Cytokine, Chemokine, and Metalloprotease Activation in the Serum of Patients with Nephropathia Epidemica from the Republic of Tatarstan and the Republic of Mordovia, Russia
Abstract
:1. Introduction
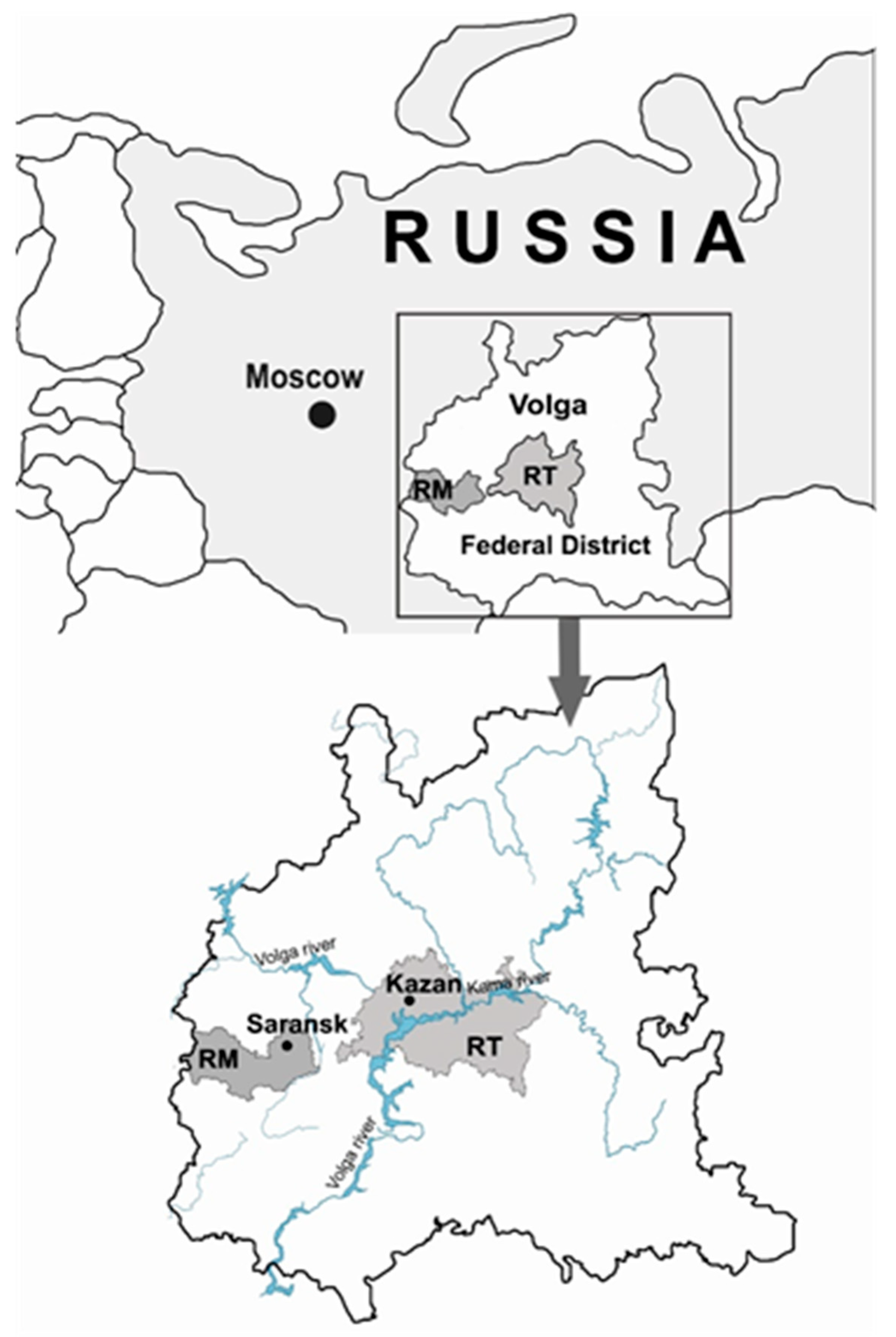
2. Materials and Methods
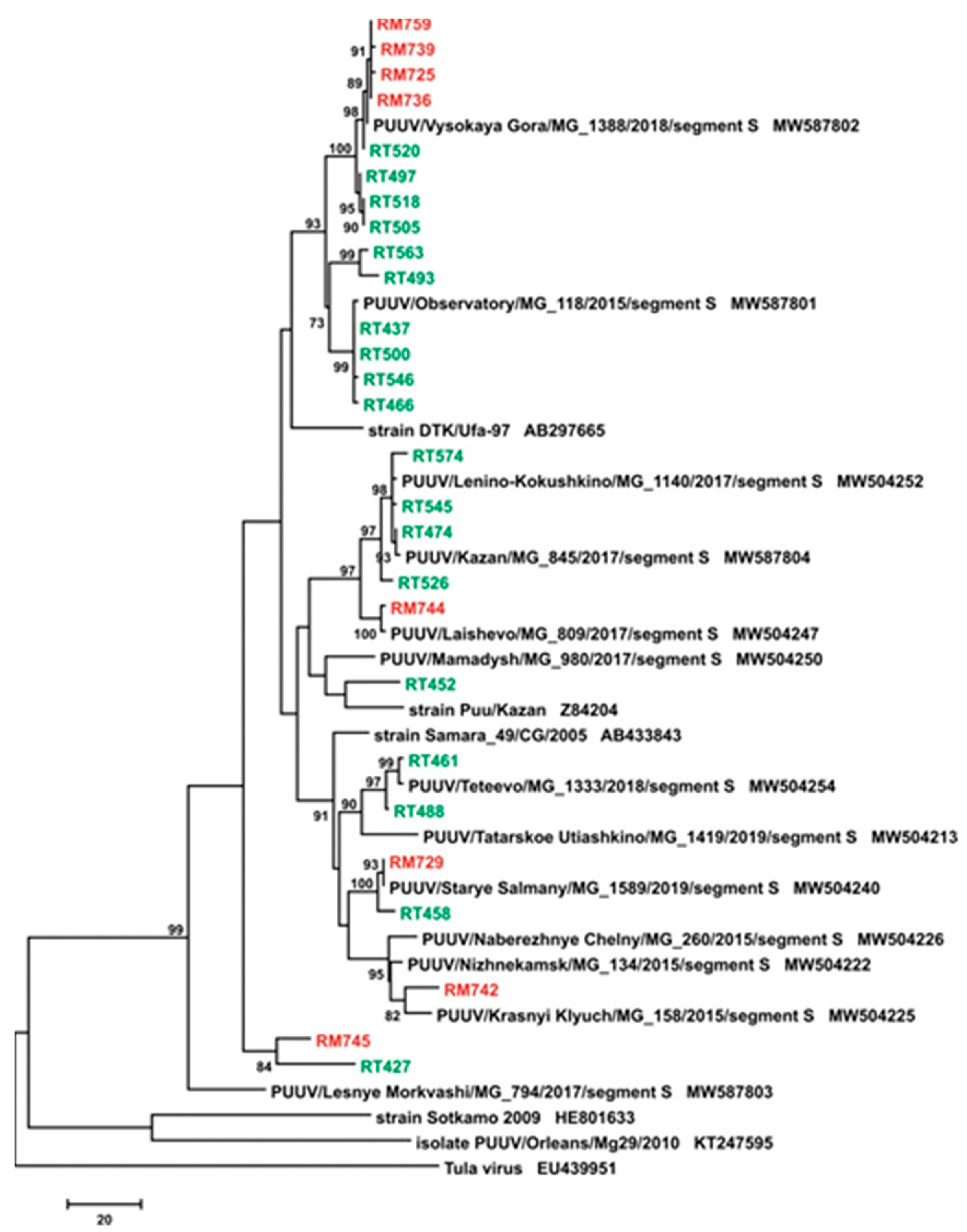
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tkachenko, E.A.; Ishmukhametov, A.A.; Dzagurova, T.K.; Bernshtein, A.D.; Morozov, V.G.; Siniugina, A.A.; Kurashova, S.S.; Balkina, A.S.; Tkachenko, P.E.; Kruger, D.H. Hemorrhagic fever with renal syndrome, Russia. Emerg. Infect. Dis. 2019, 25, 2325. [Google Scholar] [CrossRef]
- Vaheri, A.; Strandin, T.; Hepojoki, J.; Sironen, T.; Henttonen, H.; Mäkelä, S.; Mustonen, J. Uncovering the mysteries of hantavirus infections. Nat. Rev. Microbiol. 2013, 11, 539–550. [Google Scholar] [CrossRef]
- Khismatullina, N.; Karimov, M.; Khaertynov, K.; Shuralev, E.; Morzunov, S.; Khaertynova, I.; Ivanov, A.; Milova, I.; Khakimzyanova, M.; Sayfullina, G.S. Epidemiological dynamics of nephropathia epidemica in the Republic of Tatarstan, Russia, during the period of 1997–2013. Epidemiol. Infect. 2016, 144, 618–626. [Google Scholar] [CrossRef] [Green Version]
- Tkachenko, E.A.; Bernshtein, A.D.; Okulova, N.M.; Korotina, N.A.; Trankvilevskiy, D.V.; Morozov, V.G.; Yunicheva, Y.U.V.; Zavora, D.L.; Blovneva, M.V. Hemorrhagic fever with renal syndrome in Russia -problem of 21 century. Vestn. RAEN 2012, 48–54. [Google Scholar] [CrossRef]
- Ivanova, A.; Popov, N.; Pakskina, N.; Kuznecov, A.; Matrosov, A.; Shilov, M.; Moshalkin, P.; Korneev, M.; Toporcov, V. Epidemiological activity of hemorrhagic fever with renal syndrome foci in the territory of the russian federation in 2013–2017 and forecast for 2018. Probl. Part. Danger. Infect. 2018, 1, 17–21. [Google Scholar] [CrossRef] [Green Version]
- Pavelkina, V.; Uskova, Y.G. Hemorrhagic fever with renal syndrome: Clinical, pathogenetic and therapeutic aspects. Mordovia Univ. Bull. 2017, 27, 315–329. [Google Scholar] [CrossRef] [Green Version]
- Outinen, T.K.; Mäkelä, S.; Clement, J.; Paakkala, A.; Pörsti, I.; Mustonen, J. Community acquired severe acute kidney injury caused by hantavirus-induced hemorrhagic fever with renal syndrome has a favorable outcome. Nephron 2015, 130, 182–190. [Google Scholar] [CrossRef] [PubMed]
- Clement, J.; Kuypers, D.; Meijers, B.; Van Ranst, M. Hantavirus infection with renal involvement do not result in chronic renal diseases or end-stage renal failure. J. Nephrol. Renal. Dis. 2017, 1, 1. [Google Scholar]
- Mantula, P.; Tietäväinen, J.; Clement, J.; Niemelä, O.; Pörsti, I.; Vaheri, A.; Mustonen, J.; Mäkelä, S.; Outinen, T. Flash-like albuminuria in acute kidney injury caused by Puumala hantavirus infection. Pathogens 2020, 9, 615. [Google Scholar] [CrossRef]
- Jonsson, C.B.; Figueiredo, L.T.M.; Vapalahti, O. A global perspective on hantavirus ecology, epidemiology, and disease. Clin. Microbiol. Rev. 2010, 23, 412–441. [Google Scholar] [CrossRef] [Green Version]
- Brocato, R.L.; Hooper, J.W. Progress on the prevention and treatment of hantavirus disease. Viruses 2019, 11, 610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davidyuk, Y.; Kabwe, E.; Khaiboullina, S.; Ismagilova, R.; Shakirova, V.; Isaeva, G.; Pavelkina, V.; Uskova, Y.G.; Rizvanov, A.; Morzunov, S. Genetic diversity of Puumala virus isolates in the Republic of Tatarstan and the Republic of Mordovia. BioNanoScience 2017, 7, 309–312. [Google Scholar] [CrossRef]
- Reil, D.; Rosenfeld, U.M.; Imholt, C.; Schmidt, S.; Ulrich, R.G.; Eccard, J.A.; Jacob, J. Puumala hantavirus infections in bank vole populations: Host and virus dynamics in Central Europe. BMC Ecol. 2017, 17, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Mustonen, J.; Mäkelä, S.; Outinen, T.; Laine, O.; Jylhävä, J.; Arstila, P.T.; Hurme, M.; Vaheri, A. The pathogenesis of nephropathia epidemica: New knowledge and unanswered questions. Antivir. Res. 2013, 100, 589–604. [Google Scholar] [CrossRef]
- Hepojoki, J.; Vaheri, A.; Strandin, T. The fundamental role of endothelial cells in hantavirus pathogenesis. Front. Microbiol. 2014, 5, 727. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zaki, S.R. Hantavirus pulmonary syndrome: Pathogenesis of an emerging infectious disease. Am. J. Pathol. 1995, 146, 552. [Google Scholar]
- Zöller, L.; Faulde, M.; Meisel, H.; Ruh, B.; Kimmig, P.; Schelling, U.; Zeier, M.; Kulzer, P.; Becker, C.; Roggendorf, M. Seroprevalence of hantavirus antibodies in Germany as determined by a new recombinant enzyme immunoassay. Eur. J. Clin. Microbiol. Infect. Dis. 1995, 14, 305–313. [Google Scholar] [CrossRef]
- Maes, P.; Clement, J.; Groeneveld, P.H.; Colson, P.; Huizinga, T.W.; Ranst, M.V. Tumor necrosis factor-α genetic predisposing factors can influence clinical severity in nephropathia epidemica. Viral Immunol. 2006, 19, 558–564. [Google Scholar] [CrossRef]
- Hjertqvist, M.; Klein, S.L.; Ahlm, C.; Klingström, J. Mortality rate patterns for hemorrhagic fever with renal syndrome caused by Puumala virus. Emerg. Infect. Dis. 2010, 16, 1584. [Google Scholar] [CrossRef]
- Klempa, B.; Tkachenko, E.A.; Dzagurova, T.K.; Yunicheva, Y.V.; Morozov, V.G.; Okulova, N.M.; Slyusareva, G.P.; Smirnov, A.; Kruger, D.H. Hemorrhagic fever with renal syndrome caused by 2 lineages of Dobrava hantavirus, Russia. Emerg. Infect. Dis. 2008, 14, 617. [Google Scholar] [CrossRef]
- Tietäväinen, J.; Mantula, P.; Outinen, T.; Huhtala, H.; Pörsti, I.H.; Niemelä, O.; Vaheri, A.; Mäkelä, S.; Mustonen, J. Glucosuria predicts the severity of Puumala hantavirus infection. Kidney Int. Rep. 2019, 4, 1296–1303. [Google Scholar] [CrossRef] [Green Version]
- Khaiboullina, S.F.; Levis, S.; Morzunov, S.P.; Martynova, E.V.; Anokhin, V.A.; Gusev, O.A.; St Jeor, S.C.; Lombardi, V.C.; Rizvanov, A.A. Serum cytokine profiles differentiating hemorrhagic fever with renal syndrome and hantavirus pulmonary syndrome. Front. Immunol. 2017, 8, 567. [Google Scholar] [CrossRef] [Green Version]
- Maleki, K.T.; García, M.; Iglesias, A.; Alonso, D.; Ciancaglini, M.; Hammar, U.; Ljunggren, H.-G.; Schierloh, P.; Martínez, V.P.; Klingström, J. Serum markers associated with severity and outcome of hantavirus pulmonary syndrome. J. Infect. Dis. 2019, 219, 1832–1840. [Google Scholar] [CrossRef] [Green Version]
- Settergren, B.; Juto, P.; Trollfors, B.; Wadell, G.; Norrby, S. Clinical characteristics of nephropathia epidemica in Sweden: Prospective study of 74 cases. Rev. Infect. Dis. 1989, 11, 921–927. [Google Scholar] [CrossRef]
- Bren, A.F.; PavlovČIČ, S.-K.; Koselj, M.; KovaČ, J.; Kandus, A.; Kveder, R. Acute renal failure due to hemorrhagic fever with renal syndrome. Ren. Fail. 1996, 18, 635–638. [Google Scholar] [CrossRef] [PubMed]
- Mäkelä, S.; Ala-Houhala, I.; Mustonen, J.; Koivisto, A.-M.; Kouri, T.; Turjanmaa, V.; Vapalahti, O.; Vaheri, A.; Pasternack, A. Renal function and blood pressure five years after Puumala virus-induced nephropathy. Kidney Int. 2000, 58, 1711–1718. [Google Scholar] [CrossRef] [Green Version]
- Clement, J.; Leduc, J.; Mcelhinney, L.; Reynes, J.-M.; van Ranst, M.; Calisher, C. Clinical Characteristics of Ratborne Seoul Hantavirus. Emerg. Infect. Dis. 2019, 25, 387–388. [Google Scholar] [CrossRef]
- Settergren, B.; Leschinskaya, E.; Zagidullin, I.; Fazlyeva, R.; Khunafina, D.; Niklasson, B. Hemorrhagic fever with renal syndrome: Comparison of clinical course in Sweden and in the Western Soviet Union. Scand. J. Infect. Dis. 1991, 23, 549–552. [Google Scholar] [CrossRef] [PubMed]
- Peco-Antić, A.; Popović-Rolović, M.; Gligić, A.; Popović, D.; Jovanović, O.; Kostić, M. Clinical characteristics of haemorrhagic fever with renal syndrome in children. Pediatric. Nephrol. 1992, 6, 335–338. [Google Scholar] [CrossRef] [PubMed]
- Rasche, F.M.; Uhel, B.; Ulrich, R.; Krüger, D.H.; Karges, W.; Czock, D.; Hampl, W.; Keller, F.; Meisel, H.; von Müller, L. Thrombocytopenia and acute renal failure in Puumala hantavirus infections. Emerg. Infect. Dis. 2004, 10, 1420. [Google Scholar] [CrossRef] [Green Version]
- Khaiboullina, S.; Martynova, E.; Khamidullina, Z.; Lapteva, E.; Nikolaeva, I.; Anokhin, V.; Lombardi, V.; Rizvanov, A. Upregulation of IFN-γ and IL-12 is associated with a milder form of hantavirus hemorrhagic fever with renal syndrome. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 2149–2156. [Google Scholar] [CrossRef]
- Baigildina, A.A.; Khaiboullina, S.F.; Martynova, E.V.; Anokhin, V.A.; Lombardi, V.C.; Rizvanov, A.A. Inflammatory cytokines kinetics define the severity and phase of nephropathia epidemica. Biomark. Med. 2015, 9, 99–107. [Google Scholar] [CrossRef]
- Davidyuk, Y.; Shamsutdinov, A.; Kabwe, E.; Ismagilova, R.; Martynova, E.; Belyaev, A.; Shuralev, E.; Trifonov, V.; Savitskaya, T.; Isaeva, G. Prevalence of the Puumala orthohantavirus Strains in the Pre-Kama Area of the Republic of Tatarstan, Russia. Pathogens 2020, 9, 540. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tkachenko, E.; Ivanov, A.; Dzagurova, T. Immunosorbent assays for diagnosis of haemorrhagic fever with renal syndrome. Immunosorbent Assays Diagn. Haemorrh. Fever Ren. Syndr. 1981, 2, 257–258. [Google Scholar]
- Ripley, B.D. The R project in statistical computing. MSOR Connect. Newsl. LTSN Maths Stats OR Netw. 2001, 1, 23–25. [Google Scholar] [CrossRef] [Green Version]
- Dzagurova, T.; Tkachenko, E.; IuV, I.; Morozov, V.; Briukhanov, A.; Bashkirtsev, V.; Sedova, N.; Klempa, B.; Kruger, D. Discovery, clinical and etiological characteristic of hemorrhagic fever with renal syndrome in the subtropical zone of Krasnodar region. Zhurnal Mikrobiol. Epidemiol. I Immunobiol. 2008, V.1, 12–16. [Google Scholar]
- Leshchinskaia, E.; Tkachenko, E.; Ryl’tseva, E.; Petrov, V.; Ianovskiĭ, S.; Gasanova, T.; Bystrovskiĭ, V.; Mogila, T.; Koval’skiĭ, G.; Priven, V. Characteristics of endemic foci of hemorrhagic fever with renal syndrome in various regions of the USSR. Vopr. Virusol. 1990, 35, 42–45. [Google Scholar] [PubMed]
- Plyusnin, A.; Vapalahti, O.; Lankinen, H.; Lehväslaiho, H.; Apekina, N.; Myasnikov, Y.; Kallio-Kokko, H.; Henttonen, H.; Lundkvist, A.; Brummer-Korvenkontio, M. Tula virus: A newly detected hantavirus carried by European common voles. J. Virol. 1994, 68, 7833–7839. [Google Scholar] [CrossRef] [Green Version]
- Davidyuk, Y.N.; Kabwe, E.; Shakirova, V.G.; Martynova, E.V.; Ismagilova, R.K.; Khaertynova, I.M.; Khaiboullina, S.F.; Rizvanov, A.A.; Morzunov, S.P. Characterization of the Puumala orthohantavirus strains in the northwestern region of the Republic of Tatarstan in relation to the clinical manifestations in hemorrhagic fever with renal syndrome patients. Front. Pharmacol. 2019, 10, 970. [Google Scholar] [CrossRef]
- Kariwa, H.; Tkachenko, E.A.; Morozov, V.G.; Seto, T.; Tanikawa, Y.; Kolominov, S.I.; Belov, S.N.; Nakamura, I.; Hashimoto, N.; Balakiev, A.E. Epidemiological study of hantavirus infection in the Samara Region of European Russia. J. Vet. Med. Sci. 2009, 71, 1569–1578. [Google Scholar] [CrossRef] [Green Version]
- Coe, C.L.; Love, G.D.; Karasawa, M.; Kawakami, N.; Kitayama, S.; Markus, H.R.; Tracy, R.P.; Ryff, C.D. Population differences in proinflammatory biology: Japanese have healthier profiles than Americans. Brain Behav. Immun. 2011, 25, 494–502. [Google Scholar] [CrossRef] [Green Version]
- ter Meulen, J.; Sakho, M.; Koulemou, K.; Magassouba, N.F.; Bah, A.; Preiser, W.; Daffis, S.; Klewitz, C.; Bae, H.-G.; Niedrig, M. Activation of the cytokine network and unfavorable outcome in patients with yellow fever. J. Infect. Dis. 2004, 190, 1821–1827. [Google Scholar] [CrossRef]
- Yao, S.; Hong, C.-C.; Ruiz-Narváez, E.A.; Evans, S.S.; Zhu, Q.; Schaefer, B.A.; Yan, L.; Coignet, M.V.; Lunetta, K.L.; Sucheston-Campbell, L.E. Genetic ancestry and population differences in levels of inflammatory cytokines in women: Role for evolutionary selection and environmental factors. PLoS Genet. 2018, 14, e1007368. [Google Scholar] [CrossRef] [Green Version]
- Smolen, K.K.; Ruck, C.E.; Fortuno III, E.S.; Ho, K.; Dimitriu, P.; Mohn, W.W.; Speert, D.P.; Cooper, P.J.; Esser, M.; Goetghebuer, T. Pattern recognition receptor-mediated cytokine response in infants across 4 continents. J. Allergy Clin. Immunol. 2014, 133, 818–826. [Google Scholar] [CrossRef]
- Boerrigter, D.; Weickert, T.W.; Lenroot, R.; O’Donnell, M.; Galletly, C.; Liu, D.; Burgess, M.; Cadiz, R.; Jacomb, I.; Catts, V.S. Using blood cytokine measures to define high inflammatory biotype of schizophrenia and schizoaffective disorder. J. Neuroinflammation 2017, 14, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Vapalahti, O.; Mustonen, J.; Lundkvist, Å.; Henttonen, H.; Plyusnin, A.; Vaheri, A. Hantavirus infections in Europe. Lancet Infect. Dis. 2003, 3, 653–661. [Google Scholar] [CrossRef]
- Klein, S.L.; Marks, M.A.; Li, W.; Glass, G.E.; Fang, L.-Q.; Ma, J.-Q.; Cao, W.-C. Sex differences in the incidence and case fatality rates from hemorrhagic fever with renal syndrome in China, 2004–2008. Clin. Infect. Dis. 2011, 52, 1414–1421. [Google Scholar] [CrossRef] [Green Version]
- NPP. Clinical Recommendations Hemorrhagic Fever with Renal Syndrome in Adults; The Russian Archives of Internal Medicine: Izhevsk, Russia, 2014; p. 74. [Google Scholar]
- Saavedra, F.; Díaz, F.E.; Retamal-Díaz, A.; Covián, C.; González, P.A.; Kalergis, A.M. Immune response during hantavirus diseases. Implications for immunotherapies and vaccine design. Immunology 2021. [Google Scholar] [CrossRef]
- Liu, M.-M.; Lei, X.-Y.; Yu, H.; Zhang, J.-Z.; Yu, X.-J. Correlation of cytokine level with the severity of severe fever with thrombocytopenia syndrome. Virol. J. 2017, 14, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Page-McCaw, A.; Ewald, A.J.; Werb, Z. Matrix metalloproteinases and the regulation of tissue remodelling. Nat. Rev. Mol. Cell Biol. 2007, 8, 221–233. [Google Scholar] [CrossRef]
- Li, X.-D.; Zhang, X.-R.; Li, Z.-H.; Yang, Y.; Zhang, D.; Zheng, H.; Dong, S.-Y.; Chen, J.; Zeng, X.-D. Effect of matrix metallopeptidase 13 on the function of mouse bone marrow-derived dendritic cells. Chin. Med. J. 2017, 130, 717. [Google Scholar] [CrossRef]
- Abraham, M.; Shapiro, S.; Karni, A.; Weiner, H.L.; Miller, A. Gelatinases (MMP-2 and MMP-9) are preferentially expressed by Th1 vs. Th2 cells. J. Neuroimmunol. 2005, 163, 157–164. [Google Scholar] [CrossRef]
- Matsushita, H.; Hosoi, A.; Ueha, S.; Abe, J.; Fujieda, N.; Tomura, M.; Maekawa, R.; Matsushima, K.; Ohara, O.; Kakimi, K. Cytotoxic T lymphocytes block tumor growth both by lytic activity and IFNγ-dependent cell-cycle arrest. Cancer Immunol. Res. 2015, 3, 26–36. [Google Scholar] [CrossRef] [Green Version]
- Lin, F.-C.; Young, H.A. Interferons: Success in anti-viral immunotherapy. Cytokine Growth Factor Rev. 2014, 25, 369–376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dinarello, C.A. Overview of the IL-1 family in innate inflammation and acquired immunity. Immunol. Rev. 2018, 281, 8–27. [Google Scholar] [CrossRef]
- Rider, P.; Carmi, Y.; Guttman, O.; Braiman, A.; Cohen, I.; Voronov, E.; White, M.R.; Dinarello, C.A.; Apte, R.N. IL-1α and IL-1β recruit different myeloid cells and promote different stages of sterile inflammation. J. Immunol. 2011, 187, 4835–4843. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cannetti, C.A.; Leung, B.P.; Culshaw, S.; McInnes, I.B.; Cunha, F.Q.; Liew, F.Y. IL-18 enhances collagen-induced arthritis by recruiting neutrophils via TNF-α and leukotriene B4. J. Immunol. 2003, 171, 1009–1015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tominaga, K.; Yoshimoto, T.; Torigoe, K.; Kurimoto, M.; Matsui, K.; Hada, T.; Okamura, H.; Nakanishi, K. IL-12 synergizes with IL-18 or IL-1β for IFN-γ production from human T cells. Int. Immunol. 2000, 12, 151–160. [Google Scholar] [CrossRef] [Green Version]
- Gherardi, M.M.; Ramírez, J.C.; Esteban, M. IL-12 and IL-18 act in synergy to clear vaccinia virus infection: Involvement of innate and adaptive components of the immune system. J. Gen. Virol. 2003, 84, 1961–1972. [Google Scholar] [CrossRef]
- Akdis, M. Healthy immune response to allergens: T regulatory cells and more. Curr. Opin. Immunol. 2006, 18, 738–744. [Google Scholar] [CrossRef]
- Akdis, M.; Aab, A.; Altunbulakli, C.; Azkur, K.; Costa, R.A.; Crameri, R.; Duan, S.; Eiwegger, T.; Eljaszewicz, A.; Ferstl, R. Interleukins (from IL-1 to IL-38), interferons, transforming growth factor β, and TNF-α: Receptors, functions, and roles in diseases. J. Allergy Clin. Immunol. 2016, 138, 984–1010. [Google Scholar] [CrossRef] [Green Version]
- Keppel, M.P.; Saucier, N.; Mah, A.Y.; Vogel, T.P.; Cooper, M.A. Activation-specific metabolic requirements for NK Cell IFN-γ production. J. Immunol. 2015, 194, 1954–1962. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Yang, W.; Pan, M.; Scully, E.; Girardi, M.; Augenlicht, L.H.; Craft, J.; Yin, Z. γδ T cells provide an early source of interferon γ in tumor immunity. J. Exp. Med. 2003, 198, 433–442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castro, F.; Cardoso, A.P.; Gonçalves, R.M.; Serre, K.; Oliveira, M.J. Interferon-gamma at the crossroads of tumor immune surveillance or evasion. Front. Immunol. 2018, 9, 847. [Google Scholar] [CrossRef] [Green Version]
- Schroder, K.; Hertzog, P.J.; Ravasi, T.; Hume, D.A. Interferon-γ: An overview of signals, mechanisms and functions. J. Leukoc. Biol. 2004, 75, 163–189. [Google Scholar] [CrossRef]
- Boehm, U.; Klamp, T.; Groot, M.; Howard, J. Cellular responses to interferon-gamma. Annu. Rev. Immunol. 1997, 15, 749–795. [Google Scholar] [CrossRef]
- Sercan, Ö.; Hämmerling, G.J.; Arnold, B.; Schüler, T. Cutting edge: Innate immune cells contribute to the IFN-γ-dependent regulation of antigen-specific CD8+ T cell homeostasis. J. Immunol. 2006, 176, 735–739. [Google Scholar] [CrossRef] [Green Version]
- Ohta, A.; Sekimoto, M.; Sato, M.; Koda, T.; Nishimura, S.-I.; Iwakura, Y.; Sekikawa, K.; Nishimura, T. Indispensable role for TNF-α and IFN-γ at the effector phase of liver injury mediated by Th1 cells specific to hepatitis B virus surface antigen. J. Immunol. 2000, 165, 956–961. [Google Scholar] [CrossRef] [PubMed]
- Karki, R.; Sharma, B.R.; Tuladhar, S.; Williams, E.P.; Zalduondo, L.; Samir, P.; Zheng, M.; Sundaram, B.; Banoth, B.; Malireddi, R.S. Synergism of TNF-α and IFN-γ Triggers inflammatory cell death, tissue damage, and mortality in SARS-CoV-2 infection and cytokine shock syndromes. Cell 2021, 184, 149. [Google Scholar] [CrossRef]
- Bowie, M.B.; Kent, D.G.; Copley, M.R.; Eaves, C.J. Steel factor responsiveness regulates the high self-renewal phenotype of fetal hematopoietic stem cells. Blood J. Am. Soc. Hematol. 2007, 109, 5043–5048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hara, T.; Miyajima, A. Function of the IL-3 receptor system in hematopoiesis. In Gene Technology; Springer: Berlin/Heidelberg, Germany, 1996; pp. 295–307. [Google Scholar]
- Shepard, B.D.; Badley, A.D. The biology of TRAIL and the role of TRAIL-based therapeutics in infectious diseases. Anti-Infect. Agents Med. Chem. 2009, 8, 87–101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sato, K.; Hida, S.; Takayanagi, H.; Yokochi, T.; Kayagaki, N.; Takeda, K.; Yagita, H.; Okumura, K.; Tanaka, N.; Taniguchi, T. Antiviral response by natural killer cells through TRAIL gene induction by IFN–α/β. Eur. J. Immunol. 2001, 31, 3138–3146. [Google Scholar] [CrossRef]
- Gong, B.; Almasan, A. Genomic organization and transcriptional regulation of human Apo2/TRAIL gene. Biochem. Biophys. Res. Commun. 2000, 278, 747–752. [Google Scholar] [CrossRef] [PubMed]




| Name | Nested-PCR Step | Sequence, 5′→3′ | Binding Position c | Reference |
|---|---|---|---|---|
| PUUV-39S-F3 a,b | 1st | GGCCAAAACATCTATATGTATCC | 560–582 d | |
| PUUV-S-R1496 a,b | 1st | GTATAATTCCAGTTAACCCCTG | 1496–1517 d | [33] |
| PUUV-S-F704 a,b | 2nd | AACATCATGAGTCCAGTAATGGG | 682–704 d | [33] |
| PUUV-69S-B3 a,b | 2nd | GATATCTCTTTTACCTTCTGGTC | 1297–1319 d | [33] |
| DOBV-F212 a | 1st | GAAAAGAAAGGGATCCAACTGG | 191–212 e | |
| DOBV-R542 a | 1st | ATACTGGATTGTGCATTGGGC | 542–562 e | |
| DOBV-F348 a | 2nd | ATGAACCAACAGGGCAAACTG | 327–348 e | |
| DOBV-R518 a | 2nd | GACAGAAACAGGTGCTTTGGC | 518–538 e | |
| TulaV-For49 a | 1st | AAGGATCCTCTAGAAACCGCTGGTATGAGCC | 19–49 f | |
| TulaV-Rev1321 a | 1st | GTGTCTGCAGGATCCGTTGATTAGATTTTTAGTGG | 1321–1355 f | |
| TulaV-For91 a | 2nd | AGATCACCCGCCATGAACAGC | 71–91 f | |
| TulaV-Rev328 a | 2nd | CATCAAGGACATTCCCATATCTGAG | 328–352 f |
| RT | RM | p-Value | |
|---|---|---|---|
| Age (years) | 37.50 (29.75, 51.50); n = 96 | 33.50 (26.25, 48.75) n = 58 | 0.203 |
| Hospitalization (days) | 11.00 (8.00, 12.00); n = 72 | 16.00 (14.25, 16.00); n = 58 | <0.001 |
| Fever (days) | 6.00 (4.00, 7.00) n = 54 | 8.00 (7.00, 9.00); n = 58 | <0.001 |
| Duration of hemorrhage (days) | 0.00 (0.00, 0.00); n = 92 | 0.00 (0.00, 0.00); n = 58 | 0.641 |
| Systolic arterial pressure (mmHg) | 116.00 (110.00, 120.00); n = 55 | 120.00 (110.00, 137.50); n = 58 | 0.120 |
| Diastolic arterial pressure (mmHg) | 80.00 (70.00, 80.00); n = 55 | 80.00 (70.00, 90.00); n = 58 | 0.065 |
| Urea (mg/dL) | 7.10 (4.70, 12.60); n = 93 | 6.65 (4.85, 10.25); n = 58 | 0.750 |
| Creatinine (µM/L) | 131.00 (103.00, 189.00); n = 93 | 121.50 (96.00, 160.50); n = 58 | 0.172 |
| Potassium (mEq/L) | 7.10 (5.39, 10.75); n = 88 | 5.76 (3.95, 8.07); n = 58 | 0.004 |
| Thrombocytes (×103/µL) | 92.00 (67.00, 157.00); n = 93 | 78.50 (58.50, 109.75); n = 58 | 0.205 |
| Leukocytes (×103/µL) | 9.30 (6.20, 13.65); n = 55 | 7.50 (6.62, 9.73); n = 58 | 0.251 |
| vRNA +/− (+%/−%) | 72/25 (74.2/25.8); n = 96 | 10/7 (58.8/41.2); n = 17 | 0.242 |
| Hantavirus IgM +/− (+%/−%) | 75/96 (78.1/21.9); n = 96 | 33/58 (56.9/43.1); n = 58 | 0.711 |
| Hantavirus IgG +/− (+%/−%) | 86/96 (86.4/13.6); n = 96 | 50/8 (86.2/13.8); n = 58 | 0.096 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martynova, E.; Davidyuk, Y.; Kabwe, E.; Garanina, E.E.; Shakirova, V.; Pavelkina, V.; Uskova, Y.; Stott, R.J.; Foster, T.L.; Markelova, M.; et al. Cytokine, Chemokine, and Metalloprotease Activation in the Serum of Patients with Nephropathia Epidemica from the Republic of Tatarstan and the Republic of Mordovia, Russia. Pathogens 2021, 10, 527. https://doi.org/10.3390/pathogens10050527
Martynova E, Davidyuk Y, Kabwe E, Garanina EE, Shakirova V, Pavelkina V, Uskova Y, Stott RJ, Foster TL, Markelova M, et al. Cytokine, Chemokine, and Metalloprotease Activation in the Serum of Patients with Nephropathia Epidemica from the Republic of Tatarstan and the Republic of Mordovia, Russia. Pathogens. 2021; 10(5):527. https://doi.org/10.3390/pathogens10050527
Chicago/Turabian StyleMartynova, Ekaterina, Yuriy Davidyuk, Emmanuel Kabwe, Ekaterina E. Garanina, Venera Shakirova, Vera Pavelkina, Yulia Uskova, Robert J. Stott, Toshana L. Foster, Maria Markelova, and et al. 2021. "Cytokine, Chemokine, and Metalloprotease Activation in the Serum of Patients with Nephropathia Epidemica from the Republic of Tatarstan and the Republic of Mordovia, Russia" Pathogens 10, no. 5: 527. https://doi.org/10.3390/pathogens10050527









