Air Passenger Travel and International Surveillance Data Predict Spatiotemporal Variation in Measles Importations to the United States
Abstract
:1. Introduction
2. Results
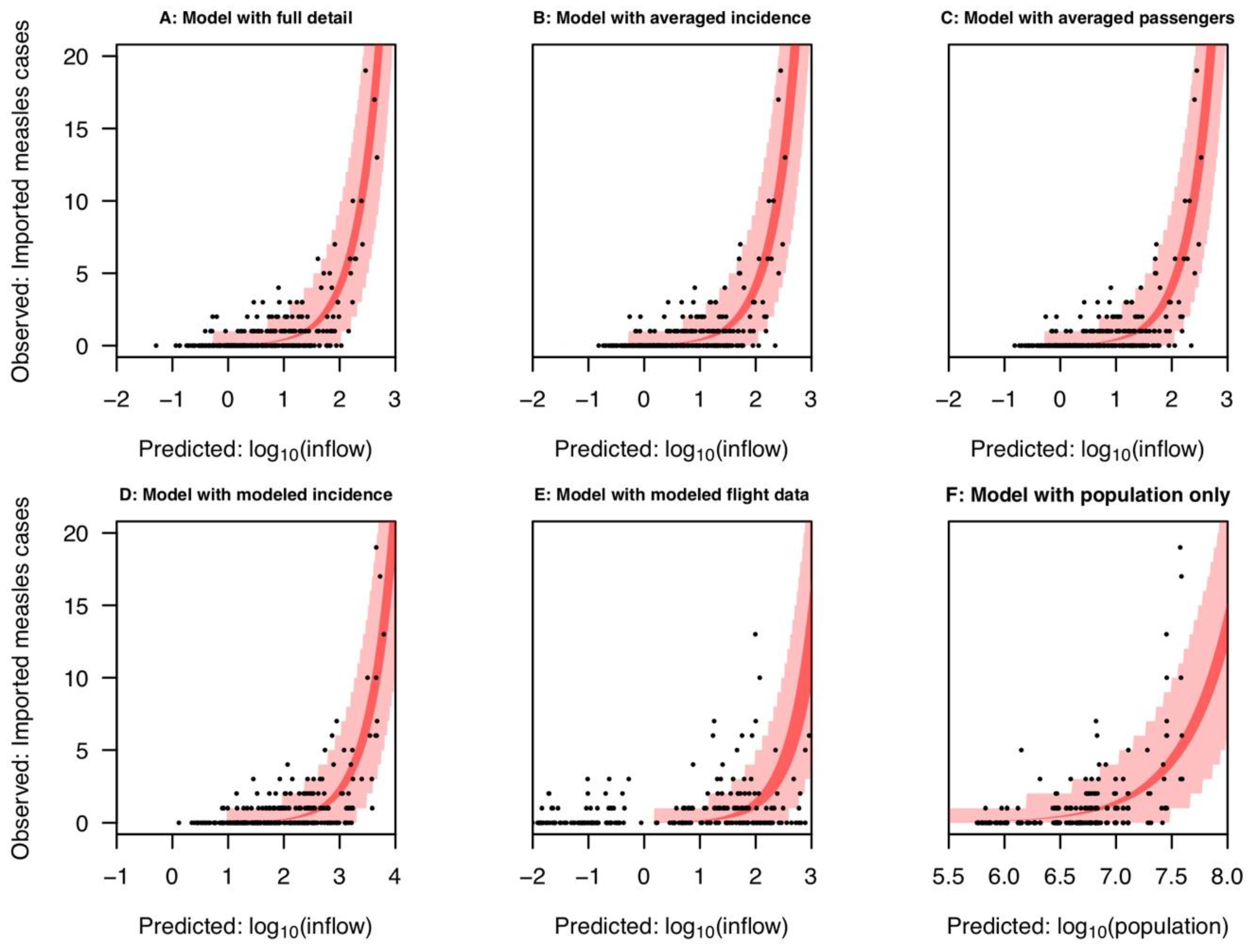
2.1. Inflow Variable Informed by Air Travel and International Surveillance Data
2.2. Comparison with Averaged Inflow Variables
2.3. Comparison with Modeled Inflow Variables
2.4. Predicting Measles Origin and Magnitude
2.5. Association between Imported and Indigenous Measles Cases
3. Discussion
4. Materials and Methods
4.1. Data
4.2. Model Description
4.3. Analyses
4.4. Alternative Inflow Variables
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gastañaduy, P.A.; Paul, P.; Fiebelkorn, A.P.; Redd, S.B.; Lopman, B.A.; Gambhir, M.; Wallace, G.S. Assessment of the Status of Measles Elimination in the United States, 2001–2014. Am. J. Epidemiol. 2017, 185, 562–569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papania, M.J.; Wallace, G.S.; Rota, P.A.; Icenogle, J.P.; Fiebelkorn, A.P.; Armstrong, G.L.; Reef, S.E.; Redd, S.B.; Abernathy, E.S.; Barskey, A.E.; et al. Elimination of Endemic Measles, Rubella, and Congenital Rubella Syndrome from the Western Hemisphere: The US Experience. JAMA Pediatr. 2014, 168, 148–155. [Google Scholar] [CrossRef] [PubMed]
- Otterman, S. Get Vaccinated or Leave School: 26,000 N.Y. Children Face a Choice; The New York Times Company: New York, NY, USA, 2019. [Google Scholar]
- McDonald, R.; Ruppert, P.S.; Souto, M. Notes from the Field: Measles Outbreaks from Imported Cases in Orthodox Jewish Communities—New York and New Jersey, 2018–2019. MMWR Morb. Mortal. Wkly. Rep. 2019, 68, 444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Measles Investigation. Clark County Public Health, 2019. Available online: https://clark.wa.gov/public-health/measles-investigation (accessed on 10 December 2020).
- Grobusch, M.P.; Rodriguez-Morales, A.J.; Wilson, M.E. Measles on the Move. Travel Med. Infect. Dis. 2017, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.D.; Clemmons, N.S.; Patel, M.; Gastañaduy, P.A. International Importations of Measles Virus into the United States During the Postelimination Era, 2001–2016. J. Infect. Dis. 2019, 219, 1616–1623. [Google Scholar] [CrossRef] [PubMed]
- Oster, N.V.; Harpaz, R.; Redd, S.B.; Papania, M.J. International Importations of Measles Virus-United States, 1993–2001. J. Infect. Dis. 2004, 189, 48–53. [Google Scholar]
- Classification of Measles Cases and Categorization of Measles Elimination Programs. Morb. Mortal. Wkly. Rep. 1983, 31, 707–711.
- Fiebelkorn, A.P.; Redd, S.B.; Gallagher, K.; Rota, P.A.; Rota, J.; Bellini, W.; Seward, J. Measles in the United States during the Postelimination Era. J. Infect. Dis. 2010, 202, 1520–1528. [Google Scholar] [CrossRef]
- Nah, K.; Otsuki, S.; Chowell, G.; Nishiura, H. Predicting the International Spread of Middle East Respiratory Syndrome (MERS). BMC Infect. Dis. 2016, 16, 356. [Google Scholar] [CrossRef] [Green Version]
- Findlater, A.; Moineddin, R.; Kain, D.; Yang, J.; Wang, X.; Lai, S.; Khan, K.; Bogoch, I.I. The Use of Air Travel Data for Predicting Dengue Importation to China: A Modelling Study. Travel Med. Infect. Dis. 2019, 101446. [Google Scholar] [CrossRef]
- Khan, K.; Bogoch, I.; Brownstein, J.S.; Miniota, J.; Nicolucci, A.; Hu, W.; Nsoesie, E.O.; Cetron, M.; Creatore, M.I.; German, M.; et al. Assessing the Origin of and Potential for International Spread of Chikungunya Virus from the Caribbean. PLoS Curr. 2014. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Hu, W.; Yakob, L.; Devine, G.J.; McGraw, E.A.; Jansen, C.C.; Faddy, H.M.; Frentiu, F.D. El Niño Southern Oscillation, Overseas Arrivals and Imported Chikungunya Cases in Australia: A Time Series Analysis. PLoS Negl. Trop. Dis. 2019, 13, e0007376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Candido, D.D.S.; Watts, A.; Abade, L.; Kraemer, M.U.G.; Pybus, O.G.; Croda, J.; de Oliveira, W.; Khan, K.; Sabino, E.C.; Faria, N.R. Routes for COVID-19 Importation in Brazil. J. Travel Med. 2020, 27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Massad, E.; Tan, S.-H.; Khan, K.; Wilder-Smith, A. Estimated Zika Virus Importations to Europe by Travellers from Brazil. Glob. Health Action 2016, 9, 31669. [Google Scholar] [CrossRef]
- Tuite, A.R.; Watts, A.G.; Khan, K.; Bogoch, I.I. Ebola Virus Outbreak in North Kivu and Ituri Provinces, Democratic Republic of Congo, and the Potential for Further Transmission through Commercial Air Travel. J. Travel Med. 2019, 26. [Google Scholar] [CrossRef]
- Tuite, A.R.; Watts, A.G.; Kraemer, M.U.G.; Khan, K.; Bogoch, I.I. Potential for Seasonal Lassa Fever Case Exportation from Nigeria. Am. J. Trop. Med. Hyg. 2019, 100, 647–651. [Google Scholar] [CrossRef] [Green Version]
- Hayman, D.T.S.; Marshall, J.C.; French, N.P.; Carpenter, T.E.; Roberts, M.G.; Kiedrzynski, T. Global Importation and Population Risk Factors for Measles in New Zealand: A Case Study for Highly Immunized Populations. Epidemiol. Infect. 2017, 145, 1875–1885. [Google Scholar] [CrossRef] [Green Version]
- Huoi, C.; Casalegno, J.S.; Bénet, T.; Neuraz, A.; Billaud, G.; Eibach, D.; Mekki, Y.; Rudigoz, R.; Massardier, J.; Huissoud, C.; et al. A Report on the Large Measles Outbreak in Lyon, France, 2010 to 2011. Eurosurveillance 2012, 17, 20264. [Google Scholar] [CrossRef]
- Chen, S.; Fricks, J.; Ferrari, M.J. Tracking Measles Infection through Non-Linear State Space Models. J. R. Stat. Soc. Ser. C (Appl. Stat.) 2012, 61, 117–134. [Google Scholar] [CrossRef]
- Muscat, M. Who Gets Measles in Europe? J. Infect. Dis. 2011, 204, S353–S365. [Google Scholar] [CrossRef] [Green Version]
- Bednarczyk, R.A.; Rebolledo, P.A.; Omer, S.B. Assessment of the Role of International Travel and Unauthorized Immigration on Measles Importation to the United States. J. Travel Med. 2016, 23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mao, L.; Wu, X.; Huang, Z.; Tatem, A.J. Modeling Monthly Flows of Global Air Travel Passengers: An Open-Access Data Resources. J. Transp. Geogr. 2015, 48, 52–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Equatorial Guinea Operations. Available online: https://www.marathonoil.com/operations/eg/ (accessed on 16 September 2020).
- Full Article: Pandemics, Tourism and Global Change: A Rapid Assessment of COVID-19. Available online: https://www.tandfonline.com/doi/full/10.1080/09669582.2020.1758708 (accessed on 16 September 2020).
- Tuite, A.R.; Bhatia, D.; Moineddin, R.; Bogoch, I.I.; Watts, A.G.; Khan, K. Global Trends in Air Travel: Implications for Connectivity and Resilience to Infectious Disease Threats. J. Travel Med. 2020, 27. [Google Scholar] [CrossRef]
- Mitruka, K.; Felsen, C.B.; Tomianovic, D.; Inman, B.; Street, K.; Yambor, P.; Reef, S.E. Measles, Rubella, and Varicella Among the Crew of a Cruise Ship Sailing From Florida, United States, 2006. J. Travel Med. 2012, 19, 233–237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Victor, D. Measles on Cruise Ship Leads to Quarantine in St. Lucia. The New York Times, 2019. [Google Scholar]
- Truelove, S.A.; Mier-y-Teran-Romero, L.; Gastanaduy, P.; Walker, A.T.; Berro, A.; Lessler, J.; Johansson, M. Epidemics, Air Travel, and Elimination in a Globalized World: The Case of Measles. medRxiv 2020. [Google Scholar] [CrossRef]
- Sarkar, S.; Zlojutro, A.; Khan, K.; Gardner, L. Measles Resurgence in the USA: How International Travel Compounds Vaccine Resistance. Lancet Infect. Dis. 2019, 19, 684–686. [Google Scholar] [CrossRef]
- Otten, M.; Kezaala, R.; Fall, A.; Masresha, B.; Martin, R.; Cairns, L.; Eggers, R.; Biellik, R.; Grabowsky, M.; Strebel, P.; et al. Public-Health Impact of Accelerated Measles Control in the WHO African Region 2000–2003. Lancet 2005, 366, 832–839. [Google Scholar] [CrossRef] [Green Version]
- Oidtman, R.J.; Lai, S.; Huang, Z.; Yang, J.; Siraj, A.S.; Reiner, R.C.; Tatem, A.J.; Perkins, T.A.; Yu, H. Inter-Annual Variation in Seasonal Dengue Epidemics Driven by Multiple Interacting Factors in Guangzhou, China. Nat. Commun. 2019, 10, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Nationally Notifiable Infectious Diseases and Conditions, United States: Annual Tables; Centers for Disease Control and Prevention: Atlanta, GA, USA, 2018.
- Morbidity and Mortality Weekly Report: Summary of Notifiable Infectious Diseases; Centers for Disease Control and Prevention: Atlanta, GA, USA, 2018.
- Sanyaolu, A.; Okorie, C.; Marinkovic, A.; Ayodele, O.; Abbasi, A.F.; Prakash, S.; Gosse, J.; Younis, S.; Mangat, J.; Chan, H. Measles Outbreak in Unvaccinated and Partially Vaccinated Children and Adults in the United States and Canada (2018–2019): A Narrative Review of Cases. Inquiry 2019, 56. [Google Scholar] [CrossRef]
- Taylor, E.M.; Burke, H.; Zhou, W. Crossing Borders: One World, Global Health. Clin. Infect. Dis. 2012, 55, v–vi. [Google Scholar]
- World Health Organization Provisional Monthly Measles and Rubella Data. Available online: https://www.who.int/teams/immunization-vaccines-and-biologicals/immunization-analysis-and-insights/surveillance/monitoring/provisional-monthly-measles-and-rubella-data (accessed on 15 May 2019).
- United Nations Department of Economic and Social Affairs. World Population Prospects; United Nations Department of Economic and Social Affairs: New York, NY, USA, 2018. [Google Scholar]
- Simons, E.; Ferrari, M.; Fricks, J.; Wannemuehler, K.; Anand, A.; Burton, A.; Strebel, P. Assessment of the 2010 Global Measles Mortality Reduction Goal: Results from a Model of Surveillance Data. Lancet 2012, 379, 2173–2178. [Google Scholar] [CrossRef]
- Perry, R.T.; Gacic-Dobo, M.; Dabbagh, A.; Mulders, M.N.; Strebel, P.M.; Okwo-Bele, J.-M.; Rota, P.A.; Goodson, J.L. Progress Toward Regional Measles Elimination—Worldwide, 2000–2013. MMWR Morb. Mortal. Wkly. Rep. 2014, 63, 1034–1038. [Google Scholar] [PubMed]
- Perry, R.T.; Murray, J.S.; Gacic-Dobo, M.; Dabbagh, A.; Mulders, M.N.; Strebel, P.M.; Okwo-Bele, J.-M.; Rota, P.A.; Goodson, J.L. Progress Toward Regional Measles Elimination—Worldwide, 2000–2014. MMWR Morb. Mortal. Wkly. Rep. 2015, 64, 1246–1251. [Google Scholar] [CrossRef] [PubMed]
- Patel, M.K.; Gacic-Dobo, M.; Strebel, P.M.; Dabbagh, A.; Mulders, M.N.; Okwo-Bele, J.-M.; Dumolard, L.; Rota, P.A.; Kretsinger, K.; Goodson, J.L. Progress Toward Regional Measles Elimination—Worldwide, 2000–2015. MMWR Morb. Mortal. Wkly. Rep. 2016, 65, 1228–1233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dabbagh, A.; Patel, M.K.; Dumolard, L.; Gacic-Dobo, M.; Mulders, M.N.; Okwo-Bele, J.-M.; Kretsinger, K.; Papania, M.J.; Rota, P.A.; Goodson, J.L. Progress toward Regional Measles Elimination—Worldwide, 2000–2016. MMWR Morb. Mortal. Wkly. Rep. 2017, 66, 1148–1153. [Google Scholar] [CrossRef] [Green Version]
- International Air Transport Association. Montreal, QC, Canada, 2018. Available online: http://www.iata.org (accessed on 10 December 2020).
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019. [Google Scholar]
- Robin, X.; Turck, N.; Hainard, A.; Tiberti, N.; Lisacek, F.; Sanches, J.-C.; Müller, M. PROC: An Open-Source Package for R and S+ to Analyze and Compare ROC Curves. BMC Bioinform. 2011, 12, 77. [Google Scholar] [CrossRef]
- State Population Totals and Components of Change: 2010–2019; United States Census Bureau: Suitland, MD, USA, 2018.

| Inflow Variable | λ | Correlation | AUC 1 |
|---|---|---|---|
| Full detail | 0.041 | 0.84 | 0.78 |
| Averaged incidence | 0.041 | 0.76 | 0.79 |
| Averaged air travel | 0.041 | 0.76 | 0.79 |
| Modeled incidence | 0.0022 | 0.77 | 0.79 |
| Modeled flight data | 0.012 | 0.61 | 0.67 |
| Population only | 4.32 × 10−5 | 0.66 | 0.58 |
| Inflow Variable | Cross-Validated λ (Mean ± SD 1) | Cross-Validated Correlation (Mean ± SD) | Cross-Validated AUC (Mean ± SD) |
|---|---|---|---|
| Full detail | 0.041 ± 0.002 | 0.78 ± 0.2 | 0.78 ± 0.06 |
| Averaged incidence | 0.041 ± 0.002 | 0.71 ± 0.2 | 0.79 ± 0.05 |
| Averaged air travel | 0.041 ± 0.002 | 0.76 ± 0.2 | 0.79 ± 0.05 |
| Modeled incidence | 0.0022 ± 1 × 10−4 | 0.77 ± 0.2 | 0.79 ± 0.06 |
| Modeled flight data | 0.012 ± 9.3 × 10−4 | 0.61 ± 0.3 | 0.67 ± 0.07 |
| Population only | 4.32 × 10−5 | 0.66 ± 0.1 | 0.58 ± 0.03 |
| Inflow Variable | Flux | Incidence |
|---|---|---|
| Full detail | Complete flight data | Complete incidence data |
| Averaged incidence | Complete flight data | Averaged incidence data |
| Averaged air travel | Averaged flight data | Complete incidence data |
| Modeled incidence | Complete flight data | Alternative incidence data |
| Modeled flight data | Alternative flight data | Complete incidence data |
| Population only | No flight data | No incidence data |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Poterek, M.L.; Kraemer, M.U.G.; Watts, A.; Khan, K.; Perkins, T.A. Air Passenger Travel and International Surveillance Data Predict Spatiotemporal Variation in Measles Importations to the United States. Pathogens 2021, 10, 155. https://doi.org/10.3390/pathogens10020155
Poterek ML, Kraemer MUG, Watts A, Khan K, Perkins TA. Air Passenger Travel and International Surveillance Data Predict Spatiotemporal Variation in Measles Importations to the United States. Pathogens. 2021; 10(2):155. https://doi.org/10.3390/pathogens10020155
Chicago/Turabian StylePoterek, Marya L., Moritz U. G. Kraemer, Alexander Watts, Kamran Khan, and T. Alex Perkins. 2021. "Air Passenger Travel and International Surveillance Data Predict Spatiotemporal Variation in Measles Importations to the United States" Pathogens 10, no. 2: 155. https://doi.org/10.3390/pathogens10020155
APA StylePoterek, M. L., Kraemer, M. U. G., Watts, A., Khan, K., & Perkins, T. A. (2021). Air Passenger Travel and International Surveillance Data Predict Spatiotemporal Variation in Measles Importations to the United States. Pathogens, 10(2), 155. https://doi.org/10.3390/pathogens10020155






