Clinical Implications of Polymicrobial Synergism Effects on Antimicrobial Susceptibility
Abstract
1. Introduction
2. Many Infections Are Polymicrobial
2.1. Skin and Soft Tissue Infections
2.2. Respiratory
2.3. Otolaryngology
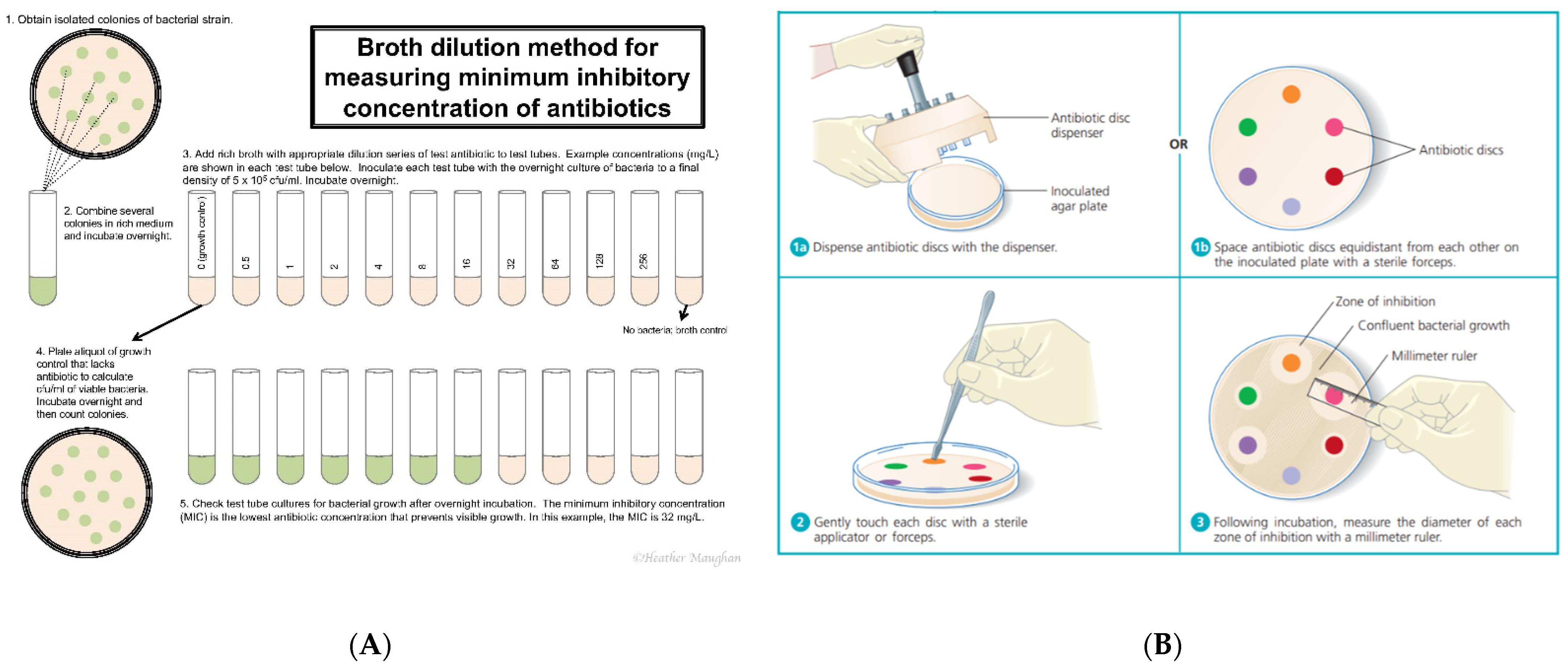
3. Clinical Determination of Antimicrobial Susceptibility
4. Mechanisms of Polymicrobial Synergism
4.1. Metabolites
4.2. Signals
4.3. Direct Contact
4.4. Host-Mediated
5. Polymicrobial Synergism and Its Impact on Antimicrobial Susceptibility
5.1. Bacteria–Bacteria Interactions
5.2. Bacteria–Fungus Interactions
5.3. Bacteria–Virus Interactions:
6. Discussion
7. Materials and Methods
Author Contributions
Funding
Conflicts of Interest
References
- Orazi, G.; O’Toole, G.A. “It Takes a Village”: Mechanisms Underlying Antimicrobial Recalcitrance of Polymicrobial Biofilms. J. Bacteriol. 2019, 202. [Google Scholar] [CrossRef] [PubMed]
- CLSI. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically, 11th ed.; CLSI: Wayne, PA, USA, 2018. [Google Scholar]
- Cummings, L.A.; Hoogestraat, D.R.; Rassoulian-Barrett, S.L.; Rosenthal, C.A.; Salipante, S.J.; Cookson, B.T.; Hoffman, N.G. Comprehensive evaluation of complex polymicrobial specimens using next generation sequencing and standard microbiological culture. Sci. Rep. 2020, 10, 5446. [Google Scholar] [CrossRef] [PubMed]
- Nair, N.; Biswas, R.; Gotz, F.; Biswas, L. Impact of Staphylococcus aureus on pathogenesis in polymicrobial infections. Infect. Immun. 2014, 82, 2162–2169. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.H.; Wang, N.Y.; Wu, A.Y.; Lin, C.C.; Lee, C.M.; Liu, C.P. Citrobacter freundii bacteremia: Risk factors of mortality and prevalence of resistance genes. J. Microbiol. Immunol. Infect. 2018, 51, 565–572. [Google Scholar] [CrossRef] [PubMed]
- Furuichi, M.; Horikoshi, Y. Sites of infection associated with Streptococcus anginosus group among children. J. Infect. Chemother. 2018, 24, 99–102. [Google Scholar] [CrossRef]
- Jaju, K.; Pichare, A.; Davane, M.; Nagoba, B. Profile and Antibiotic Susceptibility of Bacterial Pathogens Associated With Diabetic Foot Ulcers From a Rural Area. Wounds 2019, 31, 158–162. [Google Scholar]
- Zhao-Fleming, H.H.; Wilkinson, J.E.; Larumbe, E.; Dissanaike, S.; Rumbaugh, K. Obligate anaerobes are abundant in human necrotizing soft tissue infection samples—A metagenomics analysis. APMIS 2019, 127, 577–587. [Google Scholar] [CrossRef]
- Bessa, L.J.; Fazii, P.; Di Giulio, M.; Cellini, L. Bacterial isolates from infected wounds and their antibiotic susceptibility pattern: Some remarks about wound infection. Int. Wound J. 2015, 12, 47–52. [Google Scholar] [CrossRef]
- Rhoads, D.D.; Cox, S.B.; Rees, E.J.; Sun, Y.; Wolcott, R.D. Clinical identification of bacteria in human chronic wound infections: Culturing vs. 16S ribosomal DNA sequencing. BMC Infect. Dis. 2012, 12, 321. [Google Scholar] [CrossRef]
- Wolcott, R.D.; Hanson, J.D.; Rees, E.J.; Koenig, L.D.; Phillips, C.D.; Wolcott, R.A.; Cox, S.B.; White, J.S. Analysis of the chronic wound microbiota of 2,963 patients by 16S rDNA pyrosequencing. Wound Repair Regen. 2016, 24, 163–174. [Google Scholar] [CrossRef]
- Sidhu, M.S.; Cooper, G.; Jenkins, N.; Jeys, L.; Parry, M.; Stevenson, J.D. Prosthetic fungal infections. Bone Jt. J. 2019, 101, 582–588. [Google Scholar] [CrossRef] [PubMed]
- Dowd, S.E.; Delton Hanson, J.; Rees, E.; Wolcott, R.D.; Zischau, A.M.; Sun, Y.; White, J.; Smith, D.M.; Kennedy, J.; Jones, C.E. Survey of fungi and yeast in polymicrobial infections in chronic wounds. J. Wound Care 2011, 20, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Scott, J.E.; O’Toole, G.A. The Yin and Yang of Streptococcus Lung Infections in Cystic Fibrosis: A Model for Studying Polymicrobial Interactions. J. Bacteriol. 2019, 201, e00115–e00119. [Google Scholar] [CrossRef] [PubMed]
- Segura-Egea, J.J.; Gould, K.; Şen, B.H.; Jonasson, P.; Cotti, E.; Mazzoni, A.; Sunay, H.; Tjäderhane, L.; Dummer, P.M.H. Antibiotics in Endodontics: A review. Int. Endod. J. 2017, 50, 1169–1184. [Google Scholar] [CrossRef]
- Uddén, F.; Filipe, M.; Reimer, Å.; Paul, M.; Matuschek, E.; Thegerström, J.; Hammerschmidt, S.; Pelkonen, T.; Riesbeck, K. Aerobic bacteria associated with chronic suppurative otitis media in Angola. Infect. Dis Poverty 2018, 7, 42. [Google Scholar] [CrossRef]
- Dunne, E.M.; Murad, C.; Sudigdoadi, S.; Fadlyana, E.; Tarigan, R.; Indriyani, S.A.K.; Pell, C.L.; Watts, E.; Satzke, C.; Hinds, J.; et al. Carriage of Streptococcus pneumoniae, Haemophilus influenzae, Moraxella catarrhalis, and Staphylococcus aureus in Indonesian children: A cross-sectional study. PLoS ONE 2018, 13, e0195098. [Google Scholar] [CrossRef]
- Humphries, R.M.; Ambler, J.; Mitchell, S.L.; Castanheira, M.; Dingle, T.; Hindler, J.A.; Koeth, L.; Sei, K. CLSI Methods Development and Standardization Working Group Best Practices for Evaluation of Antimicrobial Susceptibility Tests. J. Clin. Microbiol. 2018, 56, e01934-17. [Google Scholar] [CrossRef]
- Syal, K.; Mo, M.; Yu, H.; Iriya, R.; Jing, W.; Guodong, S.; Wang, S.; Grys, T.E.; Haydel, S.E.; Tao, N. Current and emerging techniques for antibiotic susceptibility tests. Theranostics 2017, 7, 1795–1805. [Google Scholar] [CrossRef]
- James, G.; Cappuccino, N.S. Microbiology: A Laboratory Manual, 10th ed.; Pearson Higher Education: London, UK, 2014. [Google Scholar]
- Hudzicki, J. Kirby-Bauer Disk Diffusion Susceptibility Test Protocol; American Society for Microbiology: Washington, DC, USA, 2009. [Google Scholar]
- Wolcott, R.D. Biofilms cause chronic infections. J. Wound Care 2017, 26, 423–425. [Google Scholar] [CrossRef]
- Adamowicz, E.M.; Flynn, J.; Hunter, R.C.; Harcombe, W.R. Cross-feeding modulates antibiotic tolerance in bacterial communities. ISME J. 2018, 12, 2723–2735. [Google Scholar] [CrossRef]
- Tavernier, S.; Sass, A.; De Bruyne, M.; Baeke, F.; De Rycke, R.; Crabbé, A.; Vandecandelaere, I.; Van Nieuwerburgh, F.; Coenye, T. Decreased susceptibility of Streptococcus anginosus to vancomycin in a multispecies biofilm is due to increased thickness of the cell wall. J. Antimicrob. Chemother. 2018, 73, 2323–2330. [Google Scholar] [CrossRef] [PubMed]
- Slutzkey, S.; Moses, O.; Tal, H.; Meirowitz, A.; Matalon, S. Direct Contact Test for Evaluating Bacterial Growth on Machined and Rough Surface Implants: An: In Vitro: Study. Implant. Dent. 2017, 26, 899–903. [Google Scholar] [CrossRef] [PubMed]
- Robertson, J.; McGoverin, C.; Vanholsbeeck, F.; Swift, S. Optimisation of the Protocol for the LIVE/DEAD(®) BacLight(TM) Bacterial Viability Kit for Rapid Determination of Bacterial Load. Front. Microbiol. 2019, 10, 801. [Google Scholar] [CrossRef] [PubMed]
- Kong, E.F.; Tsui, C.; Kucharíková, S.; Van Dijck, P.; Jabra-Rizk, M.A. Modulation of Staphylococcus aureus Response to Antimicrobials by the Candida albicans Quorum Sensing Molecule Farnesol. Antimicrob. Agents Chemother. 2017, 61. [Google Scholar] [CrossRef] [PubMed]
- Salipante, S.J.; Jerome, K.R. Digital PCR—An Emerging Technology with Broad Applications in Microbiology. Clin. Chem. 2019, 66, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Flynn, J.M.; Cameron, L.C.; Wiggen, T.D.; Dunitz, J.M.; Harcombe, W.R.; Hunter, R.C. Disruption of Cross-Feeding Inhibits Pathogen Growth in the Sputa of Patients with Cystic Fibrosis. mSphere 2020, 5. [Google Scholar] [CrossRef]
- Kean, R.; Rajendran, R.; Haggarty, J.; Townsend, E.M.; Short, B.; Burgess, K.E.; Lang, S.; Millington, O.; Mackay, W.G.; Williams, C.; et al. Candida albicans Mycofilms Support Staphylococcus aureus Colonization and Enhances Miconazole Resistance in Dual-Species Interactions. Front. Microbiol. 2017, 8, 258. [Google Scholar] [CrossRef]
- Tagini, F.; Greub, G. Bacterial genome sequencing in clinical microbiology: A pathogen-oriented review. Eur. J. Clin. Microbiol. Infect. Dis. Off. Publ. Eur. Soc. Clin. Microbiol. 2017, 36, 2007–2020. [Google Scholar] [CrossRef]
- Hoffman, L.R.; Déziel, E.; D’Argenio, D.A.; Lépine, F.; Emerson, J.; McNamara, S.; Gibson, R.L.; Ramsey, B.W.; Miller, S.I. Selection for Staphylococcus aureus small-colony variants due to growth in the presence of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2006, 103, 19890–19895. [Google Scholar] [CrossRef]
- Orazi, G.; O’Toole, G.A. Pseudomonas aeruginosa Alters Staphylococcus aureus Sensitivity to Vancomycin in a Biofilm Model of Cystic Fibrosis Infection. mBio 2017, 8. [Google Scholar] [CrossRef]
- DeLeon, S.; Clinton, A.; Fowler, H.; Everett, J.; Horswill, A.R.; Rumbaugh, K.P. Synergistic Interactions of Pseudomonas aeruginosa and Staphylococcus aureus in an In Vitro Wound Model. Infect. Immun. 2014, 82, 4718–4728. [Google Scholar] [CrossRef] [PubMed]
- Vega, N.M.; Allison, K.R.; Samuels, A.N.; Klempner, M.S.; Collins, J.J. Salmonella typhimurium intercepts Escherichia coli signaling to enhance antibiotic tolerance. Proc. Natl. Acad. Sci. USA 2013, 110, 14420–14425. [Google Scholar] [CrossRef] [PubMed]
- Harriott, M.M.; Noverr, M.C. Candida albicans and Staphylococcus aureus form polymicrobial biofilms: Effects on antimicrobial resistance. Antimicrob. Agents Chemother. 2009, 53, 3914–3922. [Google Scholar] [CrossRef] [PubMed]
- Murray, J.L.; Connell, J.L.; Stacy, A.; Turner, K.H.; Whiteley, M. Mechanisms of synergy in polymicrobial infections. J. Microbiol. 2014, 52, 188–199. [Google Scholar] [CrossRef] [PubMed]
- Short, F.L.; Murdoch, S.L.; Ryan, R.P. Polybacterial human disease: The ills of social networking. Trends Microbiol. 2014, 22, 508–516. [Google Scholar] [CrossRef]
- Ramsey, M.M.; Rumbaugh, K.P.; Whiteley, M. Metabolite cross-feeding enhances virulence in a model polymicrobial infection. PLoS Pathog. 2011, 7, e1002012. [Google Scholar] [CrossRef]
- Todd, O.A.; Peters, B.M. Candida albicans and Staphylococcus aureus Pathogenicity and Polymicrobial Interactions: Lessons beyond Koch’s Postulates. J. Fungi 2019, 5, 81. [Google Scholar] [CrossRef]
- Dixon, E.F.; Hall, R.A. Noisy neighbourhoods: Quorum sensing in fungal-polymicrobial infections. Cell. Microbiol. 2015, 17, 1431–1441. [Google Scholar] [CrossRef]
- Förster, T.M.; Mogavero, S.; Dräger, A.; Graf, K.; Polke, M.; Jacobsen, I.D.; Hube, B. Enemies and brothers in arms: Candida albicans and gram-positive bacteria. Cell. Microbiol. 2016, 18, 1709–1715. [Google Scholar] [CrossRef]
- Chonmaitree, T.; Owen, M.J.; Howie, V.M. Respiratory viruses interfere with bacteriologic response to antibiotic in children with acute otitis media. J. Infect. Dis. 1990, 162, 546–549. [Google Scholar] [CrossRef]
- Lebrun, M.; de Repentigny, J.; Mathieu, L.G. Diminution of the antibacterial activity of antibiotics in cultures and in experimental mixed infections. Can. J. Microbiol. 1978, 24, 154–161. [Google Scholar] [CrossRef] [PubMed]
- Hanada, S.; Pirzadeh, M.; Carver, K.Y.; Deng, J.C. Respiratory Viral Infection-Induced Microbiome Alterations and Secondary Bacterial Pneumonia. Front. Immunol. 2018, 9, 2640. [Google Scholar] [CrossRef] [PubMed]
- Bakaletz, L.O. Viral-bacterial co-infections in the respiratory tract. Curr. Opin. Microbiol. 2017, 35, 30–35. [Google Scholar] [CrossRef] [PubMed]
- Bell, L.C.K.; Noursadeghi, M. Pathogenesis of HIV-1 and Mycobacterium tuberculosis co-infection. Nat. Reviews. Microbiol. 2018, 16, 80–90. [Google Scholar] [CrossRef] [PubMed]
- Tonoyan, L.; Vincent-Bugnas, S.; Olivieri, C.V.; Doglio, A. New Viral Facets in Oral Diseases: The EBV Paradox. Int. J. Mol. Sci. 2019, 20, 5861. [Google Scholar] [CrossRef]
- Jossart, G.H.; Canafax, D.M.; Erdmann, G.R.; Lovdahl, M.J.; Russlie, H.Q.; Juhn, S.K.; Giebink, G.S. Effect of Streptococcus pneumoniae and influenza A virus on middle ear antimicrobial pharmacokinetics in experimental otitis media. Pharm. Res. 1994, 11, 860–864. [Google Scholar] [CrossRef]
- Melvin, J.A.; Bomberger, J.M. Compromised Defenses: Exploitation of Epithelial Responses During Viral-Bacterial Co-Infection of the Respiratory Tract. PLoS Pathog. 2016, 12, e1005797. [Google Scholar] [CrossRef]
- Berger, A.K.; Mainou, B.A. Interactions between Enteric Bacteria and Eukaryotic Viruses Impact the Outcome of Infection. Viruses 2018, 10, 19. [Google Scholar] [CrossRef]
- Karst, S.M. The influence of commensal bacteria on infection with enteric viruses. Nat. Reviews. Microbiol. 2016, 14, 197–204. [Google Scholar] [CrossRef]
- Rowe, H.M.; Meliopoulos, V.A.; Iverson, A.; Bomme, P.; Schultz-Cherry, S.; Rosch, J.W. Direct interactions with influenza promote bacterial adherence during respiratory infections. Nat. Microbiol. 2019, 4, 1328–1336. [Google Scholar] [CrossRef]
- Kiedrowski, M.R.; Bomberger, J.M. Viral-Bacterial Co-infections in the Cystic Fibrosis Respiratory Tract. Front. Immunol. 2018, 9, 3067. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.Y.; Lee, H.; Wang, X.M.; Lee, T.F.; Liao, C.H.; Teng, L.J.; Hsueh, P.R. High mortality impact of Staphylococcus argenteus on patients with community-onset staphylococcal bacteraemia. Int. J. Antimicrob. Agents 2018, 52, 747–753. [Google Scholar] [CrossRef] [PubMed]
- Weinstein, M.P.; Murphy, J.R.; Reller, L.B.; Lichtenstein, K.A. The clinical significance of positive blood cultures: A comprehensive analysis of 500 episodes of bacteremia and fungemia in adults. II. Clinical observations, with special reference to factors influencing prognosis. Rev. Infect. Dis. 1983, 5, 54–70. [Google Scholar] [CrossRef] [PubMed]
- Pammi, M.; Zhong, D.; Johnson, Y.; Revell, P.; Versalovic, J. Polymicrobial bloodstream infections in the neonatal intensive care unit are associated with increased mortality: A case-control study. Bmc Infect. Dis. 2014, 14, 390. [Google Scholar] [CrossRef]
- Faix, R.G.; Kovarik, S.M. Polymicrobial sepsis among intensive care nursery infants. J. Perinatol. Off. J. Calif. Perinat. Assoc. 1989, 9, 131–136. [Google Scholar]
- Hudson, V.L.; Wielinski, C.L.; Regelmann, W.E. Prognostic implications of initial oropharyngeal bacterial flora in patients with cystic fibrosis diagnosed before the age of two years. J. Pediatrics 1993, 122, 854–860. [Google Scholar] [CrossRef]
- Maliniak, M.L.; Stecenko, A.A.; McCarty, N.A. A longitudinal analysis of chronic MRSA and Pseudomonas aeruginosa co-infection in cystic fibrosis: A single-center study. J. Cyst. Fibros. Off. J. Eur. Cyst. Fibros. Soc. 2016, 15, 350–356. [Google Scholar] [CrossRef]
- Jorge, L.S.; Fucuta, P.S.; Oliveira, M.G.L.; Nakazone, M.A.; de Matos, J.A.; Chueire, A.G.; Salles, M.J.C. Outcomes and Risk Factors for Polymicrobial Posttraumatic Osteomyelitis. J. Bone Jt. Infect. 2018, 3, 20–26. [Google Scholar] [CrossRef]
- Dowd, S.E.; Wolcott, R.D.; Kennedy, J.; Jones, C.; Cox, S.B. Molecular diagnostics and personalised medicine in wound care: Assessment of outcomes. J. Wound Care 2011, 20, 232, 234–239. [Google Scholar] [CrossRef]
- Wolcott, R.D.; Cox, S.B.; Dowd, S.E. Healing and healing rates of chronic wounds in the age of molecular pathogen diagnostics. J. Wound Care 2010, 19, 272–278, 280–281. [Google Scholar] [CrossRef]

| Citation | Organisms Studied | Observation |
|---|---|---|
| Bacteria–Bacteria | ||
| Hoffman et al. [32] | P. aeruginosa, S. aureus | 2x increase in tolerance to tobramycin |
| Orazi and O’Toole [33] | P. aeruginosa, S. aureus | Increased tolerance to B-lactam, glycopeptide, aminoglycoside, macrolide, tetracycline classes |
| Lebrun et al. [44] | P. aeruginosa, S. aureus | Increased tolerance to rifamycin, vancomycin, penicillin, cycloserine |
| DeLeon et al. [34] | P. aeruginosa, S. aureus | Increased tolerance to gentamicin, tetracycline |
| Vega et al. [35] | E. coli, S. typhimurium | Increased tolerance to ciprofloxacin |
| Adamowicz et al. [23] | E. coli, S. typhimurium | Increased tolerance to tetracycline |
| Tavernier et al. [24] | P. aeruginosa, S. aureus, S. typhimurium | Increased tolerance to vancomycin |
| Bacteria–Fungus | ||
| Harriot et al. [36] | C. albicans, S. aureus | Increase in biofilm formation |
| Todd et al. [40] | C. albicans, S. aureus | Lethality increase |
| Kean et al. [30] | C. albicans, S. aureus | 4x increased tolerance to miconazole |
| Kong et al. [27] | C. albicans, S. mutans | Increased tolerance to fluconazole, response to farnesol |
| Förster et al. [42] | C. albicans, S. epidermidis | Increased tolerance to fluconazole, “slime” production |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Little, W.; Black, C.; Smith, A.C. Clinical Implications of Polymicrobial Synergism Effects on Antimicrobial Susceptibility. Pathogens 2021, 10, 144. https://doi.org/10.3390/pathogens10020144
Little W, Black C, Smith AC. Clinical Implications of Polymicrobial Synergism Effects on Antimicrobial Susceptibility. Pathogens. 2021; 10(2):144. https://doi.org/10.3390/pathogens10020144
Chicago/Turabian StyleLittle, William, Caroline Black, and Allie Clinton Smith. 2021. "Clinical Implications of Polymicrobial Synergism Effects on Antimicrobial Susceptibility" Pathogens 10, no. 2: 144. https://doi.org/10.3390/pathogens10020144
APA StyleLittle, W., Black, C., & Smith, A. C. (2021). Clinical Implications of Polymicrobial Synergism Effects on Antimicrobial Susceptibility. Pathogens, 10(2), 144. https://doi.org/10.3390/pathogens10020144






