Atopobium vaginae and Prevotella bivia Are Able to Incorporate and Influence Gene Expression in a Pre-Formed Gardnerella vaginalis Biofilm
Abstract
1. Introduction
2. Results
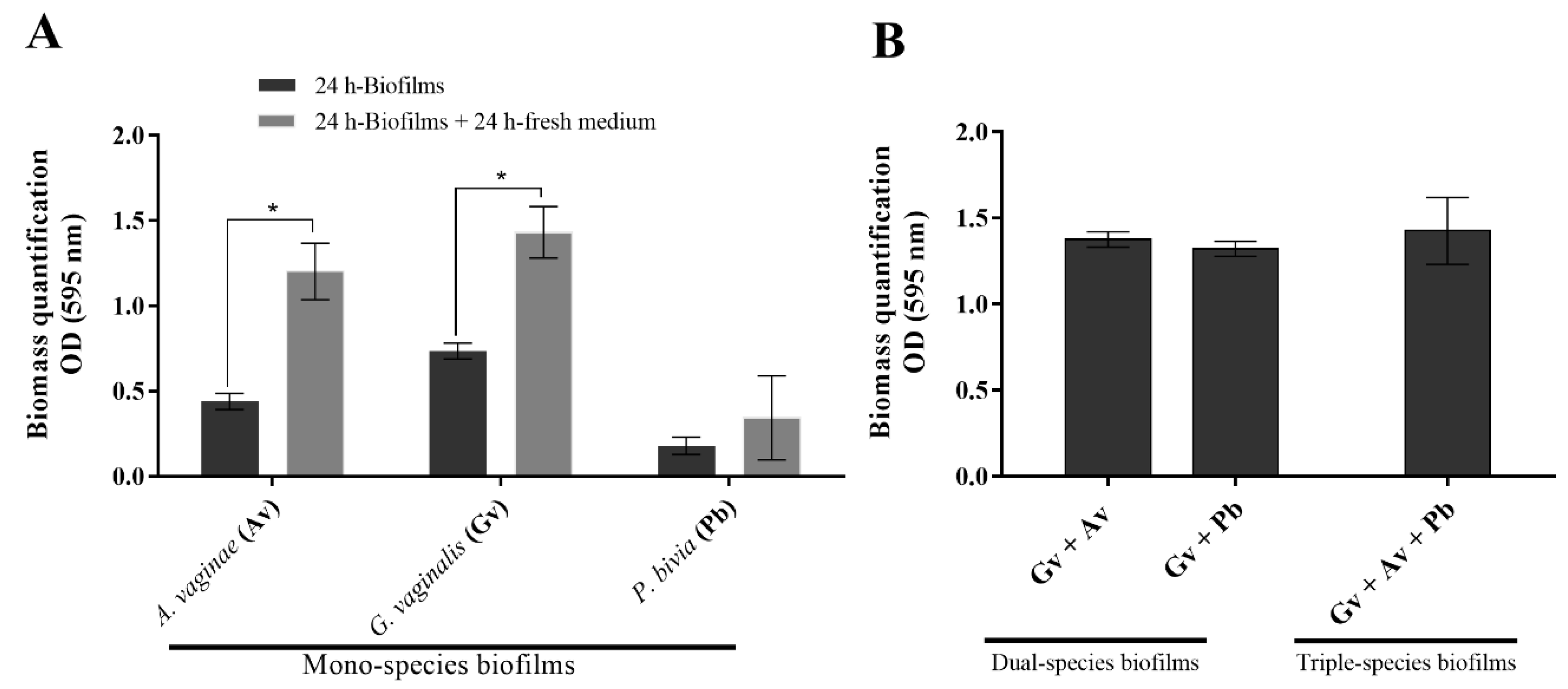
2.1. Quantification of the Biomass of Mono-, Dual-, and Triple-Species BV-Associated Biofilms
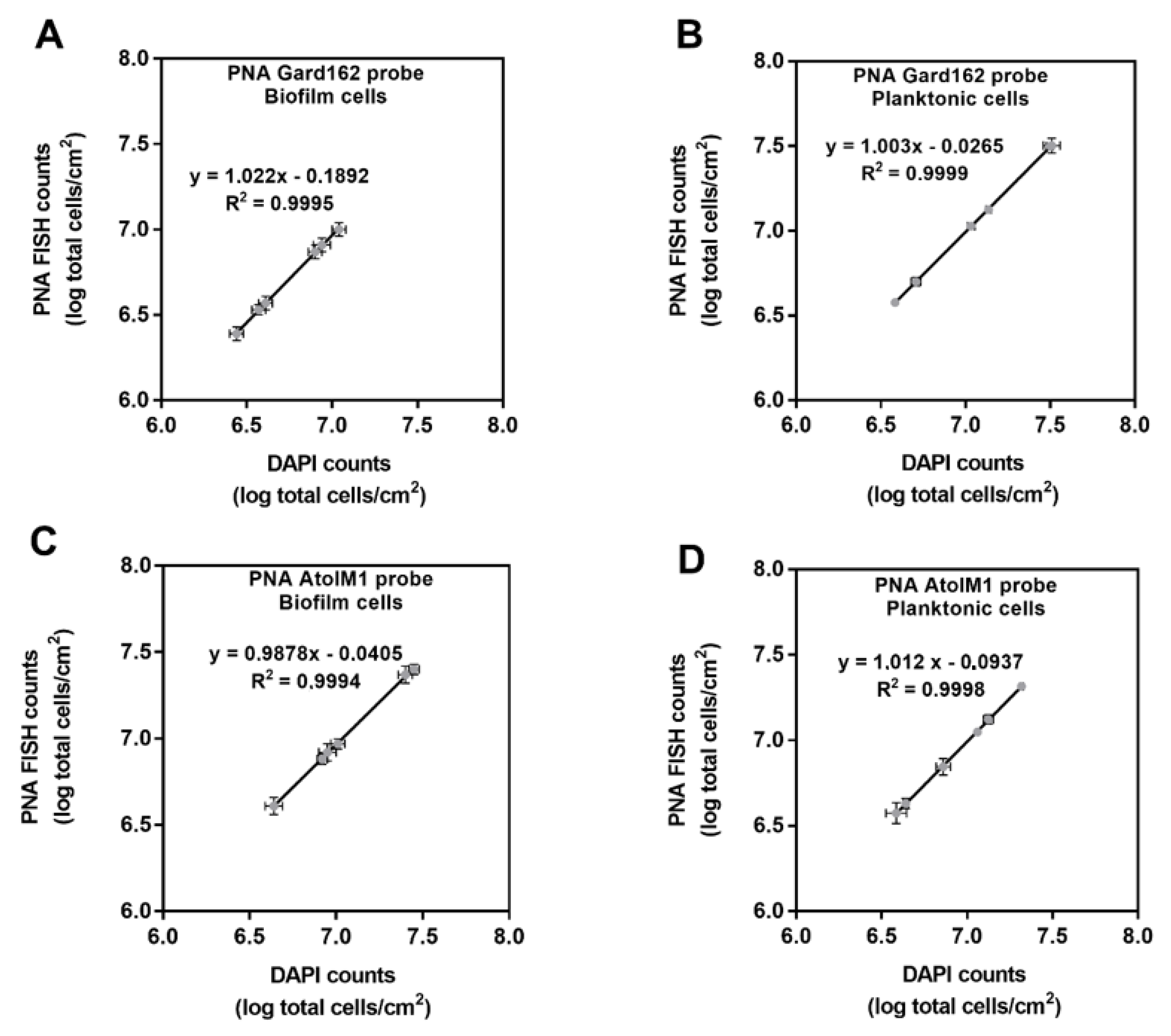
2.2. In Vitro PNA Gard162 and PNA AtoITM1 Probes Specificity and Efficiency
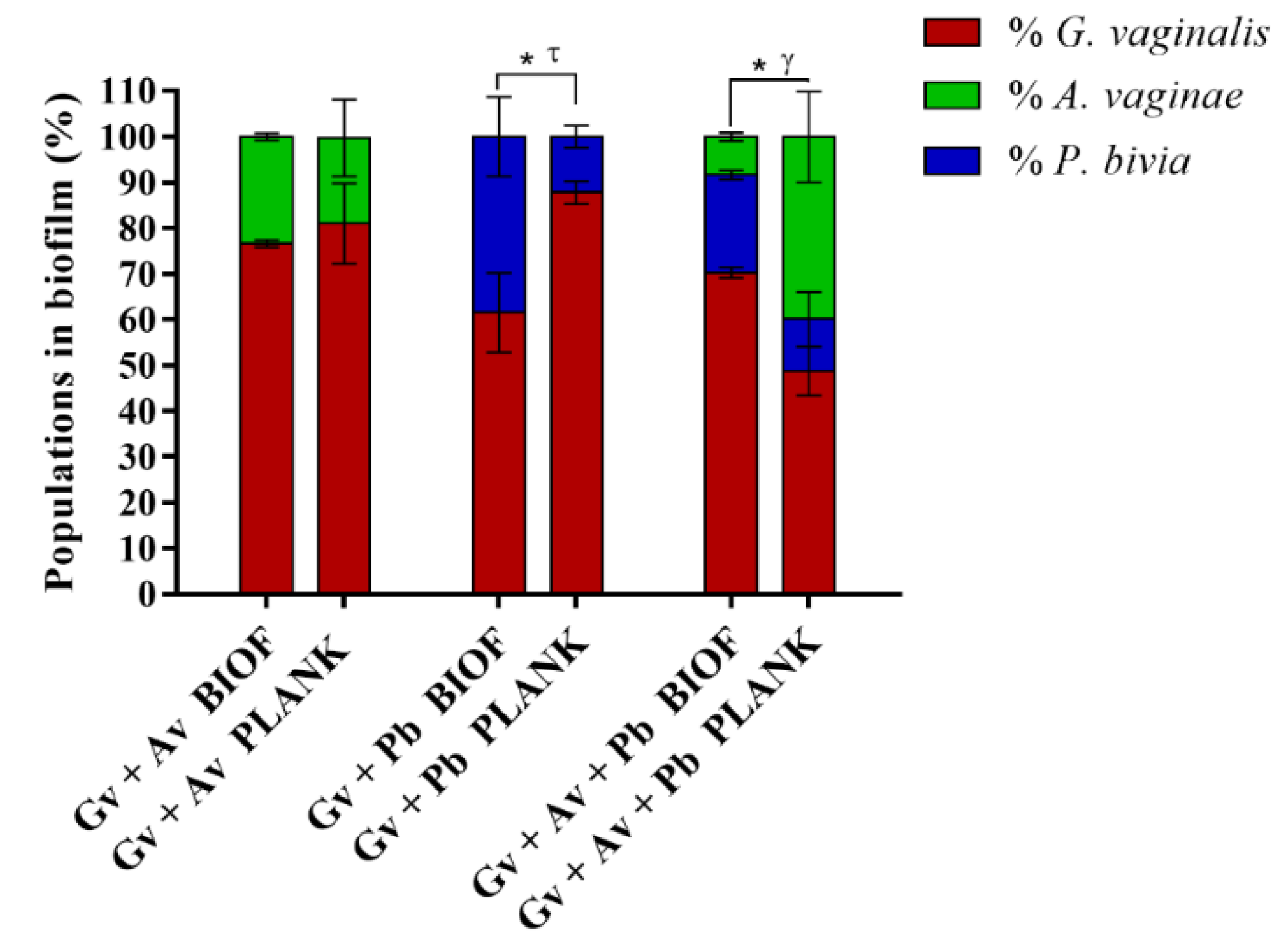
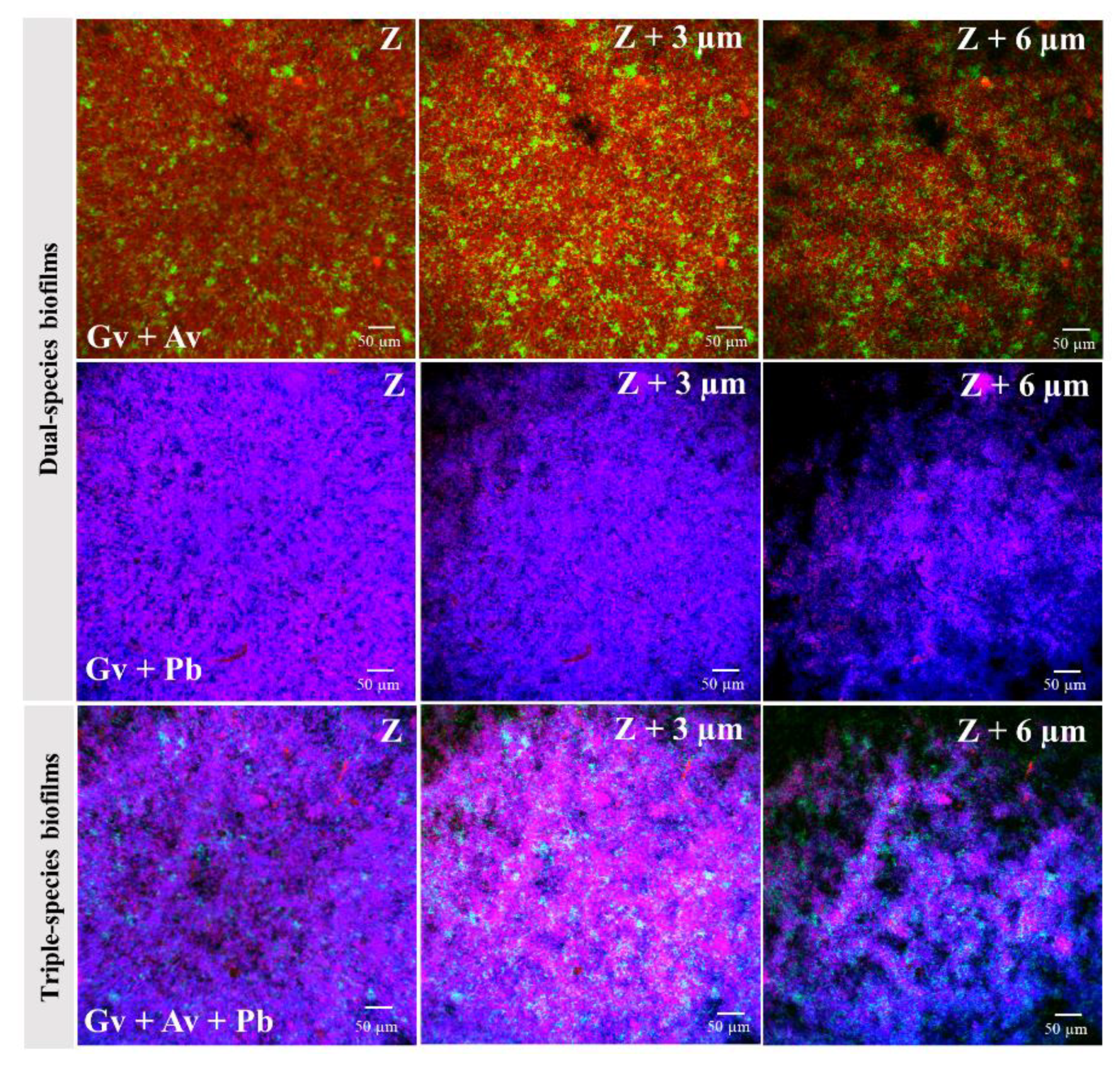
2.3. Quantification and Distribution of Bacterial Populations in Dual- and Triple-Species BV-Associated Biofilms by PNA FISH
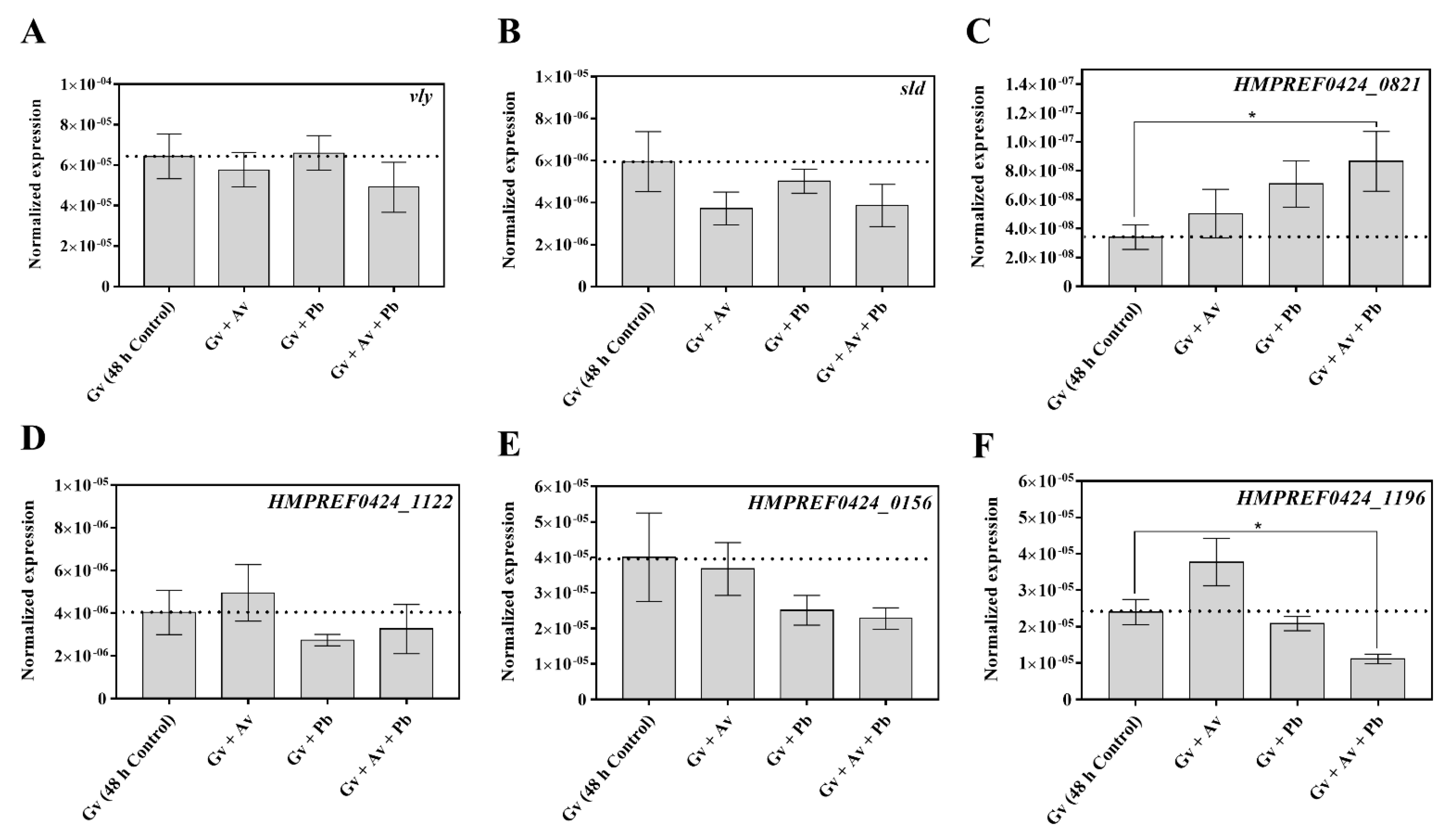
2.4. Impact of A. vaginae and P. bivia on G. vaginalis Virulence
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Culture Conditions
4.2. Biofilm Formation and Biomass Quantification by the Crystal Violet Method
4.3. In Vitro PNA Gard162 and AtoITM1 Probes Specificity and Efficiency
4.4. Quantification of Bacterial Populations in Dual- and Triple-Species Biofilms and Their Planktonic Fraction by PNA FISH
4.5. Confocal Laser Scanning Microscopy Analysis of Biofilm Bacterial Distribution
4.6. G. vaginalis Gene Expression Quantification in Mono-, Dual-, and Triple-Species Biofilms
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Rosca, A.S.; Castro, J.; Sousa, L.G.V.; Cerca, N. Gardnerella and vaginal health: The truth is out there. FEMS Microbiol. Rev. 2019, 44, 73–105. [Google Scholar] [CrossRef]
- Arif, F. Bacterial Vaginosis: Risk of Adverse Pregnancy Outcome. J. Gynecol. Res. Obstet. 2018, 4, 015–017. [Google Scholar] [CrossRef]
- Moragianni, D.; Dryllis, G.; Andromidas, P.; Kapeta-Korkouli, R.; Kouskouni, E.; Pessach, I.; Papalexis, P.; Kodonaki, A.; Athanasiou, N.; Pouliakis, A.; et al. Genital tract infection and associated factors affect the reproductive outcome in fertile females and females undergoing in vitro fertilization. Biomed. Rep. 2019, 10, 231–237. [Google Scholar] [CrossRef]
- Brusselaers, N.; Shrestha, S.; van de Wijgert, J.; Verstraelen, H. Vaginal dysbiosis and the risk of human papillomavirus and cervical cancer: Systematic review and meta-analysis. Am. J. Obstet. Gynecol. 2019, 221, 9–18.e8. [Google Scholar] [CrossRef]
- Brotman, R.M.; Klebanoff, M.A.; Nansel, T.R.; Yu, K.F.; Andrews, W.W.; Zhang, J.; Schwebke, J.R. Bacterial vaginosis assessed by Gram stain and diminished colonization resistance to incident gonococcal, chlamydial, and trichomonal genital infection. J. Infect. Dis. 2010, 202, 1907–1915. [Google Scholar] [CrossRef]
- Abbai, N.S.; Reddy, T.; Ramjee, G. Prevalent bacterial vaginosis infection—A risk factor for incident sexually transmitted infections in women in Durban, South Africa. Int. J. STD AIDS 2016, 27, 1283–1288. [Google Scholar] [CrossRef]
- Lokken, E.M.; Balkus, J.E.; Kiarie, J.; Hughes, J.P.; Jaoko, W.; Totten, P.A.; Scott McClelland, R.; Manhart, L.E. Association of Recent Bacterial Vaginosis with Acquisition of Mycoplasma genitalium. Am. J. Epidemiol. 2017, 186, 194–201. [Google Scholar] [CrossRef] [PubMed]
- Atashili, J.; Poole, C.; Ndumbe, P.M.; Adimora, A.A.; Smith, J.S. Bacterial vaginosis and HIV acquisition: A meta-analysis of published studies. Aids 2008, 22, 1493–1501. [Google Scholar] [CrossRef]
- Muzny, C.A.; Taylor, C.M.; Swords, W.E.; Tamhane, A.; Chattopadhyay, D.; Cerca, N.; Schwebke, J.R. An Updated Conceptual Model on the Pathogenesis of Bacterial Vaginosis. J. Infect. Dis. 2019, 220, 1399–1405. [Google Scholar] [CrossRef]
- Swidsinski, A.; Mendling, W.; Loening-Baucke, V.; Swidsinski, S.; Dörffel, Y.; Scholze, J.; Lochs, H.; Verstraelen, H. An adherent Gardnerella vaginalis biofilm persists on the vaginal epithelium after standard therapy with oral metronidazole. Am. J. Obstet. Gynecol. 2008, 198, 1–6. [Google Scholar] [CrossRef]
- Swidsinski, A.; Loening-Baucke, V.; Mendling, W.; Dörffel, Y.; Schilling, J.; Halwani, Z.; Jiang, X.F.; Verstraelen, H.; Swidsinski, S. Infection through structured polymicrobial Gardnerella biofilms (StPM-GB). Histol. Histopathol. 2014, 29, 567–587. [Google Scholar] [CrossRef]
- Swidsinski, A.; Verstraelen, H.; Loening-Baucke, V.; Swidsinski, S.; Mendling, W.; Halwani, Z. Presence of a Polymicrobial Endometrial Biofilm in Patients with Bacterial Vaginosis. PLoS ONE 2013, 8, 4–8. [Google Scholar] [CrossRef]
- Aroutcheva, A.A.; Simoes, J.A.; Behbakht, K.; Faro, S. Gardnerella vaginalis isolated from patients with bacterial vaginosis and from patients with healthy vaginal ecosystems. Clin. Infect. Dis. 2001, 33, 1022–1027. [Google Scholar] [CrossRef]
- Bekasiak, A.; Dammann, F.; Nader, C. A rare cause of a scrotal abscess due to the symbiotic infection of Gardnerella vaginalis and Prevotella bivia in an adult male. Pathogens 2020, 9, 93. [Google Scholar] [CrossRef]
- Vaneechoutte, M.; Guschin, A.; Van Simaey, L.; Gansemans, Y.; Van Nieuwerburgh, F.; Cools, P. Emended description of Gardnerella vaginalis and description of Gardnerella leopoldii sp. nov., Gardnerella piotii sp. nov. and Gardnerella swidsinskii sp. nov., with delineation of 13 genomic species within the genus Gardnerella. Int. J. Syst. Evol. Microbiol. 2019, 69, 679–687. [Google Scholar] [CrossRef]
- Potter, R.F.; Burnham, C.A.D.; Dantas, G. In silico analysis of Gardnerella genomospecies detected in the setting of bacterial vaginosis. Clin. Chem. 2019, 65, 1375–1387. [Google Scholar] [CrossRef]
- Pleckaityte, M.; Janulaitiene, M.; Lasickiene, R.; Zvirbliene, A. Genetic and biochemical diversity of Gardnerella vaginalis strains isolated from women with bacterial vaginosis. FEMS Immunol. Med. Microbiol. 2012, 65, 69–77. [Google Scholar] [CrossRef]
- Janulaitiene, M.; Gegzna, V.; Baranauskiene, L.; Bulavaitė, A.; Simanavicius, M.; Pleckaityte, M. Phenotypic characterization of Gardnerella vaginalis subgroups suggests differences in their virulence potential. PLoS ONE 2018, 13, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Hill, J.E.; Albert, A.Y.K.; VOGUE Research Group. Resolution and cooccurrence patterns of Gardnerella leopoldii, G. swidsinskii, G. piotii, and G. vaginalis within the vaginal microbiome. Infect. Immun. 2019, 87, e00532-19. [Google Scholar] [CrossRef]
- Schellenberg, J.J.; Patterson, M.H.; Hill, J.E. Gardnerella vaginalis diversity and ecology in relation to vaginal symptoms. Res. Microbiol. 2017, 168, 837–844. [Google Scholar] [CrossRef]
- Khan, S.; Voordouw, M.J.; Hill, J.E. Competition Among Gardnerella Subgroups From the Human Vaginal Microbiome. Front. Cell. Infect. Microbiol. 2019, 9, 1–10. [Google Scholar] [CrossRef]
- Swidsinski, A.; Mendling, W.; Loening-Baucke, V.; Ladhoff, A.; Swidsinski, S.; Hale, L.P.; Lochs, H. Adherent biofilms in bacterial vaginosis. Obstet. Gynecol. 2005, 106, 1013–1023. [Google Scholar] [CrossRef] [PubMed]
- Menard, J.P.; Mazouni, C.; Salem-Cherif, I.; Fenollar, F.; Raoult, D.; Boubli, L.; Gamerre, M.; Bretelle, F. High vaginal concentrations of Atopobium vaginae and Gardnerella vaginalis in women undergoing preterm labor. Obstet. Gynecol. 2010, 115, 134–140. [Google Scholar] [CrossRef]
- Mendling, W.; Palmeira-de-Oliveira, A.; Biber, S.; Prasauskas, V. An update on the role of Atopobium vaginae in bacterial vaginosis: What to consider when choosing a treatment? A mini review. Arch. Gynecol. Obstet. 2019, 300, 1–6. [Google Scholar] [CrossRef]
- Ferris, M.J.; Masztal, A.; Aldridge, K.E.; Fortenberry, D.; Fidel, P.L.; Martin, D.H. Association of Atopobium vaginae, a recently described metronidazole resistant anaerobe, with bacterial vaginosis. BMC Infect. Dis. 2004, 4, 1–8. [Google Scholar] [CrossRef]
- De Backer, E.; Verhelst, R.; Verstraelen, H.; Alqumber, M.A.; Burton, J.P.; Tagg, J.R.; Temmerman, M.; Vaneechoutte, M. Quantitative determination by real-time PCR of four vaginal Lactobacillus species, Gardnerella vaginalis and Atopobium vaginae indicates an inverse relationship between L. gasseri and L. iners. BMC Microbiol. 2007, 7, 115. [Google Scholar] [CrossRef] [PubMed]
- Spiegel, C.; Amsel, R.; Eschenbach, D.; Schoenknecht, F.; Holmes, K. Anaerobic bacteria in nonspecific vaginitis. N. Engl. J. Med. 1980, 303, 601–607. [Google Scholar] [CrossRef]
- Muzny, C.A.; Blanchard, E.; Taylor, C.M.; Aaron, K.J.; Talluri, R.; Griswold, M.E.; Redden, D.T.; Luo, M.; Welsh, D.A.; Van Der Pol, W.J.; et al. Identification of Key Bacteria Involved in the Induction of Incident Bacterial Vaginosis: A Prospective Study. J. Infect. Dis. 2018, 218, 966–978. [Google Scholar] [CrossRef]
- Pybus, V.; Onderdonk, A.B. Evidence for a commensal, symbiotic relationship between Gardnerella vaginalis and Prevotella bivia involving ammonia: Potential significance for bacterial vaginosis. J. Infect. Dis. 1997, 175, 406–413. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, N.M.; Lewis, W.G.; Li, G.; Sojka, D.K.; Lubin, J.B.; Lewis, A.L. Gardnerella vaginalis and Prevotella bivia Trigger Distinct and Overlapping Phenotypes in a Mouse Model of Bacterial Vaginosis. J. Infect. Dis. 2019, 220, 1099–1108. [Google Scholar] [CrossRef]
- Gibbs, R.; McDuffi Jr, R.; Kunze, M.; Barr, J.M.; Wolf, D.; Sze, C.-I.; Shikes, R.; Sherman, M. Experimental intrauterine infection with Prevotella bivia in New Zealand White rabbits. Am. J. Obs. Gynecol. 2004, 1082–1086. [Google Scholar] [CrossRef]
- Randis, T.M.; Ratner, A.J. Gardnerella and Prevotella: Co-conspirators in the Pathogenesis of Bacterial Vaginosis. J. Infect. Dis. 2019, 220, 1085–1088. [Google Scholar] [CrossRef]
- Freitas, A.C.; Bocking, A.; Hill, J.E.; Money, D.M. Increased richness and diversity of the vaginal microbiota and spontaneous preterm birth. Microbiome 2018, 6, 1–15. [Google Scholar] [CrossRef]
- Pybus, V.; Onderdonk, A.B. Microbial interactions in the vaginal ecosystem, with emphasis on the pathogenesis of bacterial vaginosis. Microbes Infect. 1999, 1, 285–292. [Google Scholar] [CrossRef]
- Castro, J.; Cerca, N. BV and non-BV associated Gardnerella vaginalis establish similar synergistic interactions with other BV-associated microorganisms in dual-species biofilms. Anaerobe 2015, 36, 56–59. [Google Scholar] [CrossRef] [PubMed]
- Castro, J.; Machado, D.; Cerca, N. Escherichia coli and Enterococcus faecalis are able to incorporate and enhance a pre-formed Gardnerella vaginal is biofilm. Pathog. Dis. 2016, 74, ftw007. [Google Scholar] [CrossRef]
- Castro, J.; Machado, D.; Cerca, N. Unveiling the role of Gardnerella vaginalis in polymicrobial Bacterial Vaginosis biofilms: The impact of other vaginal pathogens living as neighbors. ISME J. 2019, 13, 1306–1317. [Google Scholar] [CrossRef] [PubMed]
- Rosca, A.S.; Castro, J.; Cerca, N. Evaluation of different culture media to support in vitro growth and biofilm formation of bacterial vaginosis-associated anaerobes. PeerJ 2020, 8, e9917. [Google Scholar] [CrossRef]
- Machado, A.; Almeida, C.; Salgueiro, D.; Henriques, A.; Vaneechoutte, M.; Haesebrouck, F.; Vieira, M.J.; Rodrigues, L.; Azevedo, N.F.; Cerca, N. Fluorescence in situ Hybridization method using Peptide Nucleic Acid probes for rapid detection of Lactobacillus and Gardnerella spp. BMC Microbiol. 2013, 13, 82. [Google Scholar] [CrossRef]
- Hardy, L.; Jespers, V.; Dahchour, N.; Mwambarangwe, L.; Musengamana, V.; Vaneechoutte, M.; Crucitti, T. Unravelling the bacterial vaginosis-associated biofilm: A multiplex Gardnerella vaginalis and Atopobium vaginae fluorescence in situ hybridization assay using peptide nucleic acid probes. PLoS ONE 2015, 10, 1–16. [Google Scholar] [CrossRef]
- Almeida, C.; Azevedo, N.F.; Santos, S.; Keevil, C.W.; Vieira, M.J. Discriminating multi-species populations in biofilms with peptide nucleic acid fluorescence in situ hybridization (PNA FISH). PLoS ONE 2011, 6. [Google Scholar] [CrossRef] [PubMed]
- Machado, A.; Jefferson, K.K.; Cerca, N. Interactions between Lactobacillus crispatus and bacterial vaginosis (BV)-associated bacterial species in initial attachment and biofilm formation. Int. J. Mol. Sci. 2013, 14, 12004–12012. [Google Scholar] [CrossRef]
- Schwebke, J.R.; Muzny, C.A.; Josey, W.E. Role of Gardnerella vaginalis in the pathogenesis of bacterial vaginosis: A conceptual model. J. Infect. Dis. 2014, 210, 338–343. [Google Scholar] [CrossRef]
- Pleckaityte, M.; Mistiniene, E.; Lasickiene, R.; Zvirblis, G.; Zvirbliene, A. Generation of recombinant single-chain antibodies neutralizing the cytolytic activity of vaginolysin, the main virulence factor of Gardnerella vaginalis. BMC Biotechnol. 2011, 11, 100. [Google Scholar] [CrossRef]
- Zilnyte, M.; Venclovas, Č.; Zvirbliene, A.; Pleckaityte, M. The cytolytic activity of vaginolysin strictly depends on cholesterol and is potentiated by human CD59. Toxins 2015, 7, 110–128. [Google Scholar] [CrossRef]
- Lopes Dos Santos Santiago, G.; Deschaght, P.; El Aila, N.; Kiama, T.N.; Verstraelen, H.; Jefferson, K.K.; Temmerman, M.; Vaneechoutte, M. Gardnerella vaginalis comprises three distinct genotypes of which only two produce sialidase. Am. J. Obstet. Gynecol. 2011, 204, 450.e1–450.e7. [Google Scholar] [CrossRef]
- Breton, C.; Lenka, Š.; Jeanneau, C.; Ko, J.; Imberty, A. Structures and mechanisms of glycosyltransferases. Glycobiology 2006, 16, 29–37. [Google Scholar] [CrossRef]
- Yeoman, C.J.; Yildirim, S.; Thomas, S.M.; Durkin, A.S.; Torralba, M.; Sutton, G.; Buhay, C.J.; Ding, Y.; Dugan-Rocha, S.P.; Muzny, D.M.; et al. Comparative genomics of Gardnerella vaginalis strains reveals substantial differences in metabolic and virulence potential. PLoS ONE 2010, 5. [Google Scholar] [CrossRef]
- Lindahl, G.; Sta, M.; Areschoug, T. Surface Proteins of Streptococcus agalactiae and Related Proteins in Other Bacterial Pathogens. Clin. Microbiol. Rev. 2005, 18, 102–127. [Google Scholar] [CrossRef]
- Onderdonk, A.B.; Delaney, M.L.; Fichorova, N. The Human Microbiome during Bacterial Vaginosis. Clin. Microbiol. Rev. 2016, 29, 223–238. [Google Scholar] [CrossRef]
- Pekmezovic, M.; Mogavero, S.; Naglik, J.R.; Hube, B. Host–Pathogen Interactions during Female Genital Tract Infections. Trends Microbiol. 2019, 27, 982–996. [Google Scholar] [CrossRef]
- Patterson, J.L.; Stull-Lane, A.; Girerd, P.H.; Jefferson, K.K. Analysis of adherence, biofilm formation and cytotoxicity suggests a greater virulence potential of Gardnerella vaginalis relative to other bacterial-vaginosis-associated anaerobes. Microbiology 2010, 156, 392–399. [Google Scholar] [CrossRef]
- Yang, L.; Liu, Y.; Wu, H.; Høiby, N.; Molin, S.; Song, Z.J. Current understanding of multispecies biofilms. Int. J. Oral Sci. 2011, 3, 74–81. [Google Scholar] [CrossRef]
- Bai, F.; Cai, Z.; Yang, L. Recent progress in experimental and human disease-associated multi- species biofilms. Comput. Struct. Biotechnol. J. 2019, 17, 1234–1244. [Google Scholar] [CrossRef]
- Alves, P.; Castro, J.; Sousa, C.; Cereija, T.B.; Cerca, N. Gardnerella vaginalis Outcompetes 29 other bacterial species isolated from patients with bacterial vaginosis, using in an in vitro biofilm formation model. J. Infect. Dis. 2014, 210, 593–596. [Google Scholar] [CrossRef]
- Machado, A.; Salgueiro, D.; Harwich, M.; Jefferson, K.K.; Cerca, N. Quantitative analysis of initial adhesion of bacterial vaginosis-associated anaerobes to ME-180 cells. Anaerobe 2013, 23, 1–4. [Google Scholar] [CrossRef]
- Castro, J.; Alves, P.; Sousa, C.; Cereija, T.; França, Â.; Jefferson, K.K.; Cerca, N. Using an in-vitro biofilm model to assess the virulence potential of bacterial vaginosis or non-bacterial vaginosis Gardnerella vaginalis isolates. Sci. Rep. 2015, 5, 11640. [Google Scholar] [CrossRef] [PubMed]
- Bulavaitè, A.; Dalgediene, I.; Michailoviene, V.; Pleckaityte, M. Type II Restriction-Modification System from Gardnerella vaginalis ATCC 14018. Pathogens 2020, 9, 703. [Google Scholar] [CrossRef]
- Taubman, M.; Ruby, J.; Goldner, M. Nature of Symbiosis in Oral Disease. J. Dent. Res. 2007, 86, 2005–2008. [Google Scholar] [CrossRef]
- Haney, E.F.; Trimble, M.J.; Cheng, J.T.; Vallé, Q.; Hancock, R.E.W. Critical Assessment of Methods to Quantify Biofilm Growth and Evaluate Antibiofilm Activity of Host Defence Peptides. Biomolecules 2018, 8, 29. [Google Scholar] [CrossRef]
- Flemming, H.; Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. [Google Scholar] [CrossRef] [PubMed]
- Melaugh, G.; Hutchison, J.; Kasper, N.; Irie, Y.; Allen, J. Shaping the Growth Behaviour of Biofilms Initiated from Bacterial Aggregates. PLoS ONE 2016, 11, e0149683. [Google Scholar] [CrossRef]
- Cerca, N.; Pier, G.B.; Vilanova, M.; Oliveira, R.; Azeredo, J. Quantitative analysis of adhesion and biofilm formation on hydrophilic and hydrophobic surfaces of clinical isolates of Staphylococcus epidermidis. Res. Microbiol. 2005, 156, 506–514. [Google Scholar] [CrossRef]
- Roger, T.; Bhakoo, M.; Zhang, Z. Bacterial adhesion and biofilms on surfaces. Prog. Nat. Sci. 2008, 18, 1049–1056. [Google Scholar] [CrossRef]
- Dunne, W.M. Bacterial Adhesion: Seen Any Good Biofilms Lately? Clin. Microbiol. Rev. 2002, 15, 155–166. [Google Scholar] [CrossRef]
- Giaouris, E.; Heir, E.; Desvaux, M.; Hébraud, M.; Møretrø, T.; Langsrud, S.; Doulgeraki, A.; Nychas, G.-J.; Kacániová, M.; Czaczyk, K.; et al. Intra- and inter-species interactions within biofilms of important foodborne bacterial pathogens. Front. Microbiol. 2015, 6, 1–26. [Google Scholar] [CrossRef]
- Létoffé, S.; Chalabaev, S.; Dugay, J.; Stressmann, F.; Audrain, B.; Portais, J.-C.; Letisse, F.; Ghigo, J. Biofilm microenvironment induces a widespread adaptive amino-acid fermentation pathway conferring strong fitness advantage in Escherichia coli. PLoS Genet. 2017, 13, e1006800. [Google Scholar] [CrossRef]
- De Weirdt, R.; Van De Wiele, T. Micromanagement in the gut: Microenvironmental factors govern colon mucosal biofilm structure and functionality. NPJ Biofilms Microbiomes 2015, 1, 15026. [Google Scholar] [CrossRef]
- Van Der Veer, C.; Hertzberger, R.Y.; Bruisten, S.M.; Tytgat, H.L.P.; Swanenburg, J.; De Kat Angelino-Bart, A.; Schuren, F.; Molenaar, D.; Reid, G.; De Vries, H.; et al. Comparative genomics of human Lactobacillus crispatus isolates reveals genes for glycosylation and glycogen degradation: Implications for in vivo dominance of the vaginal microbiota. Microbiome 2019, 7, 1–14. [Google Scholar] [CrossRef]
- Rainey, C.; Michalek, S.M.; Wen, Z.T.; Wu, H. Glycosyltransferase-Mediated Biofilm Matrix Dynamics and Virulence of Streptococcus mutans. Appl. Environ. Microbiol. 2019, 85, 1–15. [Google Scholar] [CrossRef]
- Hardy, L.; Jespers, V.; Van den Bulck, M.; Buyze, J.; Mwambarangwe, L.; Musengamana, V.; Vaneechoutte, M.; Crucitti, T. The presence of the putative Gardnerella vaginalis sialidase A gene in vaginal specimens is associated with bacterial vaginosis biofilm. PLoS ONE 2017, 12, e0172522. [Google Scholar] [CrossRef]
- Soong, G.; Muir, A.; Gomez, M.I.; Waks, J.; Reddy, B.; Planet, P.; Singh, P.K.; Kanetko, Y.; Wolfgang, M.C.; Hsiao, Y.; et al. Bacterial neuraminidase facilitates mucosal infection by participating in biofilm production. J. Clin. Investig. 2006, 116, 2297–2305. [Google Scholar] [CrossRef] [PubMed]
- Kurukulasuriya, S.P.; Patterson, M.H.; Hill, J.E. Slipped strand mispairing in the gene encoding cell wall associated sialidase NanH3 in Gardnerella spp. Infect. Immun. 2020, IAI.00583-20. [Google Scholar] [CrossRef]
- França, A.; Cerca, N. Plasma is the main regulator of Staphylococcus epidermidis biofilms virulence genes transcription in human blood. Pathog. Dis. 2016, 74, ftv125. [Google Scholar] [CrossRef][Green Version]
- Smith, A.; Kaczmar, A.; Bamford, R.A.; Smith, C.; Frustaci, S.; Kovacs-Simon, A.; O’Neill, P.; Moore, K.; Paszkiewicz, K.; Titball, R.W.; et al. The culture environment influences both gene regulation and phenotypic heterogeneity in Escherichia coli. Front. Microbiol. 2018, 9, 1–13. [Google Scholar] [CrossRef]
- Elias, S.; Banin, E. Multi-species biofilms: Living with friendly neighbors. FEMS Microbiol. Rev. 2012, 36, 990–1004. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Wang, S.Y. Synergistic growth in bacteria depends on substrate complexity. J Microbiol. 2016, 54, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Castro, J.; Rosca, A.S.; Cools, P.; Vaneechoutte, M.; Cerca, N. Gardnerella vaginalis Enhances Atopobium vaginae Viability in an in vitro Model. Front. Microbiol. 2020, 10, 1–9. [Google Scholar] [CrossRef]
- Castro, J.; Jefferson, K.K.; Cerca, N. Innate immune components affect growth and virulence traits of bacterial-vaginosis-associated and non-bacterial-vaginosis-associated Gardnerella vaginalis strains similarly. Pathog. Dis. 2019, 76, 1–7. [Google Scholar] [CrossRef]
- Peeters, E.; Nelis, H.J.; Coenye, T. Comparison of multiple methods for quantification of microbial biofilms grown in microtiter plates. J. Microbiol. Methods 2008, 72, 157–165. [Google Scholar] [CrossRef]
- França, A.; Freitas, A.I.; Henriques, A.F.; Cerca, N. Optimizing a qPCR gene expression quantification assay for S. epidermidis biofilms: A comparison between commercial kits and a customized protocol. PLoS ONE 2012, 7. [Google Scholar] [CrossRef]
- Carvalhais, V.; Delgado-Rastrollo, M.; Melo, L.D.R.; Cerca, N. Controlled RNA contamination and degradation and its impact on qPCR gene expression in S. epidermidis biofilms. J. Microbiol. Methods 2013, 95, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Castro, J.; França, A.; Bradwell, K.R.; Serrano, M.G.; Jefferson, K.K.; Cerca, N. Comparative transcriptomic analysis of Gardnerella vaginalis biofilms vs. planktonic cultures using RNA-seq. NPJ Biofilms Microbiomes 2017, 3, 3. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M. Chapter 3: Quantification strategies in real-time PCR. In A–Z of Quantitative PCR; Ser. 5, Ch. 3; Bustin, S.A., Ed.; Int. Univ. Line: La Jolla, CA, USA, 2004; pp. 87–112. [Google Scholar] [CrossRef]
| Strains | Gard162 Probe Specificity | AtoITM1 Probe Specificity |
|---|---|---|
| G. vaginalis strain ATCC 14018T | ++++ | − |
| A. vaginae strain ATCC BAA-55T | − | ++++ |
| P. bivia strain ATCC 29303T | − | − |
| Condition | Equation | PNA Probe Efficiency (%) |
|---|---|---|
| G. vaginalis biofilm | G. vaginalis counts = (log (PNA Gard162 probe bacterial counts/area) + 0.1892)/1.022 | 92.08 |
| A. vaginae biofilm | A. vaginae biofilm cells counts = (log (PNA AtoITM1 probe bacterial counts/area) + 0.0405)/0.9878 | 91.59 |
| Planktonic fraction of G. vaginalis biofilm | G. vaginalis planktonic cells counts = (log (PNA Gard162 probe bacterial counts/area) + 0.0265)/1.003 | 98.67 |
| Planktonic fraction of A. vaginae biofilm | A. vaginae planktonic cells counts = (log (PNA AtoITM1 probe bacterial counts/area) + 0.0937)/1.012 | 98.12 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Castro, J.; Rosca, A.S.; Muzny, C.A.; Cerca, N. Atopobium vaginae and Prevotella bivia Are Able to Incorporate and Influence Gene Expression in a Pre-Formed Gardnerella vaginalis Biofilm. Pathogens 2021, 10, 247. https://doi.org/10.3390/pathogens10020247
Castro J, Rosca AS, Muzny CA, Cerca N. Atopobium vaginae and Prevotella bivia Are Able to Incorporate and Influence Gene Expression in a Pre-Formed Gardnerella vaginalis Biofilm. Pathogens. 2021; 10(2):247. https://doi.org/10.3390/pathogens10020247
Chicago/Turabian StyleCastro, Joana, Aliona S. Rosca, Christina A. Muzny, and Nuno Cerca. 2021. "Atopobium vaginae and Prevotella bivia Are Able to Incorporate and Influence Gene Expression in a Pre-Formed Gardnerella vaginalis Biofilm" Pathogens 10, no. 2: 247. https://doi.org/10.3390/pathogens10020247
APA StyleCastro, J., Rosca, A. S., Muzny, C. A., & Cerca, N. (2021). Atopobium vaginae and Prevotella bivia Are Able to Incorporate and Influence Gene Expression in a Pre-Formed Gardnerella vaginalis Biofilm. Pathogens, 10(2), 247. https://doi.org/10.3390/pathogens10020247







