Janthinobacterium tructae sp. nov., Isolated from Kidney of Rainbow Trout (Oncorhynchus mykiss)
Abstract
1. Introduction
2. Results
2.1. Bacterial Characteristics
2.1.1. Histopathology
2.1.2. Growth Characteristics
2.1.3. Morphology
2.1.4. Molecular Configuration
2.1.5. Biochemical Analysis
2.1.6. DNA-DNA Hybridization Test
2.2. Genetic Analysis
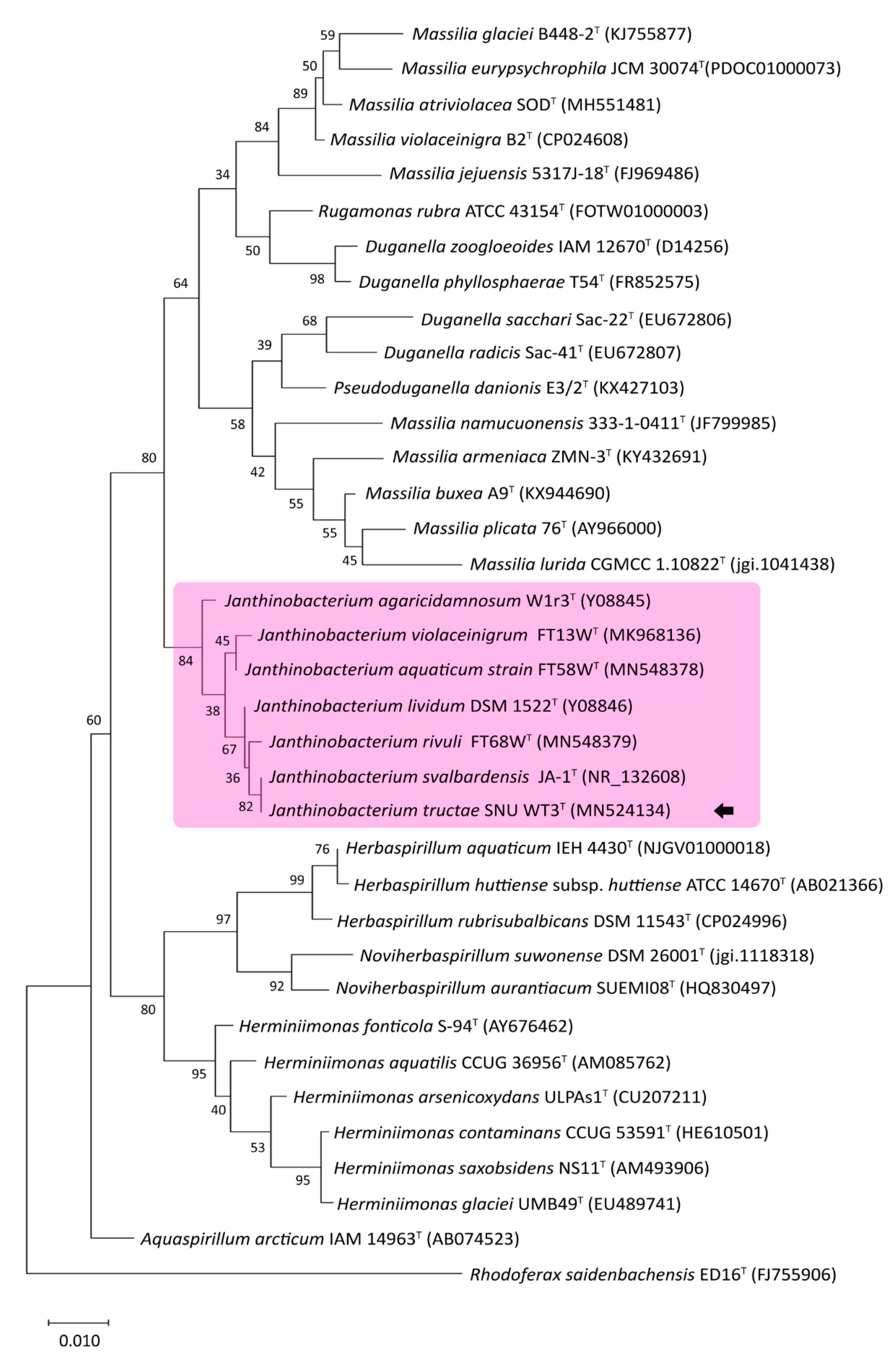
2.2.1. 16S-rRNA-Based Phylogenetic Analysis
2.2.2. Complete Genome Sequence
2.2.3. Average Nucleotide Identity and Genome-to-Genome Distance Calculator Analysis
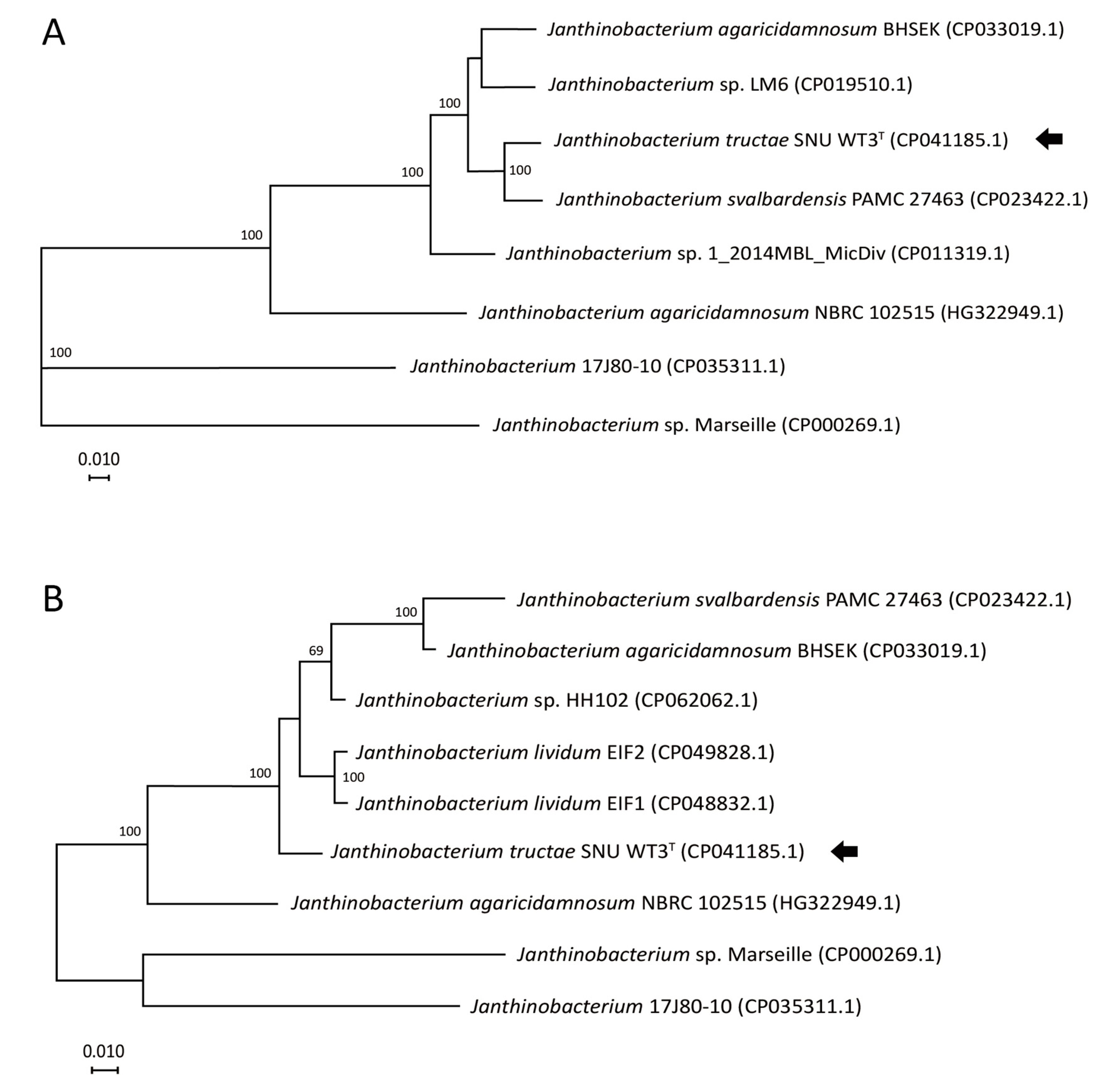
2.2.4. Core Genome Phylogeny and Multilocus Sequence Analysis (MLSA)
3. Discussion
4. Materials and Methods
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cho, Y.J.; Jung, Y.J.; Hong, S.G.; Kim, O.S. Complete Genome Sequence of a Psychrotolerant Denitrifying Bacterium, Janthinobacterium svalbardensis PAMC 27463. Microbiol. Resour. Announc. 2017, 5. [Google Scholar] [CrossRef]
- Schloss, P.D.; Allen, H.K.; Klimowicz, A.K.; Mlot, C.; Gross, J.A.; Savengsuksa, S.; Handelsman, J. Psychrotrophic strain of Janthinobacterium lividum from a cold Alaskan soil produces prodigiosin. DNA Cell Biol. 2010, 29, 533–541. [Google Scholar] [CrossRef] [PubMed]
- De Ley, J.; Segers, P.; Gillis, M. Intra-and intergeneric similarities of Chromobacterium and Janthinobacterium ribosomal ribonucleic acid cistrons. Int. J. Syst. Evol. Microbiol. 1978, 28, 154–168. [Google Scholar] [CrossRef]
- Koburger, J.A.; May, S.O. Isolation of Chromobacterium spp. from foods, soil, and water. Appl. Environ. Microbiol. 1982, 44, 1463–1465. [Google Scholar] [CrossRef] [PubMed]
- Lincoln, S.P.; Fermor, T.R.; Tindall, B.J. Janthinobacterium agaricidamnosum sp. nov., a soft rot pathogen of Agaricus bisporus. Int. J. Syst. Evol. Microbiol. 1999, 49, 1577–1589. [Google Scholar] [CrossRef] [PubMed]
- Avguštin, J.A.; Bertok, D.Ž.; Avguštin, G. Isolation and characterization of a novel violacein-like pigment producing psychrotrophic bacterial species Janthinobacterium svalbardensis sp. nov. Antonie Van Leeuwenhoek 2013, 103, 763–769. [Google Scholar] [CrossRef] [PubMed]
- Gong, X.; Skrivergaard, S.; Korsgaard, B.S.; Schreiber, L.; Marshall, I.P.; Finster, K.; Schramm, A. High quality draft genome sequence of Janthinobacterium psychrotolerans sp. nov., isolated from a frozen freshwater pond. Stand. Genom. Sci. 2017, 12, 8. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Deng, T.; Cai, Z.; Liu, F.; Yang, X.; Wang, Y.; Xu, M. Janthinobacterium violaceinigrum sp. nov., Janthinobacterium aquaticum sp. nov. and Janthinobacterium rivuli sp. nov., isolated from a subtropical stream in China. Int. J. Syst. Evol. Microbiol. 2020, 70, 2719–2725. [Google Scholar] [CrossRef]
- Patjanasoontorn, B.; Boonma, P.A.I.T.; Wilailackana, C.; Sittikesorn, J. Hospital acquired Janthinobacterium lividum septicemia in Srinagarind Hospital. J. Med. Assoc. Thail. 1992, 75, 6. [Google Scholar]
- Oh, W.T.; Giri, S.S.; Yun, S.; Kim, H.J.; Kim, S.G.; Kim, S.W.; Park, S.C. Janthinobacterium lividum as An Emerging Pathogenic Bacterium Affecting Rainbow Trout (Oncorhynchus mykiss) Fisheries in Korea. Pathogens 2019, 8, 146. [Google Scholar] [CrossRef]
- Austin, B.; Gonzalez, C.J.; Stobie, M.; Curry, J.I.; Mcloughlin, M.F. Recovery of Janthinobacterium lividum from diseased rainbow trout, Oncorhynchus mykiss (Walbaum), in Northern Ireland and Scotland. J. Fish Dis. 1992, 15, 357–359. [Google Scholar] [CrossRef]
- Wu, X.; Deutschbauer, A.M.; Kazakov, A.E.; Wetmore, K.M.; Cwick, B.A.; Walker, R.M.; Chakraborty, R. Draft genome sequences of two Janthinobacterium lividum strains, isolated from pristine groundwater collected from the Oak Ridge Field Research Center. Genome Announc. 2017, 5, 1–2. [Google Scholar] [CrossRef]
- Kumar, R.; Acharya, V.; Singh, D.; Kumar, S. Strategies for high-altitude adaptation revealed from high-quality draft genome of non-violacein producing Janthinobacterium lividum ERGS5: 01. Stand. Genom. Sci. 2018, 13, 11. [Google Scholar] [CrossRef] [PubMed]
- Pantanella, F.; Berlutti, F.; Passariello, C.; Sarli, S.; Morea, C.; Schippa, S. Violacein and biofilm production in Janthinobacterium lividum. J. Appl. Microbiol. 2007, 102, 992–999. [Google Scholar] [CrossRef]
- Harris, R.N.; Brucker, R.M.; Walke, J.B.; Becker, M.H.; Schwantes, C.R.; Flaherty, D.C.; Minbiole, K.P. Skin microbes on frogs prevent morbidity and mortality caused by a lethal skin fungus. ISME J. 2009, 3, 818. [Google Scholar] [CrossRef] [PubMed]
- Kämpfer, P.; Rossello-Mora, R.; Hermansson, M.; Persson, F.; Huber, B.; Falsen, E.; Busse, H.J. Undibacterium pigrum gen. nov., sp. nov., isolated from drinking water. Int. J. Syst. Evol. Microbiol. 2007, 57, 1510–1515. [Google Scholar] [CrossRef]
- Cho, J.C.; Tiedje, J.M. Bacterial species determination from DNA-DNA hybridization by using genome fragments and DNA microarrays. Appl. Environ. Microbiol. 2001, 67, 3677–3682. [Google Scholar] [CrossRef]
- Yoon, S.H.; Ha, S.M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA and whole genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, M.; Kishino, H.; Saitou, N. On the maximum likelihood method in molecular phylogenetics. J. Mol. Evol. 1991, 32, 443–445. [Google Scholar] [CrossRef]
- Carver, T.; Thomson, N.; Bleasby, A.; Berriman, M.; Parkhill, J. DNAPlotter: Circular and linear interactive genome visualization. Bioinformatics 2009, 25, 119–120. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Oh, H.S.; Park, S.C.; Chun, J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 2014, 64, 346–351. [Google Scholar] [CrossRef]
- Chun, J.; Oren, A.; Ventosa, A.; Christensen, H.; Arahal, D.R.; da Costa, M.S.; Trujillo, M.E. Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 2018, 68, 461–466. [Google Scholar] [CrossRef] [PubMed]
- Blom, J.; Kreis, J.; Spänig, S.; Juhre, T.; Bertelli, C.; Ernst, C.; Goesmann, A. EDGAR 2.0: An enhanced software platform for comparative gene content analyses. Nucleic Acids Res. 2016, 44, W22–W28. [Google Scholar] [CrossRef] [PubMed]
- Svenning, M.A.; Klemetsen, A.; Olsen, T. Habitat and food choice of Arctic charr in Linnévatn on Spitsbergen, Svalbard: The first year-round investigation in a High Arctic lake. Ecol. Freshw. Fish 2006, 16, 70–77. [Google Scholar] [CrossRef]
- Kämpfer, P.; Kroppenstedt, R.M. Numerical analysis of fatty acid patterns of coryneform bacteria and related taxa. Can. J. Microbiol. 1996, 42, 989–1005. [Google Scholar] [CrossRef]
- Frank, J.A.; Reich, C.I.; Sharma, S.; Weisbaum, J.S.; Wilson, B.A.; Olsen, G.J. Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl. Environ. Microbiol. 2008, 74, 2461–2470. [Google Scholar] [CrossRef]
- Sipos, R.; Székely, A.J.; Palatinszky, M.; Révész, S.; Márialigeti, K.; Nikolausz, M. Effect of primer mismatch, annealing temperature and PCR cycle number on 16S rRNA gene-targetting bacterial community analysis. FEMS Microbiol. Ecol. 2007, 60, 341–350. [Google Scholar] [CrossRef]
- Kim, O.S.; Cho, Y.J.; Lee, K.; Yoon, S.H.; Kim, M.; Na, H.; Won, S. Introducing EzTaxon-e: A prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int. J. Syst. Evol. Microbiol. 2012, 62, 716–721. [Google Scholar] [CrossRef]
- EzBioCloud 16S-Based, ID. Available online: https://www.ezbiocloud.net/identify (accessed on 10 October 2020).
- National Center for Biotechnology Information. Basic Local Alignment Search Tool. Available online: https://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed on 10 October 2020).
- Chin, C.; Alexander, D.H.; Marks, P.; Klammer, A.A.; Drake, J.; Heiner, C.; Clum, A.; Copeland, A.; Huddleston, J.; Eichler, E.E.; et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 2013, 10, 563–569. [Google Scholar] [CrossRef] [PubMed]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef]
- Haft, D.H.; DiCuccio, M.; Badretdin, A.; Brover, V.; Chetvernin, V.; O’Neill, K.; Li, W.; Chitsaz, F.; Derbyshire, M.K.; Gonzales, N.R.; et al. RefSeq: An update on prokaryotic genome annotation and curation. Nucleic Acids Res. 2018, 46, D851–D860. [Google Scholar] [CrossRef]
- Gupta, S.K.; Padmanabhan, B.R.; Diene, S.M.; Lopez-Rojas, R.; Kempf, M.; Landraud, L.; Rolain, J. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob. Agents Chemother. 2014, 58, 212–220. [Google Scholar] [CrossRef] [PubMed]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Min, S.Y. CARD 2020: Antibiotic resistome surveillance with the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef]
- Liu, B.; Zheng, D.; Jin, Q.; Chen, L.; Yang, J. VFDB 2019: A comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 2019, 47, D687–D692. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Sun, H.X.; Zhang, C.; Cheng, L.; Peng, Y.; Deng, Z.; Wang, D.; Wang, Y.; Hu, M.; Liu, W.; et al. Prophage Hunter: An integrative hunting tool for active prophages. Nucleic Acids Res. 2019, 47, W74–W80. [Google Scholar] [CrossRef]
- Bland, C.; Ramsey, T.L.; Sabree, F.; Lowe, M.; Brown, K.; Kyrpides, N.C.; Hugenholtz, P. CRISPR Recognition Tool (CRT): A tool for automatic detection of clustered regularly interspaced palindromic repeats. BMC Bioinform. 2007, 8, 1–8. [Google Scholar] [CrossRef]
- EzBioCloud OrthoANIu. Available online: https://www.ezbiocloud.net/tools/ani (accessed on 10 October 2020).
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef] [PubMed]
- Genome-to-Genome Distance Calculator 2.1. Available online: http://ggdc.dsmz.de/distcalc2.php (accessed on 10 October 2020).
- Meier-Kolthoff, J.P.; Klenk, H.P.; Göker, M. Taxonomic use of DNA G + C content and DNA-DNA hybridization in the genomic age. Int. J. Syst. Evol. Microbiol. 2014, 64, 352–356. [Google Scholar] [CrossRef] [PubMed]




| Fatty Acid | J. tructae SNU WT3T | J. lividum CCUG2344T [16] | J. svalbardensis JA-1T [8] | J. agaricidamnosum CCUG43104T [16] | J. violaceinigrum FT13WT [8] | J. aquaticum FT58WT [8] | J. rivuli FT68WT [8] |
|---|---|---|---|---|---|---|---|
| cylco-C17:0 | 41.45 | 25.0 | ND | 34.2 | ND | ND | ND |
| C16:0 | 33.86 | 30.6 | 33.9 | 34.8 | 31.3 | 29.3 | 35.1 |
| C12:0 | 5.87 | 3.9 | 3.8 | 3.0 | 8.1 | 5.6 | 6.2 |
| J. tructae SNU WT3T | J. lividum DSM 1522T [6] | J. svalbardensis JA-1T [6] | J. agaricidamnosum DSM 9628 T [6] | J. violaceinigrum FT13WT [8] | J. aquaticum FT58WT [8] | J. rivuli FT68WT [8] | J. psychrotolerans S3-2T [7] | |
|---|---|---|---|---|---|---|---|---|
| D-arabinose | − | + | − | − | − | + | + | − |
| L-arabinose | + | + | + | − | + | + | W | + |
| D-xylose | + | + | − | − | + | + | + | NP |
| Adonitol | − | − | + | − | NP | NP | NP | NP |
| Galactose | + | + | + | − | − | + | + | + |
| Sorbitol | + | + | + | − | − | + | + | − |
| N-acetylglucosamine | − | − | + | − | NP | NP | NP | − |
| Arbutin | − | + | + | − | NP | NP | NP | NP |
| Salicin | − | + | − | − | NP | NP | NP | + |
| Cellobiose | + | + | − | − | − | + | − | + |
| Maltose | + | + | + | − | − | − | + | − |
| Trehalose | + | − | + | + | − | − | - | − |
| Xylitol | + | − | + | − | NP | NP | NP | NP |
| β gentiobiose | − | − | − | + | NP | NP | NP | NP |
| D-lyxose | + | + | + | − | − | + | - | NP |
| L-fucose | − | + | − | − | − | + | + | + |
| 2-ketogluconate | − | + | − | − | NP | NP | NP | + |
| Rhamnose | − | − | − | − | NP | NP | NP | + |
| Inulin | − | − | − | − | NP | NP | NP | NP |
| D-raffinose | − | − | − | − | NP | NP | NP | + |
| Janthinobacterium tructae SNU WT3 (ANI Value) | Janthinobacterium tructae SNU WT3 (GGDC Value) | |
|---|---|---|
| Janthinobacterium svalbardensis PAMC 27463 | 95.0% | 60.6% |
| Janthinobacterium lividum EIF1 | 91.7% | 45.7% |
| Janthinobacterium lividum EIF2 | 91.7% | 45.7% |
| Janthinobacterium agaricidamnosum BHSEK | 91.7% | 45.9% |
| Janthinobacterium sp. 1_2014MBL_MicDiv | 89.4% | 38.5% |
| Janthinobacterium sp. 17J80-10 | 91.8% | 46.1% |
| Janthinobacterium sp. B9-8 | 67.1% | 20.3% |
| Janthinobacterium sp. LM6 | 74.5% | 20.6% |
| Janthinobacterium sp. Marseille | 73.0% | 31% |
| Herbaspirillum rubrisubalbicans DSM 11543 | 74.0% | 20.6% |
| Herminiimonas arsenicoxydans | 73.6% | 20.3% |
| Massilia armeniaca ZMN-3 | 76.6% | 21.0% |
| Massilia plicata DSM 17505 | 76.3% | 21.1% |
| Massilia violaceinigra B2 | 76.6% | 21.3% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jung, W.J.; Kim, S.W.; Giri, S.S.; Kim, H.J.; Kim, S.G.; Kang, J.W.; Kwon, J.; Lee, S.B.; Oh, W.T.; Jun, J.W.; et al. Janthinobacterium tructae sp. nov., Isolated from Kidney of Rainbow Trout (Oncorhynchus mykiss). Pathogens 2021, 10, 229. https://doi.org/10.3390/pathogens10020229
Jung WJ, Kim SW, Giri SS, Kim HJ, Kim SG, Kang JW, Kwon J, Lee SB, Oh WT, Jun JW, et al. Janthinobacterium tructae sp. nov., Isolated from Kidney of Rainbow Trout (Oncorhynchus mykiss). Pathogens. 2021; 10(2):229. https://doi.org/10.3390/pathogens10020229
Chicago/Turabian StyleJung, Won Joon, Sang Wha Kim, Sib Sankar Giri, Hyoun Joong Kim, Sang Guen Kim, Jeong Woo Kang, Jun Kwon, Sung Bin Lee, Woo Taek Oh, Jin Woo Jun, and et al. 2021. "Janthinobacterium tructae sp. nov., Isolated from Kidney of Rainbow Trout (Oncorhynchus mykiss)" Pathogens 10, no. 2: 229. https://doi.org/10.3390/pathogens10020229
APA StyleJung, W. J., Kim, S. W., Giri, S. S., Kim, H. J., Kim, S. G., Kang, J. W., Kwon, J., Lee, S. B., Oh, W. T., Jun, J. W., & Park, S. C. (2021). Janthinobacterium tructae sp. nov., Isolated from Kidney of Rainbow Trout (Oncorhynchus mykiss). Pathogens, 10(2), 229. https://doi.org/10.3390/pathogens10020229








