 2209–2218. [Google Scholar] [CrossRef]
- Zhang, M.; Hill, J.E.; Fernando, C.; Alexander, T.W.; Timsit, E.; van der Meer, F.; Huang, Y. Respiratory viruses identified in western Canadian beef cattle by metagenomic sequencing and their association with bovine respiratory disease. Transbound. Emerg. Dis. 2019, 66, 1379–1386. [Google Scholar] [CrossRef]
- Zhang, M.; Hill, J.E.; Godson, D.L.; Ngeleka, M.; Fernando, C.; Huang, Y. The pulmonary virome, bacteriological and histopathological findings in bovine respiratory disease from western Canada. Transbound. Emerg. Dis. 2019, 67, 924–934. [Google Scholar] [CrossRef]
- Yilmaz, A.; Umar, S.; Turan, N.; Aydin, O.; Tali, H.E.; Oguzoglu, T.C.; Yilmaz, H.; Richt, J.A.; Ducatez, M.F. First report of influenza D virus infection in Turkish cattle with respiratory disease. Res. Vet. Sci. 2020, 130, 98–102. [Google Scholar] [CrossRef]
- Trombetta, C.M.; Montomoli, E.; Di Bartolo, I.; Ostanello, F.; Chiapponi, C.; Marchi, S. Detection of antibodies against influenza D virus in swine veterinarians in Italy in 2004. J. Med Virol. 2021. [Google Scholar] [CrossRef]
- O'Donovan, T.; Donohoe, L.; Ducatez, M.F.; Meyer, G.; Ryan, E. Seroprevalence of influenza D virus in selected sample groups of Irish cattle, sheep and pigs. Ir. Vet. J. 2019, 72, 11. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Porter, E.P.; Lohman, M.; Lu, N.; Peddireddi, L.; Hanzlicek, G.; Marthaler, D.; Liu, X.; Bai, J. Complete Genome Sequence of an Influenza C Virus Strain Identified from a Sick Calf in the United States. Microbiol. Resour. Announc. 2018, 7, e00828-18. [Google Scholar] [CrossRef] [Green Version]
- Ferguson, L.; Luo, K.; Olivier, A.K.; Cunningham, F.L.; Blackmon, S.; Hanson-Dorr, K.; Sun, H.; Baroch, J.; Lutman, M.W.; Quade, B.; et al. Influenza D Virus Infection in Feral Swine Populations, United States. Emerg. Infect. Dis. 2018, 24, 1020–1028. [Google Scholar] [CrossRef] [Green Version]
- Lauterbach, S.E.; Nelson, S.W.; Robinson, M.E.; Lorbach, J.N.; Nolting, J.M.; Bowman, A.S. Assessing exhibition swine as potential disseminators of infectious disease through the detection of five respiratory pathogens at agricultural exhibitions. Vet. Res. 2019, 50, 63. [Google Scholar] [CrossRef] [Green Version]
- Farrell, A.S.; Bui, V.N.; Dao, T.D.; Hoang, T.D.; Gray, G.C. No influenza D virus detected among pigs, northern Vietnam. Influenza Other Respir. Viruses 2021, 15, 315–317. [Google Scholar] [CrossRef]
- Anderson, B.D.; Yondon, M.; Bailey, E.S.; Duman, E.K.; Simmons, R.A.; Greer, A.G.; Gray, G.C. Environmental bioaerosol surveillance as an early warning system for pathogen detection in North Carolina swine farms: A pilot study. Transbound. Emerg. Dis. 2021, 68, 361–367. [Google Scholar] [CrossRef]
- More, G.D.; Cave, N.J.; Biggs, P.J.; Acke, E.; Dunowska, M. A molecular survey of canine respiratory viruses in New Zealand. N. Z. Vet. J. 2021, 69, 224–233. [Google Scholar] [CrossRef]
- Blouse, L.E.; Buddingh, G.J. Experimental pneumococcal infection in embryonated eggs previously infected with influenza C virus. Infect. Immun. 1972, 5, 688–694. [Google Scholar] [CrossRef] [Green Version]
- Buddingh, G.J. Bacterial Dynamics in Combined Infection. A study of the population dynamics of strains of hemophilus influenzae type b in combined infection with influenza c virus in embryonated eggs. Am. J. Pathol. 1963, 43, 407–418. [Google Scholar]
- Buddingh, G.J. Experimental combined viral and bacterial infection (influenza C and hemophilus influenzae, type B) in embryonated eggs. J. Exp. Med. 1956, 104, 947–958. [Google Scholar] [CrossRef] [Green Version]
- Buddingh, G.J.; Buff, E.E. Inhibition of Influenza G Virus by Hemophilus influenzae in Embryonated Eggs. Proc. Soc. Exp. Biol. Med. 1965, 118, 94–96. [Google Scholar] [CrossRef] [PubMed]
- Glezen, W.P. Influenza C virus infection. Arch. Intern. Med. 1980, 140, 1278. [Google Scholar] [CrossRef] [PubMed]
- Inoue, H.; Horio, S.; Ichinose, M.; Ida, S.; Hida, W.; Takishima, T.; Ohwada, K.; Homma, M. Changes in bronchial reactivity to acetylcholine with type C influenza virus infection in dogs. Am. Rev. Respir. Dis. 1986, 133, 367–371. [Google Scholar] [CrossRef] [PubMed]
- Ohwada, K.; Kitame, F.; Homma, M. Experimental infections of dogs with type C influenza virus. Microbiol. Immunol. 1986, 30, 451–460. [Google Scholar] [CrossRef] [Green Version]
- Miura, M.; Inoue, H.; Ichinose, M.; Shimura, S.; Katsumata, U.; Kimura, K.; Shindoh, Y.; Tanno, Y.; Takishima, T. Increase in luminal mast cell and epithelial damage may account for increased airway responsiveness after viral infection in dogs. Am. Rev. Respir. Dis. 1989, 140, 1738–1744. [Google Scholar] [CrossRef]
- Takiguchi, K.; Tashiro, M.; Nakamura, K. Influenza C virus infection in rats. Microbiol. Immunol. 1990, 34, 35–44. [Google Scholar] [CrossRef]
- Takiguchi, K.; Sugawara, K.; Hongo, S.; Nishimura, H.; Kitame, F.; Nakamura, K. Protective effect of serum antibody on respiratory infection of influenza C virus in rats. Arch. Virol. 1992, 122, 1–11. [Google Scholar] [CrossRef]
- Parker, M.S.; O'Callaghan, R.J.; Smith, D.E.; Spence, H.A. The effect of influenza C virus on the Purkinje cells of chick embryo cerebellum. Int. J. Dev. Neurosci. 1994, 12, 461–470. [Google Scholar] [CrossRef]
- Joosting, A.C.; Head, B.; Bynoe, M.L.; Tyrrell, D.A. Production of common colds in human volunteers by influenza C virus. Br. Med. J. 1968, 4, 153–154. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.; Wang, L.; Palinski, R.; Walsh, T.; He, D.; Li, Y.; Wu, R.; Lang, Y.; Sunwoo, S.Y.; Richt, J.A.; et al. Comparison of Pathogenicity and Transmissibility of Influenza B and D Viruses in Pigs. Viruses 2019, 11, 905. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hause, B.M.; Huntimer, L.; Falkenberg, S.; Henningson, J.; Lechtenberg, K.; Halbur, T. An inactivated influenza D virus vaccine partially protects cattle from respiratory disease caused by homologous challenge. Vet. Microbiol. 2017, 199, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Sreenivasan, C.; Thomas, M.; Sheng, Z.; Hause, B.M.; Collin, E.A.; Knudsen, D.E.; Pillatzki, A.; Nelson, E.; Wang, D.; Kaushik, R.S.; et al. Replication and Transmission of the Novel Bovine Influenza D Virus in a Guinea Pig Model. J. Virol. 2015, 89, 11990–12001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oliva, J.; Mettier, J.; Sedano, L.; Delverdier, M.; Bourges-Abella, N.; Hause, B.; Loupias, J.; Pardo, I.; Bleuart, C.; Bordignon, P.J.; et al. A murine model for the study of influenza D virus. J. Virol. 2019, 94, e01662-19. [Google Scholar] [CrossRef] [Green Version]
- Kauppila, J.; Ronkko, E.; Juvonen, R.; Saukkoriipi, A.; Saikku, P.; Bloigu, A.; Vainio, O.; Ziegler, T. Influenza C virus infection in military recruits--symptoms and clinical manifestation. J. Med Virol. 2014, 86, 879–885. [Google Scholar] [CrossRef]
- Pfeifer, J.B.; Compans, R.W. Structure and variation of the influenza C glycoprotein. Vaccine 1985, 3, 189–194. [Google Scholar] [CrossRef]
- Katagiri, S.; Ohizumi, A.; Ohyama, S.; Homma, M. Follow-up study of type C influenza outbreak in a children’s home. Microbiol. Immunol. 1987, 31, 337–343. [Google Scholar] [CrossRef]
- Calvo, C.; Garcia-Garcia, M.L.; Centeno, M.; Perez-Brena, P.; Casas, I. Influenza C virus infection in children, Spain. Emerg. Infect. Dis. 2006, 12, 1621–1622. [Google Scholar] [CrossRef]
- Shimizu, Y.; Abiko, C.; Ikeda, T.; Mizuta, K.; Matsuzaki, Y. Influenza C Virus and Human Metapneumovirus Infections in Hospitalized Children With Lower Respiratory Tract Illness. Pediatric Infect. Dis. J. 2015, 34, 1273–1275. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, L.; Olivier, A.K.; Genova, S.; Epperson, W.B.; Smith, D.R.; Schneider, L.; Barton, K.; McCuan, K.; Webby, R.J.; Wan, X.F. Pathogenesis of Influenza D Virus in Cattle. J. Virol. 2016, 90, 5636–5642. [Google Scholar] [CrossRef] [Green Version]
- Kaplan, B.S.; Falkenberg, S.; Dassanayake, R.; Neill, J.; Velayudhan, B.; Li, F.; Vincent, A.L. Virus strain influenced the interspecies transmission of influenza D virus between calves and pigs. Transbound. Emerg. Dis. 2020. [Google Scholar] [CrossRef]
- Saegerman, C.; Bianchini, J.; Snoeck, C.J.; Moreno, A.; Chiapponi, C.; Zohari, S.; Ducatez, M.F. First expert elicitation of knowledge on drivers of emergence of influenza D in Europe. Transbound. Emerg. Dis. 2021, 68, 3349–3359. [Google Scholar] [CrossRef]
- Nakada, S.; Creager, R.S.; Krystal, M.; Aaronson, R.P.; Palese, P. Influenza C virus hemagglutinin: Comparison with influenza A and B virus hemagglutinins. J. Virol. 1984, 50, 118–124. [Google Scholar] [CrossRef] [Green Version]
- Pfeifer, J.B.; Compans, R.W. Structure of the influenza C glycoprotein gene as determined from cloned DNA. Virus Res. 1984, 1, 281–296. [Google Scholar] [CrossRef]
- Njouom, R.; Monamele, G.C.; Ermetal, B.; Tchatchouang, S.; Moyo-Tetang, S.; McCauley, J.W.; Daniels, R.S. Detection of Influenza C Virus Infection among Hospitalized Patients, Cameroon. Emerg. Infect. Dis. 2019, 25, 607–609. [Google Scholar] [CrossRef] [Green Version]
- Kawamura, H.; Tashiro, M.; Kitame, F.; Homma, M.; Nakamura, K. Genetic variation among human strains of influenza C virus isolated in Japan. Virus Res. 1986, 4, 275–288. [Google Scholar] [CrossRef]
- Adachi, K.; Kitame, F.; Sugawara, K.; Nishimura, H.; Nakamura, K. Antigenic and genetic characterization of three influenza C strains isolated in the Kinki district of Japan in 1982–1983. Virology 1989, 172, 125–133. [Google Scholar] [CrossRef]
- Fritsch, A.; Schweiger, B.; Biere, B. Influenza C virus in pre-school children with respiratory infections: Retrospective analysis of data from the national influenza surveillance system in Germany, 2012 to 2014. Euro Surveill 2019, 24, 1800174. [Google Scholar] [CrossRef] [Green Version]
- Thielen, B.K.; Friedlander, H.; Bistodeau, S.; Shu, B.; Lynch, B.; Martin, K.; Bye, E.; Como-Sabetti, K.; Boxrud, D.; Strain, A.K.; et al. Detection of Influenza C Viruses Among Outpatients and Patients Hospitalized for Severe Acute Respiratory Infection, Minnesota, 2013-2016. Clin. Infect. Dis. 2018, 66, 1092–1098. [Google Scholar] [CrossRef] [PubMed]
- Potdar, V.A.; Hinge, D.D.; Dakhave, M.R.; Manchanda, A.; Jadhav, N.; Kulkarni, P.B.; Chadha, M.S. Molecular detection and characterization of Influenza ‘C’ viruses from western India. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2017, 54, 466–477. [Google Scholar] [CrossRef] [PubMed]
- Ting, P.J.; Seah, S.G.; Lim, E.A.; Liaw, J.C.; Boon-Huan, T. Genetic characterisation of influenza C viruses detected in Singapore in 2006. Influenza Other Respir. Viruses 2016, 10, 27–33. [Google Scholar] [CrossRef]
- Anton, A.; Marcos, M.A.; Codoner, F.M.; de Molina, P.; Martinez, A.; Cardenosa, N.; Godoy, P.; Torner, N.; Martinez, M.J.; Ramon, S.; et al. Influenza C virus surveillance during the first influenza A (H1N1) 2009 pandemic wave in Catalonia, Spain. Diagn. Microbiol. Infect. Dis. 2011, 69, 419–427. [Google Scholar] [CrossRef]
- Jelley, L.; Levy, A.; Deng, Y.M.; Spirason, N.; Lang, J.; Buettner, I.; Druce, J.; Blyth, C.; Effler, P.; Smith, D.; et al. Influenza C infections in Western Australia and Victoria from 2008 to 2014. Influenza Other Respir. Viruses 2016, 10, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Mekata, H.; Yamamoto, M.; Hamabe, S.; Tanaka, H.; Omatsu, T.; Mizutani, T.; Hause, B.M.; Okabayashi, T. Molecular epidemiological survey and phylogenetic analysis of bovine influenza D virus in Japan. Transbound. Emerg. Dis. 2018, 65, e355–e360. [Google Scholar] [CrossRef]
- Hause, B.M.; Collin, E.A.; Liu, R.; Huang, B.; Sheng, Z.; Lu, W.; Wang, D.; Nelson, E.A.; Li, F. Characterization of a novel influenza virus in cattle and Swine: Proposal for a new genus in the Orthomyxoviridae family. mBio 2014, 5, e00031-14. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.; Yu, J.; Hause, B.M.; Park, J.Y.; Sreenivasan, C.; Uprety, T.; Sheng, Z.; Wang, D.; Li, F. Emergence of new phylogenetic lineage of Influenza D virus with broad antigenicity in California, United States. Emerg. Microbes. Infect. 2021, 10, 739–742. [Google Scholar] [CrossRef]
- Meier-Ewert, H.; Petri, T.; Bishop, D.H. Oligonucleotide fingerprint analyses of influenza C virion RNA recovered from five different isolates. Arch. Virol. 1981, 67, 141–147. [Google Scholar] [CrossRef]
- Zhang, W.; Zhang, L.; He, W.; Zhang, X.; Wen, B.; Wang, C.; Xu, Q.; Li, G.; Zhou, J.; Veit, M.; et al. Genetic Evolution and Molecular Selection of the HE Gene of Influenza C Virus. Viruses 2019, 11, 167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Furuse, Y.; Matsuzaki, Y.; Nishimura, H.; Oshitani, H. Analyses of Evolutionary Characteristics of the Hemagglutinin-Esterase Gene of Influenza C Virus during a Period of 68 Years Reveals Evolutionary Patterns Different from Influenza A and B Viruses. Viruses 2016, 8, 321. [Google Scholar] [CrossRef] [Green Version]
- Matsuzaki, Y.; Sugawara, K.; Furuse, Y.; Shimotai, Y.; Hongo, S.; Oshitani, H.; Mizuta, K.; Nishimura, H. Genetic Lineage and Reassortment of Influenza C Viruses Circulating between 1947 and 2014. J. Virol. 2016, 90, 8251–8265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsuzaki, Y.; Mizuta, K.; Sugawara, K.; Tsuchiya, E.; Muraki, Y.; Hongo, S.; Suzuki, H.; Nishimura, H. Frequent reassortment among influenza C viruses. J. Virol. 2003, 77, 871–881. [Google Scholar] [CrossRef] [Green Version]
- Saegerman, C.; Gaudino, M.; Savard, C.; Broes, A.; Ariel, O.; Meyer, G.; Ducatez, M.F. Influenza D virus in respiratory disease in Canadian, province of Québec, cattle: Relative importance and evidence of new reassortment between different clades. Transbound. Emerg. Dis. 2021. [Google Scholar] [CrossRef]
- Xu, Y.; Liang, H.; Wen, H. Frequent reassortment and potential recombination shape the genetic diversity of influenza D viruses. J. Infect. 2021, 82, e36–e38. [Google Scholar] [CrossRef]
- Wan, X.F.; Ferguson, L.; Oliva, J.; Rubrum, A.; Eckard, L.; Zhang, X.; Woolums, A.R.; Lion, A.; Meyer, G.; Murakami, S.; et al. Limited Cross-Protection Provided by Prior Infection Contributes to High Prevalence of Influenza D Viruses in Cattle. J. Virol. 2020, 94, e00240-20. [Google Scholar] [CrossRef]
- Ishida, H.; Murakami, S.; Kamiki, H.; Matsugo, H.; Takenaka-Uema, A.; Horimoto, T.; Heise, M.T. Establishment of a Reverse Genetics System for Influenza D Virus. J. Virol. 2020, 94, e01767-19. [Google Scholar] [CrossRef] [PubMed]
- Su, S.; Fu, X.; Li, G.; Kerlin, F.; Veit, M. Novel Influenza D virus: Epidemiology, pathology, evolution and biological characteristics. Virulence 2017, 8, 1580–1591. [Google Scholar] [CrossRef] [Green Version]
- Nakatsu, S.; Murakami, S.; Shindo, K.; Horimoto, T.; Sagara, H.; Noda, T.; Kawaoka, Y. Influenza C and D Viruses Package Eight Organized Ribonucleoprotein Complexes. J. Virol. 2018, 92, e02084-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Green, I.J.; Lieberman, M.; Mogabgab, W.J.; Peterkin, W.G.; Phillips, I.A. The Behavior of Influenza Viruses in Various Tissue Culture Systems. J. Immunol. 1957, 78, 233. [Google Scholar] [PubMed]
- Mogabgab, W.J. Influenza C virus in monkey kidney tissue cultures. J. Bacteriol. 1962, 83, 209–210. [Google Scholar] [CrossRef] [Green Version]
- Takemoto, K.K.; Lynt, R.K.; Rowe, W.P.; Huebner, R.J.; Bell, J.A.; Mellin, G.W.; Davis, D.J. Primary Isolation of Influenza A, B, and G Viruses in Monkey Kidney Tissue Cultures. Proc. Soc. Exp. Biol. Med. 1955, 89, 308–311. [Google Scholar] [CrossRef]
- Chakraverty, P. The detection and multiplication of influenza C virus in tissue culture. J. Gen. Virol. 1974, 25, 421–425. [Google Scholar] [CrossRef]
- Austin, M.A.; Monto, A.S.; Maassab, H.F. Growth characteristics of influenza virus type C in avian hosts. Brief report. Arch. Virol. 1978, 58, 349–353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nerome, K.; Ishida, M. The multiplication of an influenza C virus in an established line of canine kidney (MDCK) cells. J. Gen. Virol. 1978, 39, 179–181. [Google Scholar] [CrossRef] [PubMed]
- Nerome, K.; Nakayama, M.; Ishida, M. Established cell line sensitive to influenza C virus. J. Gen. Virol. 1979, 43, 257–259. [Google Scholar] [CrossRef]
- Spence, H.A.; O'Callaghan, R.J. Induction of chick embryo feather malformations by an influenza C virus. Teratology 1985, 32, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, H.; Sugawara, K.; Kitame, F.; Nakamura, K.; Katsushima, N.; Moriuchi, H.; Numazaki, Y. A human melanoma cell line highly susceptible to influenza C virus. J. Gen. Virol. 1989, 70 Pt 7, 1653–1661. [Google Scholar] [CrossRef]
- Umetsu, Y.; Sugawara, K.; Nishimura, H.; Hongo, S.; Matsuzaki, M.; Kitame, F.; Nakamura, K. Selection of antigenically distinct variants of influenza C viruses by the host cell. Virology 1992, 189, 740–744. [Google Scholar] [CrossRef]
- Wang, M.; Veit, M. Hemagglutinin-esterase-fusion (HEF) protein of influenza C virus. Protein cell 2016, 7, 28–45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uprety, T.; Sreenivasan, C.C.; Bhattarai, S.; Wang, D.; Kaushik, R.S.; Li, F. Isolation and development of bovine primary respiratory cells as model to study influenza D virus infection. Virology 2021, 559, 89–99. [Google Scholar] [CrossRef] [PubMed]
- Strässle, M.; Laloli, L.; Gultom, M.; V'Kovski, P.; Stoffel, M.H.; Crespo Pomar, S.; Chanfon Bätzner, A.; Ebert, N.; Labroussaa, F.; Dijkman, R.; et al. Establishment of caprine airway epithelial cells grown in an air-liquid interface system to study caprine respiratory viruses and bacteria. Vet. Microbiol. 2021, 257, 109067. [Google Scholar] [CrossRef] [PubMed]
- Sreenivasan, C.C.; Thomas, M.; Antony, L.; Wormstadt, T.; Hildreth, M.B.; Wang, D.; Hause, B.; Francis, D.H.; Li, F.; Kaushik, R.S. Development and characterization of swine primary respiratory epithelial cells and their susceptibility to infection by four influenza virus types. Virology 2019, 528, 152–163. [Google Scholar] [CrossRef]
- Nogales, A.; Aydillo, T.; Ávila-Pérez, G.; Escalera, A.; Chiem, K.; Cadagan, R.; DeDiego, M.L.; Li, F.; García-Sastre, A.; Martínez-Sobrido, L. Functional Characterization and Direct Comparison of Influenza A, B, C, and D NS1 Proteins in vitro and in vivo. Front. Microbiol. 2019, 10, 2862. [Google Scholar] [CrossRef] [PubMed]
- Mazzetto, E.; Bortolami, A.; Fusaro, A.; Mazzacan, E.; Maniero, S.; Vascellari, M.; Beato, M.S.; Schiavon, E.; Chiapponi, C.; Terregino, C.; et al. Replication of Influenza D Viruses of Bovine and Swine Origin in Ovine Respiratory Explants and Their Attachment to the Respiratory Tract of Bovine, Sheep, Goat, Horse, and Swine. Front. Microbiol. 2020, 11, 1136. [Google Scholar] [CrossRef] [PubMed]
- Nemanichvili, N.; Berends, A.J.; Wubbolts, R.W.; Gröne, A.; Rijks, J.M.; de Vries, R.P.; Verheije, M.H. Tissue Microarrays to Visualize Influenza D Attachment to Host Receptors in the Respiratory Tract of Farm Animals. Viruses 2021, 13, 586. [Google Scholar] [CrossRef] [PubMed]
- Ritchey, M.B.; Palese, P.; Kilbourne, E.D. RNAs of influenza A, B, and C viruses. J. Virol. 1976, 18, 738–744. [Google Scholar] [CrossRef] [Green Version]
- Cox, N.J.; Kendal, A.P. Presence of a segmented single-stranded RNA genome in influenza C virus. Virology 1976, 74, 239–241. [Google Scholar] [CrossRef]
- Hirst, G.K. The relationship of the receptors of a new strain of virus to those of the mumps-ndv-influenza group. J. Exp. Med. 1950, 91, 177–184. [Google Scholar] [CrossRef] [Green Version]
- Nerome, K.; Ishida, M.; Nakayama, M. Absence of neuraminidase from influenza C virus. Arch. Virol. 1976, 50, 241–244. [Google Scholar] [CrossRef]
- Martin, M.L.; Palmer, E.L.; Kendal, A.P. Lack of characteristic hexagonal surface structure on a newly isolated influenza C virus. J. Clin. Microbiol. 1977, 6, 84–86. [Google Scholar] [CrossRef]
- Waterson, A.P.; Hurrell, J.M.; Jensen, K.E. The fine structure of influenza A, B and C viruses. Arch Gesamte Virusforsch 1963, 12, 487–495. [Google Scholar] [CrossRef]
- Badham, M.D.; Rossman, J.S. Filamentous Influenza Viruses. Curr. Clin. Microbiol. Rep. 2016, 3, 155–161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petri, T.; Meier-Ewert, H.; Crumpton, W.M.; Dimmock, N.J. RNA'S of influenza C virus strains. Arch. Virol. 1979, 61, 239–243. [Google Scholar] [CrossRef] [PubMed]
- Yokota, M.; Nakamura, K.; Sugawara, K.; Homma, M. The synthesis of polypeptides in influenza C virus-infected cells. Virology 1983, 130, 105–117. [Google Scholar] [CrossRef]
- Elliott, R.M.; Yuanji, G.; Desselberger, U. Polypeptide synthesis in MDCK cells infected with human and pig influenza C viruses. J. Gen. Virol. 1984, 65, 1873–1880. [Google Scholar] [CrossRef] [PubMed]
- Govorkova, E.A.; Ivanova, V.T.; Zakstel'skaia, L.; Miller, G.G. Sedimentation characteristics of the virions and subviral particles of influenza C viruses. Vopr. Virusol 1984, 29, 687–691. [Google Scholar]
- Veit, M.; Herrler, G.; Schmidt, M.F.; Rott, R.; Klenk, H.D. The hemagglutinating glycoproteins of influenza B and C viruses are acylated with different fatty acids. Virology 1990, 177, 807–811. [Google Scholar] [CrossRef]
- Halldorsson, S.; Sader, K.; Turner, J.; Calder, L.J.; Rosenthal, P.B. In situ structure and organization of the influenza C virus surface glycoprotein. Nat. Commun. 2021, 12, 1694. [Google Scholar] [CrossRef] [PubMed]
- Saletti, D.; Radzimanowski, J.; Effantin, G.; Midtvedt, D.; Mangenot, S.; Weissenhorn, W.; Bassereau, P.; Bally, M. The Matrix protein M1 from influenza C virus induces tubular membrane invaginations in an in vitro cell membrane model. Sci. Rep. 2017, 7, 40801. [Google Scholar] [CrossRef] [Green Version]
- Sakai, T.; Takagi, H.; Muraki, Y.; Saito, M. Unique Directional Motility of Influenza C Virus Controlled by Its Filamentous Morphology and Short-Range Motions. J. Virol. 2018, 92. [Google Scholar] [CrossRef] [Green Version]
- Goto, T.; Shimotai, Y.; Matsuzaki, Y.; Muraki, Y.; Sho, R.; Sugawara, K.; Hongo, S. Effect of Phosphorylation of CM2 Protein on Influenza C Virus Replication. J. Virol. 2017, 91. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Ludwig, K.; Bottcher, C.; Veit, M. The role of stearate attachment to the hemagglutinin-esterase-fusion glycoprotein HEF of influenza C virus. Cell. Microbiol. 2016, 18, 692–704. [Google Scholar] [CrossRef]
- Elliott, R.M.; Yuanji, G.; Desselberger, U. Protein and nucleic acid analyses of influenza C viruses isolated from pigs and man. Vaccine 1985, 3, 182–188. [Google Scholar] [CrossRef]
- Lapatschek, M.S.; Marschall, M.; Meier-Ewert, H. The persistent variant of influenza C virus carries one characteristic point mutation in RNA segment 1. Virus Res. 1995, 39, 47–54. [Google Scholar] [CrossRef]
- Hengrung, N.; El Omari, K.; Serna Martin, I.; Vreede, F.T.; Cusack, S.; Rambo, R.P.; Vonrhein, C.; Bricogne, G.; Stuart, D.I.; Grimes, J.M.; et al. Crystal structure of the RNA-dependent RNA polymerase from influenza C virus. Nature 2015, 527, 114–117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamashita, M.; Krystal, M.; Palese, P. Comparison of the three large polymerase proteins of influenza A, B, and C viruses. Virology 1989, 171, 458–466. [Google Scholar] [CrossRef]
- Sugawara, K.; Kitame, F.; Homma, M.; Nakamura, K. An assay for the receptor-destroying activity of influenza C virus. Microbiol. Immunol. 1985, 29, 1207–1217. [Google Scholar] [CrossRef]
- Vlasak, R.; Krystal, M.; Nacht, M.; Palese, P. The influenza C virus glycoprotein (HE) exhibits receptor-binding (hemagglutinin) and receptor-destroying (esterase) activities. Virology 1987, 160, 419–425. [Google Scholar] [CrossRef]
- Formanowski, F.; Meier-Ewert, H. Isolation of the influenza C virus glycoprotein in a soluble form by bromelain digestion. Virus Res. 1988, 10, 177–191. [Google Scholar] [CrossRef]
- Herrler, G.; Dürkop, I.; Becht, H.; Klenk, H.D. The glycoprotein of influenza C virus is the haemagglutinin, esterase and fusion factor. J. Gen. Virol. 1988, 69, 839–846. [Google Scholar] [CrossRef]
- Herrler, G.; Szepanski, S.; Schultze, B. 9-O-acetylated sialic acid, a receptor determinant for influenza C virus and coronaviruses. Behring Inst. Mitt. 1991, 89, 177–184. [Google Scholar]
- Pleschka, S.; Klenk, H.D.; Herrler, G. The catalytic triad of the influenza C virus glycoprotein HEF esterase: Characterization by site-directed mutagenesis and functional analysis. J. Gen. Virol. 1995, 76 Pt 10, 2529–2537. [Google Scholar] [CrossRef]
- Wagaman, P.C.; Spence, H.A.; O'Callaghan, R.J. Detection of influenza C virus by using an in situ esterase assay. J. Clin. Microbiol. 1989, 27, 832–836. [Google Scholar] [CrossRef] [Green Version]
- Takashita, E.; Muraki, Y.; Sugawara, K.; Asao, H.; Nishimura, H.; Suzuki, K.; Tsuji, T.; Hongo, S.; Ohara, Y.; Kawaoka, Y.; et al. Intrinsic temperature sensitivity of influenza C virus hemagglutinin-esterase-fusion protein. J. Virol. 2012, 86, 13108–13111. [Google Scholar] [CrossRef] [Green Version]
- Tang, Y.S.; Lo, C.Y.; Mok, C.K.; Chan, P.K.; Shaw, P.C. The Extended C-Terminal Region of Influenza C Virus Nucleoprotein Is Important for Nuclear Import and Ribonucleoprotein Activity. J. Virol. 2019, 93, e02048-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Donchet, A.; Oliva, J.; Labaronne, A.; Tengo, L.; Miloudi, M.; Gerard, F.C.; Mas, C.; Schoehn, G.; Ruigrok, R.W.; Ducatez, M.; et al. The structure of the nucleoprotein of Influenza D shows that all Orthomyxoviridae nucleoproteins have a similar NPCORE, with or without a NPTAIL for nuclear transport. Sci. Rep. 2019, 9, 600. [Google Scholar] [CrossRef] [Green Version]
- Hongo, S. Type C influenza. Nihon Rinsho Jpn. J. Clin. Med. 2006, 64, 1942–1949. [Google Scholar]
- Gregoriades, A.; Guzman, G.G.; Paoletti, E. The phosphorylation of the integral membrane (M1) protein of influenza virus. Virus Res. 1990, 16, 27–41. [Google Scholar] [CrossRef]
- Muraki, Y.; Murata, T.; Takashita, E.; Matsuzaki, Y.; Sugawara, K.; Hongo, S. A mutation on influenza C virus M1 protein affects virion morphology by altering the membrane affinity of the protein. J. Virol. 2007, 81, 8766–8773. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muraki, Y.; Washioka, H.; Sugawara, K.; Matsuzaki, Y.; Takashita, E.; Hongo, S. Identification of an amino acid residue on influenza C virus M1 protein responsible for formation of the cord-like structures of the virus. J. Gen. Virol. 2004, 85, 1885–1893. [Google Scholar] [CrossRef]
- Yamashita, M.; Krystal, M.; Palese, P. Evidence that the matrix protein of influenza C virus is coded for by a spliced mRNA. J. Virol. 1988, 62, 3348–3355. [Google Scholar] [CrossRef] [Green Version]
- Shimotai, Y.; Goto, T.; Matsuzaki, Y.; Muraki, Y.; Sugawara, K.; Hongo, S. The effect of the cytoplasmic tail of influenza C virus CM2 protein on its biochemical properties and intracellular processing. Biochem. Biophys. Rep. 2015, 3, 1–6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stewart, S.M.; Pekosz, A. The influenza C virus CM2 protein can alter intracellular pH, and its transmembrane domain can substitute for that of the influenza A virus M2 protein and support infectious virus production. J. Virol. 2012, 86, 1277–1281. [Google Scholar] [CrossRef] [Green Version]
- Furukawa, T.; Muraki, Y.; Noda, T.; Takashita, E.; Sho, R.; Sugawara, K.; Matsuzaki, Y.; Shimotai, Y.; Hongo, S. Role of the CM2 protein in the influenza C virus replication cycle. J. Virol. 2011, 85, 1322–1329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okuwa, T.; Muraki, Y.; Himeda, T.; Ohara, Y. Glycosylation of CM2 is important for efficient replication of influenza C virus. Virology 2012, 433, 167–175. [Google Scholar] [CrossRef] [Green Version]
- Muraki, Y.; Okuwa, T.; Furukawa, T.; Matsuzaki, Y.; Sugawara, K.; Himeda, T.; Hongo, S.; Ohara, Y. Palmitoylation of CM2 is dispensable to influenza C virus replication. Virus Res. 2011, 157, 99–105. [Google Scholar] [CrossRef] [PubMed]
- Nakada, S.; Graves, P.N.; Desselberger, U.; Creager, R.S.; Krystal, M.; Palese, P. Influenza C virus RNA 7 codes for a nonstructural protein. J. Virol. 1985, 56, 221–226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakada, S.; Graves, P.N.; Palese, P. The influenza C virus NS gene: Evidence for a spliced mRNA and a second NS gene product (NS2 protein). Virus Res. 1986, 4, 263–273. [Google Scholar] [CrossRef]
- Pachler, K.; Vlasak, R. Influenza C virus NS1 protein counteracts RIG-I-mediated IFN signalling. Virol. J. 2011, 8, 48. [Google Scholar] [CrossRef] [Green Version]
- Kohno, Y.; Muraki, Y.; Matsuzaki, Y.; Takashita, E.; Sugawara, K.; Hongo, S. Intracellular localization of influenza C virus NS2 protein (NEP) in infected cells and its incorporation into virions. Arch. Virol. 2009, 154, 235–243. [Google Scholar] [CrossRef]
- Pease, C.M. An evolutionary epidemiological mechanism, with applications to type A influenza. Theor. Popul. Biol. 1987, 31, 422–452. [Google Scholar] [CrossRef]
- Webster, R.G.; Bean, W.J.; Gorman, O.T.; Chambers, T.M.; Kawaoka, Y. Evolution and ecology of influenza A viruses. Microbiol. Rev. 1992, 56, 152–179. [Google Scholar] [CrossRef]
- Gatherer, D. Tempo and mode in the molecular evolution of influenza C. PLoS Curr. 2010, 2, RRN1199. [Google Scholar] [CrossRef] [Green Version]
- Yamashita, M.; Krystal, M.; Fitch, W.M.; Palese, P. Influenza B virus evolution: Co-circulating lineages and comparison of evolutionary pattern with those of influenza A and C viruses. Virology 1988, 163, 112–122. [Google Scholar] [CrossRef]
- Kesinger, E.; Liu, J.; Jensen, A.; Chia, C.P.; Demers, A.; Moriyama, H. Influenza D virus M2 protein exhibits ion channel activity in Xenopus laevis oocytes. PLoS ONE 2018, 13, e0199227. [Google Scholar] [CrossRef]
- O'Callaghan, R.J.; Loughlin, M.; Labat, D.D.; Howe, C. Properties of influenza C virus grown in cell culture. J. Virol. 1977, 24, 875–882. [Google Scholar] [CrossRef] [Green Version]
- Barclay, G.R.; Leader-Williams, L.K.; Flewett, T.H. Aspects of influenza C virus replication. J. Hyg. 1971, 69, 587–592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herrler, G.; Compans, R.W.; Meier-Ewert, H. A precursor glycoprotein in influenza C virus. Virology 1979, 99, 49–56. [Google Scholar] [CrossRef]
- Sugawara, K.; Ohuchi, M.; Nakamura, K.; Homma, M. Effects of various proteases on the glycoprotein composition and the infectivity of influenza C virus. Arch. Virol. 1981, 68, 147–151. [Google Scholar] [CrossRef] [PubMed]
- Racaniello, V.R.; Palese, P. Isolation of influenza C virus recombinants. J. Virol. 1979, 32, 1006–1014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kitame, F.; Sugawara, K.; Ohwada, K.; Homma, M. Proteolytic activation of hemolysis and fusion by influenza C virus. Arch. Virol. 1982, 73, 357–361. [Google Scholar] [CrossRef] [PubMed]
- Ohuchi, M.; Ohuchi, R.; Mifune, K. Demonstration of hemolytic and fusion activities of influenza C virus. J. Virol. 1982, 42, 1076–1079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Formanowski, F.; Wharton, S.A.; Calder, L.J.; Hofbauer, C.; Meier-Ewert, H. Fusion characteristics of influenza C viruses. J. Gen. Virol. 1990, 71, 1181–1188. [Google Scholar] [CrossRef]
- Marschall, M.; Böswald, C.; Schuler, A.; Youzbashi, E.; Meier-Ewert, H. Productive and non-productive phases during long-term persistence of influenza C virus. J. Gen. Virol. 1993, 74, 2019–2023. [Google Scholar] [CrossRef]
- Mironova, N.I.; Grigor'ev, V.B.; Zhirnov, O.P. Biochemical features of protein matrix M1 of the influenza C virus. Mol. Biol. 1993, 27, 1113–1125. [Google Scholar]
- Herrler, G.; Gross, H.J.; Imhof, A.; Brossmer, R.; Milks, G.; Paulson, J.C. A synthetic sialic acid analogue is recognized by influenza C virus as a receptor determinant but is resistant to the receptor-destroying enzyme. J. Biol. Chem. 1992, 267, 12501–12505. [Google Scholar] [CrossRef]
- Serrão, V.H.B.; Cook, J.D.; Lee, J.E. Snapshot of an influenza virus glycoprotein fusion intermediate. Cell Rep. 2021, 35, 109152. [Google Scholar] [CrossRef] [PubMed]
- Strobl, B.; Vlasak, R. The receptor-destroying enzyme of influenza C virus is required for entry into target cells. Virology 1993, 192, 679–682. [Google Scholar] [CrossRef]
- Matsuzaki, Y.; Matsuzaki, M.; Muraki, Y.; Sugawara, K.; Hongo, S.; Kitame, F.; Nakamura, K. Comparison of receptor-binding properties among influenza C virus isolates. Virus Res. 1995, 38, 291–296. [Google Scholar] [CrossRef]
- Hongo, S.; Sugawara, K.; Homma, M.; Nakamura, K. Effects of glycosylation on the conformation and antigenicity of influenza C viral glycoproteins. Vaccine 1985, 3, 223–226. [Google Scholar] [CrossRef]
- Sugawara, K.; Nishimura, H.; Kitame, F.; Nakamura, K. Antigenic variation among human strains of influenza C virus detected with monoclonal antibodies to gp88 glycoprotein. Virus Res. 1986, 6, 27–32. [Google Scholar] [CrossRef]
- Hongo, S.; Sugawara, K.; Homma, M.; Nakamura, K. The functions of oligosaccharide chains associated with influenza C viral glycoproteins. I. The formation of influenza C virus particles in the absence of glycosylation. Arch. Virol. 1986, 89, 171–187. [Google Scholar] [CrossRef]
- Herrler, G.; Klenk, H.D. Structure and function of the HEF glycoprotein of influenza C virus. Adv. Virus Res. 1991, 40, 213–234. [Google Scholar] [CrossRef]
- Yu, J.; Huang, C.; Sheng, Z.; Wang, Z.; Li, F.; Wang, D. Identification of One Critical Amino Acid Residue of the Nucleoprotein as a Determinant for In Vitro Replication Fitness of Influenza D Virus. J. Virol. 2021, 95, e0097121. [Google Scholar] [CrossRef]
- Herrler, G.; Rott, R.; Klenk, H.D.; Müller, H.P.; Shukla, A.K.; Schauer, R. The receptor-destroying enzyme of influenza C virus is neuraminate-O-acetylesterase. Embo. J. 1985, 4, 1503–1506. [Google Scholar] [CrossRef]
- Rogers, G.N.; Herrler, G.; Paulson, J.C.; Klenk, H.D. Influenza C virus uses 9-O-acetyl-N-acetylneuraminic acid as a high affinity receptor determinant for attachment to cells. J. Biol. Chem. 1986, 261, 5947–5951. [Google Scholar] [CrossRef]
- Herrler, G.; Klenk, H.D. The surface receptor is a major determinant of the cell tropism of influenza C virus. Virology 1987, 159, 102–108. [Google Scholar] [CrossRef]
- Luther, P.; Cushley, W.; Hölzer, C.; Desselberger, U.; Oxford, J.S. Acetylated galactosamine is a receptor for the influenza C virus glycoprotein. Arch Virol. 1988, 101, 247–254. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, H.; Sugawara, K.; Kitame, F.; Nakamura, K. Attachment of influenza C virus to human erythrocytes. J. Gen. Virol. 1988, 69, 2545–2553. [Google Scholar] [CrossRef] [PubMed]
- Minuse, E.; Quilligan, J.J., Jr.; Francis, T., Jr. Type C influenza virus. I. Studies of the virus and its distribution. J. Lab. Clin. Med. 1954, 43, 31–42. [Google Scholar]
- Schauer, R.; Reuter, G.; Stoll, S.; Posadas del Rio, F.; Herrler, G.; Klenk, H.D. Isolation and characterization of sialate 9(4)-O-acetylesterase from influenza C virus. Biol. Chem. Hoppe Seyler 1988, 369, 1121–1130. [Google Scholar] [CrossRef] [PubMed]
- Mayr, J.; Haselhorst, T.; Langereis, M.A.; Dyason, J.C.; Huber, W.; Frey, B.; Vlasak, R.; de Groot, R.J.; von Itzstein, M. Influenza C virus and bovine coronavirus esterase reveal a similar catalytic mechanism: New insights for drug discovery. Glycoconj. J. 2008, 25, 393–399. [Google Scholar] [CrossRef] [Green Version]
- Garcia-Sastre, A.; Villar, E.; Manuguerra, J.C.; Hannoun, C.; Cabezas, J.A. Activity of influenza C virus O-acetylesterase with O-acetyl-containing compounds. Biochem. J. 1991, 273 Pt 2, 435–441. [Google Scholar] [CrossRef] [Green Version]
- Herrler, G.; Gross, H.J.; Milks, G.; Paulson, J.C.; Klenk, H.D.; Brossmer, R. Use of a sialic acid analogue to analyze the importance of the receptor-destroying enzyme for the interaction of influenza C virus with cells. Acta. Histochem. Suppl. 1990, 40, 39–41. [Google Scholar]
- Zimmer, G.; Reuter, G.; Schauer, R. Use of influenza C virus for detection of 9-O-acetylated sialic acids on immobilized glycoconjugates by esterase activity. Eur. J. Biochem. 1992, 204, 209–215. [Google Scholar] [CrossRef] [PubMed]
- Szepanski, S.; Gross, H.J.; Brossmer, R.; Klenk, H.D.; Herrler, G. A single point mutation of the influenza C virus glycoprotein (HEF) changes the viral receptor-binding activity. Virology 1992, 188, 85–92. [Google Scholar] [CrossRef]
- Liu, R.; Sreenivasan, C.; Yu, H.; Sheng, Z.; Newkirk, S.J.; An, W.; Smith, D.F.; Chen, X.; Wang, D.; Li, F. Influenza D virus diverges from its related influenza C virus in the recognition of 9-O-acetylated N-acetyl- or N-glycolyl-neuraminic acid-containing glycan receptors. Virology 2020, 545, 16–23. [Google Scholar] [CrossRef]
- Klein, A.; Krishna, M.; Varki, N.M.; Varki, A. 9-O-acetylated sialic acids have widespread but selective expression: Analysis using a chimeric dual-function probe derived from influenza C hemagglutinin-esterase. Proc. Natl. Acad. Sci. USA 1994, 91, 7782–7786. [Google Scholar] [CrossRef] [Green Version]
- Wasik, B.R.; Barnard, K.N.; Ossiboff, R.J.; Khedri, Z.; Feng, K.H.; Yu, H.; Chen, X.; Perez, D.R.; Varki, A.; Parrish, C.R.; et al. Distribution of O-Acetylated Sialic Acids among Target Host Tissues for Influenza Virus. mSphere 2017, 2, e00379-16. [Google Scholar] [CrossRef] [Green Version]
- Hana, L.; Styk, B. Some properties of influenza C virus haemagglutination inhibitor from rat serum. Acta. Virol. 1959, 3, 85–90. [Google Scholar]
- Styk, B. Nonspecific inhibitors in normal rat serum against influenza virus C; correlation between influenza C, Newcastle disease, and epidemic parotitis viruses. Ceskoslov. Biol. 1955, 4, 263–268. [Google Scholar] [PubMed]
- O'Callaghan, R.J.; Labat, D.D. Evidence of a soluble substrate for the receptor-destroying enzyme of influenza C virus. Infect. Immun. 1983, 39, 305–310. [Google Scholar] [CrossRef] [Green Version]
- Herrler, G.; Geyer, R.; Müller, H.P.; Stirm, S.; Klenk, H.D. Rat alpha 1 macroglobulin inhibits hemagglutination by influenza C virus. Virus Res. 1985, 2, 183–192. [Google Scholar] [CrossRef]
- Herrler, G.; Rott, R.; Klenk, H.D. Neuraminic acid is involved in the binding of influenza C virus to erythrocytes. Virology 1985, 141, 144–147. [Google Scholar] [CrossRef]
- Kitame, F.; Nakamura, K.; Saito, A.; Suzuki, Y.; Sinohara, H.; Homma, M. Effects of alpha-macroglobulins and murinoglobulins on the hemagglutination by influenza C virus. Biochem. Int. 1986, 12, 649–652. [Google Scholar] [PubMed]
- Kitame, F.; Nakamura, K.; Saito, A.; Sinohara, H.; Homma, M. Isolation and characterization of influenza C virus inhibitor in rat serum. Virus Res. 1985, 3, 231–244. [Google Scholar] [CrossRef]
- Marschall, M.; Zach, A.; Hechtfischer, A.; Foerst, G.; Meier-Ewert, H.; Haller, O. Inhibition of influenza C viruses by human MxA protein. Virus Res. 2000, 67, 179–188. [Google Scholar] [CrossRef]
- Petri, T.; Meier-Ewert, H.; Compans, R.W. Inhibition of influenza C virus replication by actinomycin D, alpha-amanitin, and UV irradiation. J. Virol. 1979, 32, 1037–1040. [Google Scholar] [CrossRef] [Green Version]
- Mishin, V.P.; Patel, M.C.; Chesnokov, A.; De La Cruz, J.; Nguyen, H.T.; Lollis, L.; Hodges, E.; Jang, Y.; Barnes, J.; Uyeki, T.; et al. Susceptibility of Influenza A, B, C, and D Viruses to Baloxavir (1). Emerg. Infect. Dis. 2019, 25, 1969–1972. [Google Scholar] [CrossRef] [Green Version]
- Zhao, L.; Xia, H.; Huang, J.; Zheng, Y.; Liu, C.; Su, J.; Ping, J. Features of Nuclear Export Signals of NS2 Protein of Influenza D Virus. Viruses 2020, 12, 1100. [Google Scholar] [CrossRef]






| Year | Country | Species | Seropositivity | References | |
|---|---|---|---|---|---|
| Age group | Prevalence | ||||
| 1978 | USA | Human (n = 334) | 0–5 years | 64% | [62] |
| 6–10 years | 96% | ||||
| 11–15 years | 100% | ||||
| 16–25 years | 98% | ||||
| 1980 | USA | Human (n = 237) | 1–2 years | 36% | [63] |
| 2–5 years | 47.2% | ||||
| 20–30 years | 96% | ||||
| 65–85 years | 66.7% | ||||
| 1981 | China | Pigs | 3% | [35] | |
| 1983 | Japan | Human (n = 653) | 2–4 years | 50% | [66] |
| ≥5 years | 90–100% | ||||
| 1984 | Russia | Human | Not reported | [67] | |
| 1985 | Russia | Human (n = 975) | adults (20–60 years) | 85.7% | [68] |
| 1987 | Japan | Dogs (n = 112) | Not reported | 4.5% | [21] |
| 1987 | Japan | Pigs (n = 269) | Not reported | 38% | [21] |
| 1991 | Japan | Pig (n = 240) | Not reported | 19% | [36] |
| 1992 | France | Dogs (n = 134) | 6 months–16 years | 32% | [20] |
| 1992 | France | Humans (n = 301) | 4 months–88 years | 61% (High in 16–30 years group) | [69] |
| 1993 | Spain | Dogs | Not reported | 56.3% | [70] |
| 1996 | Bavaria | Dogs (n = 150) and pigs (n = 240) | Not reported | Dogs—50.6% Pigs—49.9% | [71] |
| 2000 | Brazil | Humans (n = 67) | Not reported | 56.7% | [72] |
| 2020 | Japan | Humans | 21–66 years | 17.5% | [64] |
| Country | Year | Bovine | Small Ruminant | Swine | Equine | Camelid | Human | Ref. |
|---|---|---|---|---|---|---|---|---|
| USA # | 2013 | 9.5 | * 1.3 | [82] | ||||
| 2016 | ** 94–95 | [26] | ||||||
| 2015 | S: 5.2, | [27] | ||||||
| G: 8.8 | ||||||||
| 2017 | 11–12 | [46] | ||||||
| China # | 2017 | DC: 7.8, | G: 33.8 | [45] | ||||
| B: 5.9 | ||||||||
| France # | 2014–18 | 47.2 | 1.5 | [80] | ||||
| Japan # | 2016 | 30.5 | [41] | |||||
| Togo | 2009,2015 2017–20 | 10.4 6.3 | S: 2; G:4.4 | [32,81] | ||||
| Morocco | 2012–15 | 35 | [81] | |||||
| Kenya | 2015 | 99–100 | [32] | |||||
| Cote-d’Ivoire | 2017–19 | 7.2 | S: 4.1, G:3.7 | [81] | ||||
| Benin | 2017–19 | 3.9 | [81] | |||||
| Uganda | 2017–19 | 20.9 | 20.5 | [81] | ||||
| Ethiopia | 2019 | 55.2 | [42] | |||||
| Italy # | 2004 | 4.88 | [93] | |||||
| 2005–17 | 5.1–46 | [23] | ||||||
| 2016 | 11.7 | [78] | ||||||
| Ireland # | 2015, 2017 | 94.6 | 4.5 | 5.8 | [40,94] | |||
| Mexico # | 2015 | [87] | ||||||
| Luxembourg | 2016 | 80.2 | [43] | |||||
| 2014–15 | 5.9 | |||||||
| Argentina | 2013 | 68 | [39] | |||||
| Turkey # | 2018–19 | [92] | ||||||
| Western Canada | 2018–19 | 7.1 | [89] |
| Segment | Protein | hICV | bIDV | Function | ||
|---|---|---|---|---|---|---|
| nt # | aa * | nt | aa | |||
| 1 | PB2 | 2365 | 774 | 2364 | 772 | Cap-binding, form heterotrimeric polymerase complex for replication and mRNA synthesis |
| 2 | PB1 | 2363 | 754 | 2330 | 753 | Contains Polymerase active site, form heterotrimeric polymerase complex for replication and mRNA synthesis |
| 3 | P3 | 2183 | 709 | 2195 | 710 | Contains endonuclease domain for cap-snatching, form heterotrimeric polymerase complex for replication and mRNA synthesis |
| 4 | HEF | 2073 | 655 | 2049 | 664 | Spike protein, receptor binding, fusion, and receptor destroying functions |
| 5 | NP | 1807 | 565 | 1775 | 552 | Forms viral ribonucleoprotein complex (vRNPs) together with polymerases and vRNA |
| 6 | P42 | 1180 | 374 | 1219 | 387 | |
| M1 | 1125 | 242 | 246 | Structural protein that lines the viral membrane inside | ||
| M2 | 420 | 139 | 148 | Transmembrane protein with ion proton-channel activity | ||
| 7 | NS | 935 | 286 | 868 | ||
| NS1 | 740 | 246 | 732 | 243 | Interferon antagonist | |
| NS2/NEP | 549 | 182 | 555 | 184 | Nuclear export of RNPs | |
| Gene | bICV vs. hICV | swICV vs. hICV | bICV vs. swICV | bIDV vs. hICV | bIDV vs. bICV | swIDV vs. swICV | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| nt | aa | nt | aa | nt | aa | nt | aa | nt | aa | nt | aa | |
| PB2 | 94.5 | 98.97 | 98.8 | 99.87 | 95.10 | 99.1 | 65.07 | 52.59 | 66.45 | 52.59 | 66.28 | 52.72 |
| PB1 | 94.25 | 98.67 | 98.62 | 99.34 | 94.86 | 98.81 | 69.69 | 72.44 | 70.15 | 72.84 | 69.48 | 72.57 |
| P3 | 96.43 | 98.59 | 96.03 | 98.59 | 95.61 | 98.03 | 65.42 | 49.86 | 65.48 | 50.14 | 74.74 | 49.86 |
| HEF | 89.99 | 92.6 | 94.69 | 96.15 | 91.33 | 93.84 | 67.44 | 52.64 | 67.22 | 52.85 | 67.46 | 53.5 |
| NP | 95.83 | 97.7 | 96.92 | 99.29 | 95.32 | 97.52 | 85.29 | 39.33 | 85.29 | 39.25 | 77.78 | 39.92 |
| M | 95.99 | 97.06 | 99.38 | 100.0 | 95.95 | 97.06 | 95.65 | 35.94 | - * | 36.20 | - * | 37.76 |
| NS | 94.83 | 94.72 | 98.18 | 97.97 | 95.69 | 95.93 | 100 | 33.04 | 85.71 | 32.46 | 85.71 | 32.89 |
| Viruses | Accession No. | No: of Sites | Glycosylation Sites |
|---|---|---|---|
| D/bovine/Oklahoma/660/2013 | AGS48804.1 | 7 | N(28)ES, N(54)VT, N(146)WT, N(249)KT, N(346)AT, N(513)DT, N(613)GS |
| D/swine/Oklahoma/1334/2011 | YP_009449559.1 | 6 | N(28)ES, N(54)VT, N(146)WT, N(346)AT, N(513)DT, N(613)GS |
| D/bovine/Ibaraki/7768/2016 | BAV17997.1 | 6 | N(28)ES, N(54)VT, N(146)WT, N(346)AT, N(513)DT, N(613)GS |
| D/bovine/Yamagata/1/2019 | BBM60897.1 | 8 | N(28)ES, N(54)VT, N(146)WT, N(249)KT, N(346)AT, N(390)DT, N(513)DT, N(613)GS |
| D/bovine/Califonia/0363/2019 | QOF88712.1 | 7 | N(28)ES, N(54)VT, N(146)WT, N(249)RT, N(346)AT, N(513)DT, N(613)GS |
| Species | Tissue | Sialic Acid | Linkages | ||||
|---|---|---|---|---|---|---|---|
| 9-O-Ac | 7,9-O-Ac | 5,9-O-Ac | 4-O-Ac | α2-3-linked Sias | α2-6-linked Sias | ||
| Human | Trachea Lung | ++ +++ | ++ ++ | ND | _ _ | +++ +++ | +++ ++ |
| Mouse | Trachea Lung Saliva RBC | +++ +++ ND ND | ++ +++ ND ND | ND ND +++ +++ | + + ND ND | + +++ ND ND | + + ND ND |
| Ferret | Trachea Lung | _ + | _ _ | ND ND | _ _ | + + | + a + |
| Guinea pig | Trachea Lung | + + | _ _ | ND ND | +++ b +++ | ++ ++ | + ++ |
| Pig | Trachea Lung | + _ | ++ _ | ND ND | _ _ | +++ +++ | +++ +++ |
| Horse | Trachea Lung Saliva RBC | ++ + ND ND | + - ND ND | ND ND +++ _ | +++ +++ ND ND | +++ +++ ND ND | +++ +++ ND ND |
| Dog | Trachea Lung | +++ _ | +++ _ | ND ND | _ _ | +++ ++ | ++ +++ |
| Cow | Saliva RBC | ND ND | ND ND | +++ _ | +++ ND | ND ND | ND ND |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sreenivasan, C.C.; Sheng, Z.; Wang, D.; Li, F. Host Range, Biology, and Species Specificity of Seven-Segmented Influenza Viruses—A Comparative Review on Influenza C and D. Pathogens 2021, 10, 1583. https://doi.org/10.3390/pathogens10121583
Sreenivasan CC, Sheng Z, Wang D, Li F. Host Range, Biology, and Species Specificity of Seven-Segmented Influenza Viruses—A Comparative Review on Influenza C and D. Pathogens. 2021; 10(12):1583. https://doi.org/10.3390/pathogens10121583
Chicago/Turabian StyleSreenivasan, Chithra C., Zizhang Sheng, Dan Wang, and Feng Li. 2021. "Host Range, Biology, and Species Specificity of Seven-Segmented Influenza Viruses—A Comparative Review on Influenza C and D" Pathogens 10, no. 12: 1583. https://doi.org/10.3390/pathogens10121583







