Refining Host-Pathogen Interactions: Organ-on-Chip Side of the Coin
Abstract
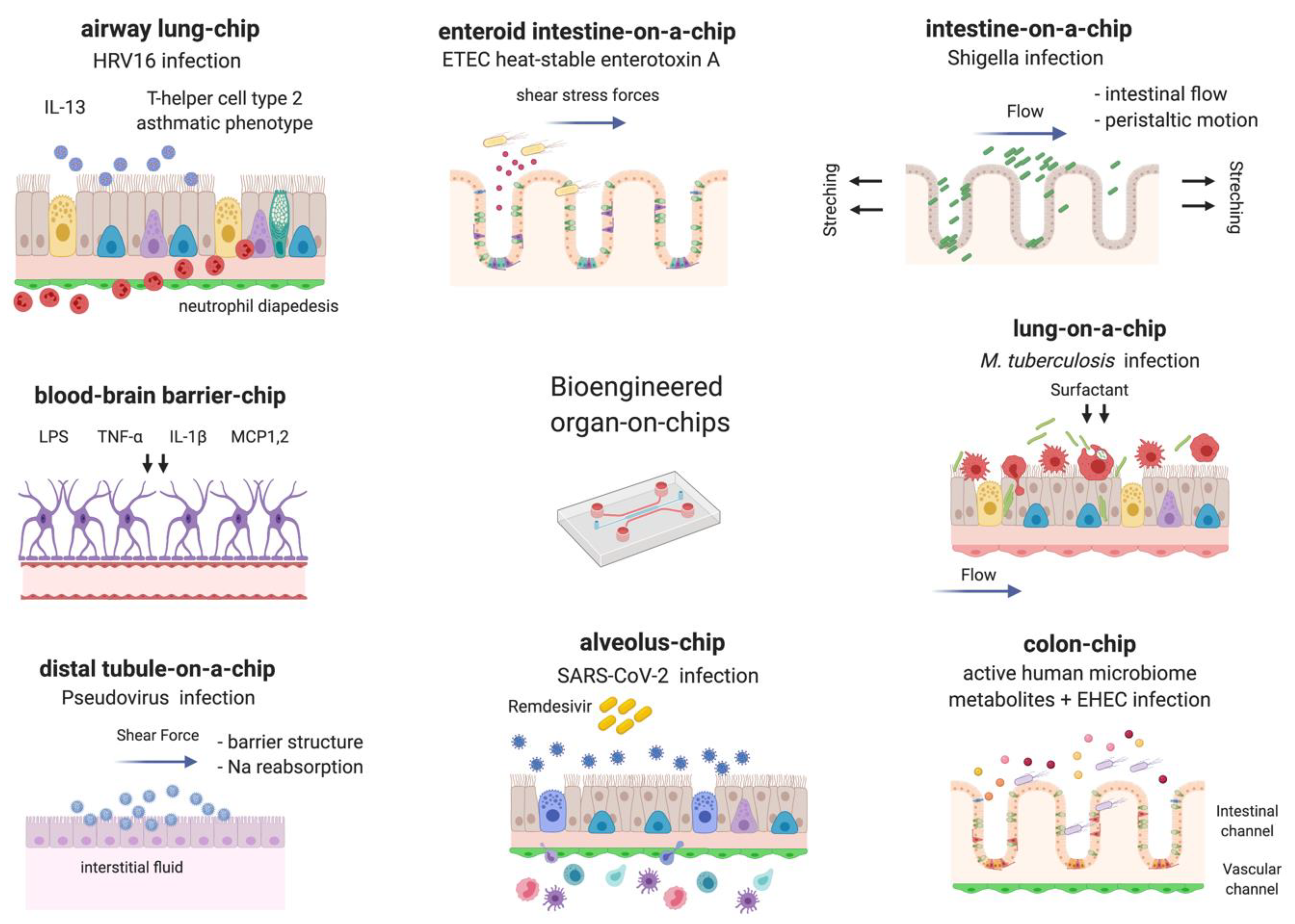
1. Introduction
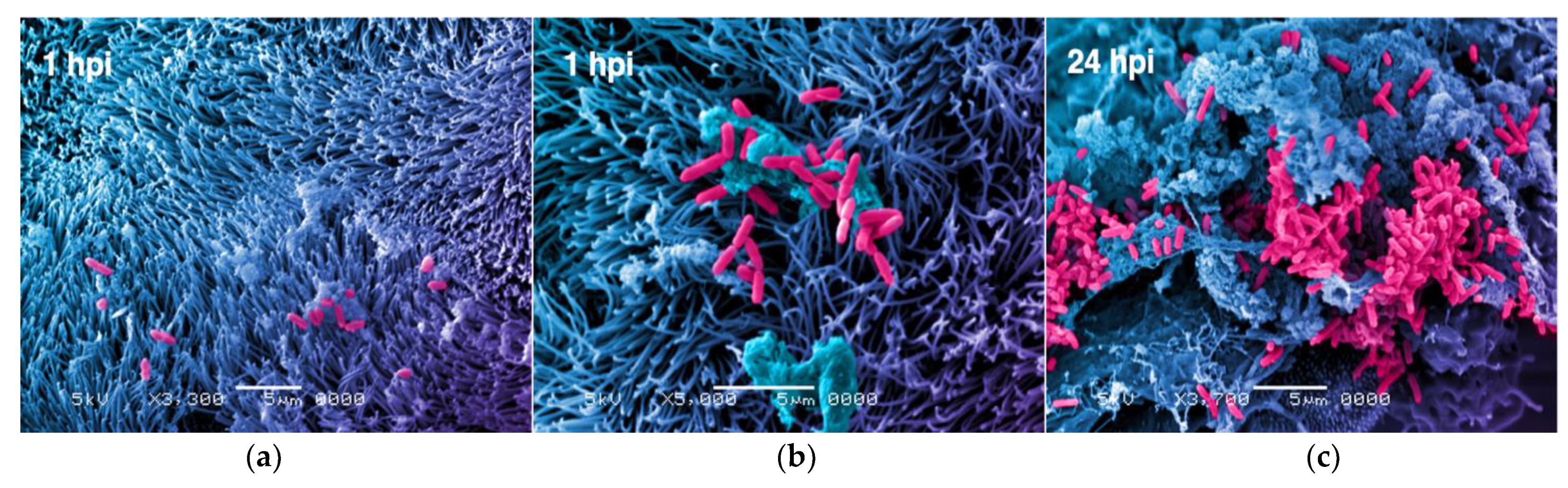
2. Bacteria
3. Viruses
4. Fungi and Parasites
5. Microbiome
6. Moving Forward: What’s Next on the Chip?
7. Limitations
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Sin, A.; Chin, K.C.; Jamil, M.F.; Kostov, Y.; Rao, G.; Shuler, M.L. The Design and Fabrication of Three-Chamber Microscale Cell Culture Analog Devices with Integrated Dissolved Oxygen Sensors. Biotechnol. Prog. 2004, 20, 338–345. [Google Scholar] [CrossRef]
- Viravaidya, K.; Sin, A.; Shuler, M.L. Development of a Microscale Cell Culture Analog to Probe Naphthalene Toxicity. Biotechnol. Prog. 2004, 20, 316–323. [Google Scholar] [CrossRef]
- Huh, D.; Matthews, B.D.; Mammoto, A.; Montoya-Zavala, M.; Yuan Hsin, H.; Ingber, D.E. Reconstituting organ-level lung functions on a chip. Science 2010, 328, 1662–1668. [Google Scholar] [CrossRef] [PubMed]
- Huh, D.; Fujioka, H.; Tung, Y.C.; Futai, N.; Paine, R.; Grotberg, J.B.; Takayama, S. Acoustically detectable cellular-level lung injury induced by fluid mechanical stresses in microfluidic airway systems. Proc. Natl. Acad. Sci. USA 2007, 104, 18886–18891. [Google Scholar] [CrossRef] [PubMed]
- Huh, D.; Kim, H.J.; Fraser, J.P.; Shea, D.E.; Khan, M.; Bahinski, A.; Hamilton, G.A.; Ingber, D.E. Microfabrication of human organs-on-chips. Nat. Protoc. 2013, 8, 2135–2157. [Google Scholar] [CrossRef]
- Mahler, G.J.; Esch, M.B.; Glahn, R.P.; Shuler, M.L. Characterization of a gastrointestinal tract microscale cell culture analog used to predict drug toxicity. Biotechnol. Bioeng. 2009, 104, 193–205. [Google Scholar] [CrossRef] [PubMed]
- Kimura, H.; Yamamoto, T.; Sakai, H.; Sakai, Y.; Fujii, T. An integrated microfluidic system for long-term perfusion culture and on-line monitoring of intestinal tissue models. Lab Chip 2008, 8, 741–746. [Google Scholar] [CrossRef]
- Lee, P.J.; Hung, P.J.; Lee, L.P. An artificial liver sinusoid with a microfluidic endothelial-like barrier for primary hepatocyte culture. Biotechnol. Bioeng. 2007, 97, 1340–1346. [Google Scholar] [CrossRef]
- Carraro, A.; Hsu, W.M.; Kulig, K.M.; Cheung, W.S.; Miller, M.L.; Weinberg, E.J.; Swart, E.F.; Kaazempur-Mofrad, M.; Borenstein, J.T.; Vacanti, J.P.; et al. In Vitro analysis of a hepatic device with intrinsic microvascular-based channels. Biomed. Microdevices 2008, 10, 795–805. [Google Scholar] [CrossRef]
- Jang, K.J.; Suh, K.Y. A multi-layer microfluidic device for efficient culture and analysis of renal tubular cells. Lab Chip 2010, 10, 36–42. [Google Scholar] [CrossRef]
- Harris, S.G.; Shuler, M.L. Growth of endothelial cells on microfabricated silicon nitride membranes for an In Vitro model of the blood-brain barrier. Biotechnol. Bioprocess Eng. 2003, 8, 246–251. [Google Scholar] [CrossRef]
- Booth, R.; Kim, H. Characterization of a microfluidic in vitro model of the blood-brain barrier (μBBB). Lab Chip 2012, 12, 1784–1792. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, A.; Goss, J.A.; Cho, A.; McCain, M.L.; Parker, K.K. Microfluidic heart on a chip for higher throughput pharmacological studies. Lab Chip 2013, 13, 3599–3608. [Google Scholar] [CrossRef]
- Hou, L.; Hagen, J.; Wang, X.; Papautsky, I.; Naik, R.; Kelley-Loughnane, N.; Heikenfeld, J. Artificial microfluidic skin for in vitro perspiration simulation and testing. Lab Chip 2013, 13, 1868. [Google Scholar] [CrossRef] [PubMed]
- Song, J.W.; Gu, W.; Futai, N.; Warner, K.A.; Nor, J.E.; Takayama, S. Computer-controlled microcirculatory support system for endothelial cell culture and shearing. Anal. Chem. 2005, 77, 3993–3999. [Google Scholar] [CrossRef] [PubMed]
- Jang, K.; Sato, K.; Igawa, K.; Chung, U.-I.; Kitamori, T. Development of an osteoblast-based 3D continuous-perfusion microfluidic system for drug screening. Anal. Bioanal. Chem. 2008, 390, 825–832. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Kongsuphol, P.; Gourikutty, S.B.N.; Ramadan, Q. Human adipocyte differentiation and characterization in a perfusion-based cell culture device. Biomed. Microdevices 2017, 19, 84. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.S.; Romero, R.; Han, Y.M.; Kim, H.C.; Kim, C.J.; Hong, J.S.; Huh, D. Placenta-on-A-chip: A novel platform to study the biology of the human placenta. J. Matern. Fetal Neonatal Med. 2016, 29, 1046–1054. [Google Scholar] [CrossRef]
- Zervantonakis, I.K.; Hughes-Alford, S.K.; Charest, J.L.; Condeelis, J.S.; Gertler, F.B.; Kamm, R.D. Three-dimensional microfluidic model for tumor cell intravasation and endothelial barrier function. Proc. Natl. Acad. Sci. USA 2012, 109, 13515–13520. [Google Scholar] [CrossRef]
- Shirure, V.S.; Bi, Y.; Curtis, M.B.; Lezia, A.; Goedegebuure, M.M.; Goedegebuure, S.P.; Aft, R.; Fields, R.C.; George, S.C. Tumor-on-a-chip platform to investigate progression and drug sensitivity in cell lines and patient-derived organoids. Lab Chip 2018, 18, 3687–3702. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.S.; Davoudi, F.; Walch, P.; Manbachi, A.; Luo, X.; Dell’Erba, V.; Miri, A.K.; Albadawi, H.; Arneri, A.; Li, X.; et al. Bioprinted thrombosis-on-a-chip. Lab Chip 2016, 16, 4097–4105. [Google Scholar] [CrossRef]
- Park, J.; Lee, B.K.; Jeong, G.S.; Hyun, J.K.; Lee, C.J.; Lee, S.H. Three-dimensional brain-on-a-chip with an interstitial level of flow and its application as an in vitro model of Alzheimer’s disease. Lab Chip 2015, 15, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Warrick, J.W.; Haubert, K.; Beebe, D.J.; Yin, J. Infection on a chip: A microscale platform for simple and sensitive cell-based virus assays. Biomed. Microdevices 2009, 11, 565–570. [Google Scholar] [CrossRef]
- Wu, Q.; Liu, J.; Wang, X.; Feng, L.; Wu, J.; Zhu, X.; Wen, W.; Gong, X. Organ-on-a-chip: Recent breakthroughs and future prospects. Biomed. Eng. Online 2020, 19, 9. [Google Scholar] [CrossRef] [PubMed]
- Ergir, E.; Bachmann, B.; Redl, H.; Forte, G.; Ertl, P. Small force, big impact: Next generation organ-on-a-chip systems incorporating biomechanical cues. Front. Physiol. 2018, 9, 1417. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Korolj, A.; Lai, B.F.L.; Radisic, M. Advances in organ-on-a-chip engineering. Nat. Rev. Mater. 2018, 3, 257–278. [Google Scholar] [CrossRef]
- Sosa-Hernández, J.E.; Villalba-Rodríguez, A.M.; Romero-Castillo, K.D.; Aguilar-Aguila-Isaías, M.A.; García-Reyes, I.E.; Hernández-Antonio, A.; Ahmed, I.; Sharma, A.; Parra-Saldívar, R.; Iqbal, H.M.N. Organs-on-a-chip module: A review from the development and applications perspective. Micromachines 2018, 9, 536. [Google Scholar] [CrossRef] [PubMed]
- Yi, H.G.; Lee, H.; Cho, D.W. 3D printing of organs-on-chips. Bioengineering 2017, 4, 10. [Google Scholar] [CrossRef]
- Barrila, J.; Crabbé, A.; Yang, J.; Franco, K.; Nydam, S.D.; Forsyth, R.J.; Davis, R.R.; Gangaraju, S.; Mark Ott, C.; Coyne, C.B.; et al. Modeling host-pathogen interactions in the context of the microenvironment: Three-dimensional cell culture comes of age. Infect. Immun. 2018, 86. [Google Scholar] [CrossRef]
- Ingber, D.E. Is it Time for Reviewer 3 to Request Human Organ Chip Experiments Instead of Animal Validation Studies? Adv. Sci. 2020, 7. [Google Scholar] [CrossRef]
- Fasciano, A.C.; Mecsas, J.; Isberg, R.R. New Age Strategies To Reconstruct Mucosal Tissue Colonization and Growth in Cell Culture Systems. Microbiol. Spectr. 2019, 7, 59–68. [Google Scholar] [CrossRef]
- Mounier, J.; Vasselon, T.; Hellio, R.; Lesourd, M.; Sansonetti, P.J. Shigella flexneri enters human colonic Caco-2 epithelial cells through the basolateral pole. Infect. Immun. 1992, 60, 237–248. [Google Scholar] [CrossRef] [PubMed]
- Koestler, B.J.; Ward, C.M.; Fisher, C.R.; Rajan, A.; Maresso, A.W.; Payne, S.M. Human intestinal enteroids as a model system of shigella pathogenesis. Infect. Immun. 2019, 87, e00733-18. [Google Scholar] [CrossRef] [PubMed]
- Ranganathan, S.; Doucet, M.; Grassel, C.L.; Delaine-Elias, B.O.; Zachos, N.C.; Barry, E.M. Evaluating shigella flexneri pathogenesis in the human enteroid model. Infect. Immun. 2019, 87, e00740-18. [Google Scholar] [CrossRef]
- Kasendra, M.; Tovaglieri, A.; Sontheimer-Phelps, A.; Jalili-Firoozinezhad, S.; Bein, A.; Chalkiadaki, A.; Scholl, W.; Zhang, C.; Rickner, H.; Richmond, C.A.; et al. Development of a primary human Small Intestine-on-a-Chip using biopsy-derived organoids. Sci. Rep. 2018, 8, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Jalili-Firoozinezhad, S.; Prantil-Baun, R.; Jiang, A.; Potla, R.; Mammoto, T.; Weaver, J.C.; Ferrante, T.C.; Kim, H.J.; Cabral, J.M.S.; Levy, O.; et al. Modeling radiation injury-induced cell death and countermeasure drug responses in a human Gut-on-a-Chip article. Cell Death Dis. 2018, 9, 1–14. [Google Scholar] [CrossRef]
- Guo, Y.; Li, Z.; Su, W.; Wang, L.; Zhu, Y.; Qin, J. A Biomimetic Human Gut-on-a-Chip for Modeling Drug Metabolism in Intestine. Artif. Organs 2018, 42, 1196–1205. [Google Scholar] [CrossRef]
- Grassart, A.; Malardé, V.; Gobba, S.; Sartori-Rupp, A.; Kerns, J.; Karalis, K.; Marteyn, B.; Sansonetti, P.; Sauvonnet, N. Bioengineered Human Organ-on-Chip Reveals Intestinal Microenvironment and Mechanical Forces Impacting Shigella Infection. Cell Host Microbe 2019, 26, 435–444.e4. [Google Scholar] [CrossRef]
- Sunuwar, L.; Yin, J.; Kasendra, M.; Karalis, K.; Kaper, J.; Fleckenstein, J.; Donowitz, M. Mechanical stimuli affect Escherichia coli heat-stable enterotoxin-cyclic GMP signaling in a human enteroid intestine-chip model. Infect. Immun. 2020, 88, e00866-19. [Google Scholar] [CrossRef]
- Torrelles, J.B.; Schlesinger, L.S. Integrating Lung Physiology, Immunology, and Tuberculosis. Trends Microbiol. 2017, 25, 688–697. [Google Scholar] [CrossRef]
- Thacker, V.V.; Dhar, N.; Sharma, K.; Barrile, R.; Karalis, K.; McKinney, J.D. A lung-on-chip model of early M. tuberculosis infection reveals an essential role for alveolar epithelial cells in controlling bacterial growth. eLife 2020, 9, e59961. [Google Scholar] [CrossRef]
- Deinhardt-Emmer, S.; Rennert, K.; Schicke, E.; Cseresnyés, Z.; Windolph, M.; Nietzsche, S.; Heller, R.; Siwczak, F.; Haupt, K.F.; Carlstedt, S.; et al. Co-infection with Staphylococcus aureus after primary influenza virus infection leads to damage of the endothelium in a human alveolus-on-a-chip model. Biofabrication 2020, 12, 025012. [Google Scholar] [CrossRef] [PubMed]
- Shi, D.; Mi, G.; Wang, M.; Webster, T.J. In Vitro and Ex Vivo systems at the forefront of infection modeling and drug discovery. Biomaterials 2019, 198, 228–249. [Google Scholar] [CrossRef]
- Terry, J.; Neethirajan, S. A novel microfluidic wound model for testing antimicrobial agents against Staphylococcus pseudintermedius biofilms. J. Nanobiotechnol. 2014, 12, 1. [Google Scholar] [CrossRef]
- Wright, E.; Neethirajan, S.; Weng, X. Microfluidic wound model for studying the behaviors of Pseudomonas aeruginosa in polymicrobial biofilms. Biotechnol. Bioeng. 2015, 112, 2351–2359. [Google Scholar] [CrossRef]
- Tang, H.; Abouleila, Y.; Si, L.; Ortega-Prieto, A.M.; Mummery, C.L.; Ingber, D.E.; Mashaghi, A. Human Organs-on-Chips for Virology. Trends Microbiol. 2020, 28, 934–946. [Google Scholar] [CrossRef]
- Benam, K.H.; Villenave, R.; Lucchesi, C.; Varone, A.; Hubeau, C.; Lee, H.H.; Alves, S.E.; Salmon, M.; Ferrante, T.C.; Weaver, J.C.; et al. Small airway-on-a-chip enables analysis of human lung inflammation and drug responses in vitro. Nat. Methods 2016, 13, 151–157. [Google Scholar] [CrossRef]
- Ortega-Prieto, A.M.; Cherry, C.; Gunn, H.; Dorner, M. In Vivo Model Systems for Hepatitis B Virus Research. ACS Infect. Dis. 2019, 5, 688–702. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.B.A.; Sodunke, T.R.; Lamontagne, J.; Cirillo, J.; Rajiv, C.; Bouchard, M.J.; Noh, M. Liver sinusoid on a chip: Long-term layered co-culture of primary rat hepatocytes and endothelial cells in microfluidic platforms. Biotechnol. Bioeng. 2015, 112, 2571–2582. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.B.; Rawat, S.; Duchemin, N.; Bouchard, M.; Noh, M. Human liver sinusoid on a chip for hepatitis B virus replication study. Micromachines 2017, 8, 27. [Google Scholar] [CrossRef]
- Ortega-Prieto, A.M.; Skelton, J.K.; Wai, S.N.; Large, E.; Lussignol, M.; Vizcay-Barrena, G.; Hughes, D.; Fleck, R.A.; Thursz, M.; Catanese, M.T.; et al. 3D microfluidic liver cultures as a physiological preclinical tool for hepatitis B virus infection. Nat. Commun. 2018, 9, 1–15. [Google Scholar] [CrossRef]
- Villenave, R.; Wales, S.Q.; Hamkins-Indik, T.; Papafragkou, E.; Weaver, J.C.; Ferrante, T.C.; Bahinski, A.; Elkins, C.A.; Kulka, M.; Ingber, D.E. Human gut-on-a-chip supports polarized infection of coxsackie B1 virus in vitro. PLoS ONE 2017, 12, e0169412. [Google Scholar] [CrossRef]
- Miccoli, B.; Braeken, D.; Li, Y.-C.E. Brain-on-a-chip Devices for Drug Screening and Disease Modeling Applications. Curr. Pharm. Des. 2019, 24, 5419–5436. [Google Scholar] [CrossRef] [PubMed]
- Johnson, B.N.; Lancaster, K.Z.; Hogue, I.B.; Meng, F.; Kong, Y.L.; Enquist, L.W.; McAlpine, M.C. 3D printed nervous system on a chip. Lab Chip 2016, 16, 1393–1400. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, C.; Xu, N.; Liu, Z.-F.; Pang, D.-W.; Zhang, Z.-L. A virus-induced kidney disease model based on organ-on-a-chip: Pathogenesis exploration of virus-related renal dysfunctions. Biomaterials 2019, 219, 119367. [Google Scholar] [CrossRef] [PubMed]
- Nawroth, J.C.; Lucchesi, C.; Cheng, D.; Shukla, A.; Ngyuen, J.; Shroff, T.; Varone, A.; Karalis, K.; Lee, H.H.; Alves, S.; et al. A microengineered airway lung chip models key features of viral-induced exacerbation of asthma. Am. J. Respir. Cell Mol. Biol. 2020, 63, 591–600. [Google Scholar] [CrossRef] [PubMed]
- Si, L.; Prantil-Baun, R.; Benam, K.H.; Bai, H.; Rodas, M.; Burt, M.; Ingber, D.E. Discovery of influenza drug resistance mutations and host therapeutic targets using a human airway chip. bioRxiv 2019. [Google Scholar] [CrossRef]
- Steel, J.; Lowen, A.C. Influenza a virus reassortment. Curr. Top. Microbiol. Immunol. 2014, 385, 377–401. [Google Scholar] [CrossRef]
- Si, L.; Bai, H.; Rodas, M.; Cao, W.; Oh, C.Y.; Jiang, A.; Nurani, A.; Zhu, D.; Goyal, G.; Gilpin, S.; et al. Human organs-on-chips as tools for repurposing approved drugs as potential influenza and COVID19 therapeutics in viral pandemics. bioRxiv 2020. [Google Scholar] [CrossRef]
- Zhang, M.; Wang, P.; Luo, R.; Wang, Y.; Li, Z.; Guo, Y.; Yao, Y.; Li, M.; Tao, T.; Chen, W.; et al. Biomimetic Human Disease Model of SARS-CoV-2 Induced Lung Injury and Immune Responses on Organ Chip System. Adv. Sci. 2020, 8, 2002928. [Google Scholar] [CrossRef]
- Junaid, A.; Tang, H.; van Reeuwijk, A.; Abouleila, Y.; Wuelfroth, P.; van Duinen, V.; Stam, W.; van Zonneveld, A.J.; Hankemeier, T.; Mashaghi, A. Ebola Hemorrhagic Shock Syndrome-on-a-Chip. iScience 2020, 23, 100765. [Google Scholar] [CrossRef] [PubMed]
- Henry, B.; Roussel, C.; Carucci, M.; Brousse, V.; Ndour, P.A.; Buffet, P. The Human Spleen in Malaria: Filter or Shelter? Trends Parasitol. 2020, 36, 435–446. [Google Scholar] [CrossRef] [PubMed]
- Rigat-Brugarolas, L.G.; Elizalde-Torrent, A.; Bernabeu, M.; De Niz, M.; Martin-Jaular, L.; Fernandez-Becerra, C.; Homs-Corbera, A.; Samitier, J.; Del Portillo, H.A. A functional microengineered model of the human splenon-on-a-chip. Lab Chip 2014, 14, 1715–1724. [Google Scholar] [CrossRef] [PubMed]
- Introini, V.; Carciati, A.; Tomaiuolo, G.; Cicuta, P.; Guido, S. Endothelial glycocalyx regulates cytoadherence in Plasmodium falciparum malaria. J. R. Soc. Interface 2018, 15, 20180773. [Google Scholar] [CrossRef]
- Arakawa, C.; Gunnarsson, C.; Howard, C.; Bernabeu, M.; Phong, K.; Yang, E.; DeForest, C.A.; Smith, J.D.; Zheng, Y. Biophysical and biomolecular interactions of malaria-infected erythrocytes in engineered human capillaries. Sci. Adv. 2020, 6, eaay7243. [Google Scholar] [CrossRef]
- Picot, J.; Ndour, P.A.; Lefevre, S.D.; El Nemer, W.; Tawfik, H.; Galimand, J.; Da Costa, L.; Ribeil, J.A.; de Montalembert, M.; Brousse, V.; et al. A biomimetic microfluidic chip to study the circulation and mechanical retention of red blood cells in the spleen. Am. J. Hematol. 2015, 90, 339–345. [Google Scholar] [CrossRef] [PubMed]
- Burton, A. Gaining ground against cerebral malaria. Lancet Neurol. 2017, 16, 345–346. [Google Scholar] [CrossRef]
- Baydoun, M.; Vanneste, S.B.; Creusy, C.; Guyot, K.; Gantois, N.; Chabe, M.; Delaire, B.; Mouray, A.; Baydoun, A.; Forzy, G.; et al. Three-dimensional (3D) culture of adult murine colon as an in vitro model of cryptosporidiosis: Proof of concept. Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Gunasekera, S.; Zahedi, A.; O’dea, M.; King, B.; Monis, P.; Thierry, B.; Carr, J.M.; Ryan, U. Organoids and bioengineered intestinal models: Potential solutions to the Cryptosporidium culturing dilemma. Microorganisms 2020, 8, 715. [Google Scholar] [CrossRef]
- Duque-Correa, M.A.; Maizels, R.M.; Grencis, R.K.; Berriman, M. Organoids—New Models for Host–Helminth Interactions. Trends Parasitol. 2020, 36, 170–181. [Google Scholar] [CrossRef]
- Asghar, W.; Sher, M.; Khan, N.S.; Vyas, J.M.; Demirci, U. Microfluidic Chip for Detection of Fungal Infections. ACS Omega 2019, 4, 7474–7481. [Google Scholar] [CrossRef] [PubMed]
- Qiang, L.; Guo, J.; Han, Y.; Jiang, J.; Su, X.; Liu, H.; Qi, Q.; Han, L. A novel anti Candida albicans drug screening system based on high-throughput microfluidic chips. Sci. Rep. 2019, 9, 8087. [Google Scholar] [CrossRef] [PubMed]
- Barkal, L.J.; Walsh, N.M.; Botts, M.R.; Beebe, D.J.; Hull, C.M. Leveraging a high resolution microfluidic assay reveals insights into pathogenic fungal spore germination. Integr. Biol. 2016, 8, 603–615. [Google Scholar] [CrossRef]
- May, S.; Evans, S.; Parry, L. Organoids, organs-on-chips and other systems, and microbiota. Emerg. Top. Life Sci. 2017, 1, 385–400. [Google Scholar]
- Kim, H.J.; Huh, D.; Hamilton, G.; Ingber, D.E. Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab Chip 2012, 12, 2165–2174. [Google Scholar] [CrossRef] [PubMed]
- Dai, C.; Zhao, D.H.; Jiang, M. VSL#3 probiotics regulate the intestinal epithelial barrier in Vivo and in Vitro via the p38 and ERK signaling pathways. Int. J. Mol. Med. 2012, 29, 202–208. [Google Scholar] [CrossRef]
- Kim, H.J.; Li, H.; Collins, J.J.; Ingber, D.E. Contributions of microbiome and mechanical deformation to intestinal bacterial overgrowth and inflammation in a human gut-on-a-chip. Proc. Natl. Acad. Sci. USA 2016, 113, E7–E15. [Google Scholar] [CrossRef]
- Shin, W.; Kim, H.J. Intestinal barrier dysfunction orchestrates the onset of inflammatory host-microbiome cross-talk in a human gut inflammation-on-a-chip. Proc. Natl. Acad. Sci. USA 2018, 115, E10539–E10547. [Google Scholar] [CrossRef]
- Fukuda, S.; Toh, H.; Hase, K.; Oshima, K.; Nakanishi, Y.; Yoshimura, K.; Tobe, T.; Clarke, J.M.; Topping, D.L.; Suzuki, T.; et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature 2011, 469, 543–547. [Google Scholar] [CrossRef]
- Jacobson, A.; Lam, L.; Rajendram, M.; Tamburini, F.; Honeycutt, J.; Pham, T.; Van Treuren, W.; Pruss, K.; Stabler, S.R.; Lugo, K.; et al. A Gut Commensal-Produced Metabolite Mediates Colonization Resistance to Salmonella Infection. Cell Host Microbe 2018, 24, 296–307.e7. [Google Scholar] [CrossRef] [PubMed]
- Tovaglieri, A.; Sontheimer-Phelps, A.; Geirnaert, A.; Prantil-Baun, R.; Camacho, D.M.; Chou, D.B.; Jalili-Firoozinezhad, S.; De Wouters, T.; Kasendra, M.; Super, M.; et al. Species-specific enhancement of enterohemorrhagic E. coli pathogenesis mediated by microbiome metabolites. Microbiome 2019. [Google Scholar] [CrossRef]
- Ramadan, Q.; Jafarpoorchekab, H.; Huang, C.; Silacci, P.; Carrara, S.; Koklü, G.; Ghaye, J.; Ramsden, J.; Ruffert, C.; Vergeres, G.; et al. NutriChip: Nutrition analysis meets microfluidics. Lab Chip 2013, 13, 196–203. [Google Scholar] [CrossRef] [PubMed]
- Shah, P.; Fritz, J.V.; Glaab, E.; Desai, M.S.; Greenhalgh, K.; Frachet, A.; Niegowska, M.; Estes, M.; Jäger, C.; Seguin-Devaux, C.; et al. A microfluidics-based in vitro model of the gastrointestinal human-microbe interface. Nat. Commun. 2016, 7, 11535. [Google Scholar] [CrossRef] [PubMed]
- Trujillo-de Santiago, G.; Lobo-Zegers, M.J.; Montes-Fonseca, S.L.; Zhang, Y.S.; Alvarez, M.M. Gut-microbiota-on-a-chip: An enabling field for physiological research. Microphysiol. Syst. 2018, 2. [Google Scholar] [CrossRef]
- Barr, J.J.; Auro, R.; Sam-Soon, N.; Kassegne, S.; Peters, G.; Bonilla, N.; Hatay, M.; Mourtada, S.; Bailey, B.; Youle, M.; et al. Subdiffusive motion of bacteriophage in mucosal surfaces increases the frequency of bacterial encounters. Proc. Natl. Acad. Sci. USA 2015, 112, 13675. [Google Scholar] [CrossRef] [PubMed]
- Barr, J.J. Missing a Phage: Unraveling Tripartite Symbioses within the Human Gut. mSystems 2019, 4, e00105-19. [Google Scholar] [CrossRef]
- Ingber, D.E. Developmentally inspired human ‘organs on chips’. Development 2018, 145, dev156125. [Google Scholar] [CrossRef]
- Wong, I.; Ho, C.M. Surface molecular property modifications for poly(dimethylsiloxane) (PDMS) based microfluidic devices. Microfluid. Nanofluid. 2009, 7, 291–306. [Google Scholar] [CrossRef] [PubMed]
- Fritschen, A.; Blaeser, A. Biosynthetic, biomimetic, and self-assembled vascularized Organ-on-a-Chip systems. Biomaterials 2021, 268, 120556. [Google Scholar] [CrossRef]
- Quan, Y.; Sun, M.; Tan, Z.; Eijkel, J.C.T.; Van Den Berg, A.; Van Der Meer, A.; Xie, Y. Organ-on-a-chip: The next generation platform for risk assessment of radiobiology. RSC Adv. 2020, 10, 39521–39530. [Google Scholar] [CrossRef]
- Wnorowski, A.; Yang, H.; Wu, J.C. Progress, obstacles, and limitations in the use of stem cells in organ-on-a-chip models. Adv. Drug Deliv. Rev. 2019, 140, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Allwardt, V.; Ainscough, A.J.; Viswanathan, P.; Sherrod, S.D.; McLean, J.A.; Haddrick, M.; Pensabene, V. Translational roadmap for the organs-on-a-chip industry toward broad adoption. Bioengineering 2020, 7, 112. [Google Scholar] [CrossRef] [PubMed]
- Bassi, G.; Grimaudo, M.A.; Panseri, S.; Montesi, M. Advanced multi-dimensional cellular models as emerging reality to reproduce In Vitro the human body complexity. Int. J. Mol. Sci. 2021, 22, 1195. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baddal, B.; Marrazzo, P. Refining Host-Pathogen Interactions: Organ-on-Chip Side of the Coin. Pathogens 2021, 10, 203. https://doi.org/10.3390/pathogens10020203
Baddal B, Marrazzo P. Refining Host-Pathogen Interactions: Organ-on-Chip Side of the Coin. Pathogens. 2021; 10(2):203. https://doi.org/10.3390/pathogens10020203
Chicago/Turabian StyleBaddal, Buket, and Pasquale Marrazzo. 2021. "Refining Host-Pathogen Interactions: Organ-on-Chip Side of the Coin" Pathogens 10, no. 2: 203. https://doi.org/10.3390/pathogens10020203
APA StyleBaddal, B., & Marrazzo, P. (2021). Refining Host-Pathogen Interactions: Organ-on-Chip Side of the Coin. Pathogens, 10(2), 203. https://doi.org/10.3390/pathogens10020203






