Regulation of Expression of the TIR-Containing Protein C Gene of the Uropathogenic Escherichia coli Strain CFT073
Abstract
1. Introduction
2. Results
2.1. Overexpression of TcpC Stops Growth and Provokes Filamentation of the Uropathogenic E. coli Strain CFT073
2.2. Endogenous Promoters Controlling the tcpC Gene
2.3. Glucose Induces the tcpC Promoter
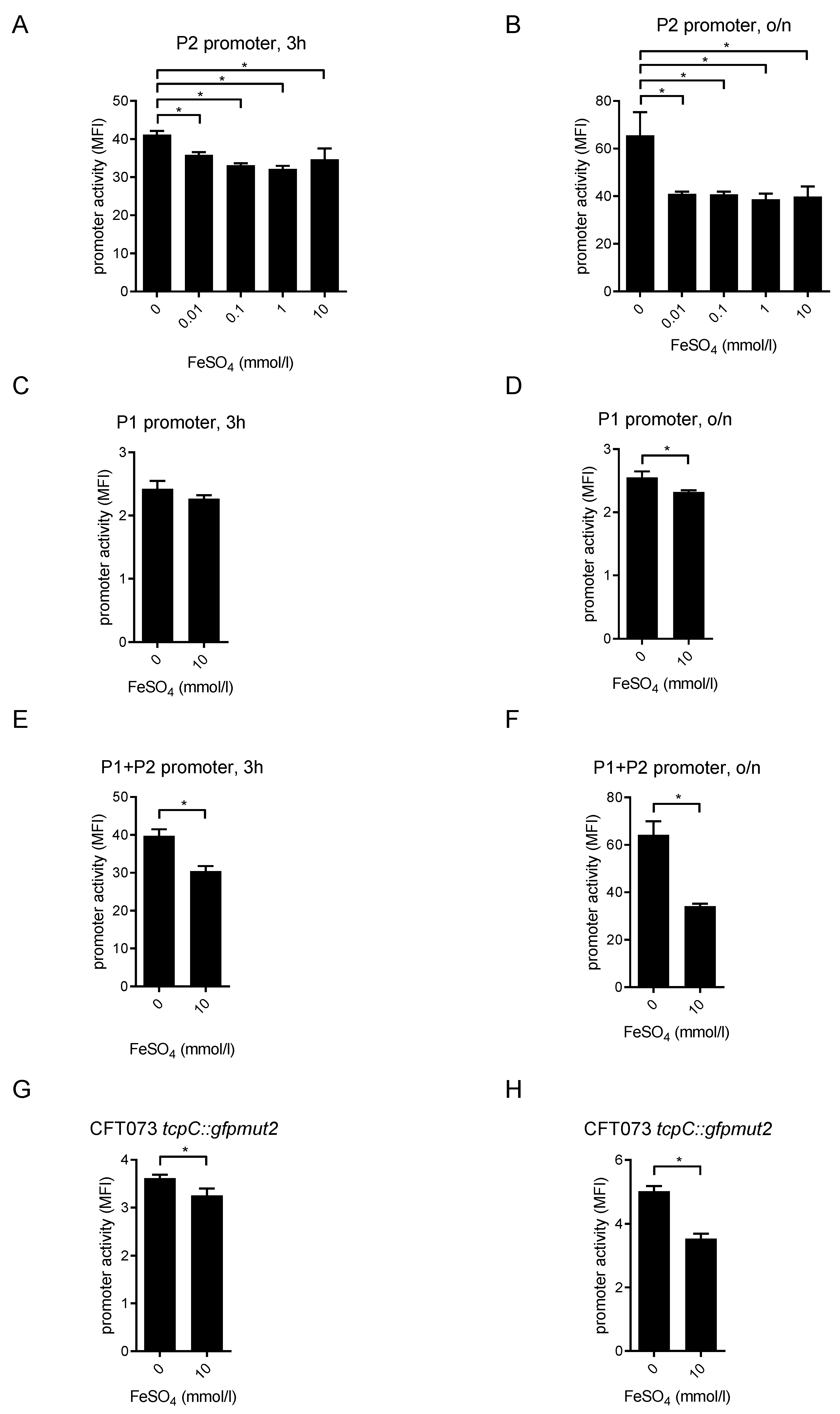
2.4. FeSO4 Dampens tcpC Promoter Activity
2.5. Human Urine Induces the tcpC Promoter
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains
4.2. Plasmids
4.3. Reagents and Culture Media
4.4. Culture of Bacteria
4.5. Growth Determination
4.6. Reporter Assays
4.7. Reverse Transcription PCR and 5′ RACE PCR
4.8. Statistics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cirl, C.; Wieser, A.; Yadav, M.; Duerr, S.; Schubert, S.; Fischer, H.; Stappert, D.; Wantia, N.; Rodriguez, N.; Wagner, H.; et al. Subversion of Toll-like receptor signaling by a unique family of bacterial Toll/interleukin-1 receptor domain-containing proteins. Nat. Med. 2008, 14, 399–406. [Google Scholar] [CrossRef]
- Snyder, G.A.; Cirl, C.; Jiang, J.; Chen, K.; Waldhuber, A.; Smith, P.; Römmler, F.; Snyder, N.; Fresquez, T.; Durr, S.; et al. Molecular mechanisms for the subversion of MyD88 signaling by TcpC from virulent uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 2013, 110, 6985–6990. [Google Scholar] [CrossRef]
- Newman, R.M.; Salunkhe, P.; Godzik, A.; Reed, J.C. Identification and characterization of a novel bacterial virulence factor that shares homology with mammalian Toll/interleukin-1 receptor family proteins. Infect. Immun. 2006, 74, 594–601. [Google Scholar] [CrossRef]
- Norenberg, D.; Wieser, A.; Magistro, G.; Hoffmann, C.; Meyer, C.; Messerer, M.; Schubert, S. Molecular analysis of a novel Toll/interleukin-1 receptor (TIR)-domain containing virulence protein of Y. pseudotuberculosis among Far East scarlet-like fever serotype I strains. Int. J. Med. Microbiol. 2013, 303, 583–594. [Google Scholar] [CrossRef] [PubMed]
- Salcedo, S.P.; Marchesini, M.I.; Lelouard, H.; Fugier, E.; Jolly, G.; Balor, S.; Muller, A.; Lapaque, N.; Demaria, O.; Alexopoulou, L.; et al. Brucella control of dendritic cell maturation is dependent on the TIR-containing protein Btp1. PLoS Pathog. 2008, 4, e21. [Google Scholar] [CrossRef]
- Askarian, F.; van Sorge, N.M.; Sangvik, M.; Beasley, F.C.; Henriksen, J.R.; Sollid, J.U.; van Strijp, J.A.; Nizet, V.; Johannessen, M. A Staphylococcus aureus TIR domain protein virulence factor blocks TLR2-mediated NF-kappaB signaling. J. Innate Immun. 2014, 6, 485–498. [Google Scholar] [CrossRef] [PubMed]
- Kraemer, T.D.; Haro, O.D.Q.; Domann, E.; Chakraborty, T.; Tchatalbachev, S. The TIR domain containing locus of Enterococcus faecalis is predominant among urinary tract infection isolates and downregulates host inflammatory response. Int. J. Microbiol. 2014, 2014, 918143. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Baghdayan, A.S.; Payne, S.J.; Shankar, N. A TIR domain protein from E. faecalis attenuates MyD88-mediated signaling and NF-kappaB activation. PLoS ONE 2014, 9, e112010. [Google Scholar] [CrossRef] [PubMed]
- Low, L.Y.; Mukasa, T.; Reed, J.C.; Pascual, J. Characterization of a TIR-like protein from Paracoccus denitrificans. Biochem. Biophys. Res. Commun. 2007, 356, 481–486. [Google Scholar] [CrossRef]
- Chan, S.L.; Low, L.Y.; Hsu, S.; Li, S.; Liu, T.; Santelli, E.; le Negrate, G.; Reed, J.C.; Woods, V.L., Jr.; Pascual, J. Molecular mimicry in innate immunity: Crystal structure of a bacterial TIR domain. J. Biol. Chem. 2009, 284, 21386–21392. [Google Scholar] [CrossRef]
- Waldhuber, A.; Snyder, G.A.; Rommler, F.; Cirl, C.; Muller, T.; Xiao, T.S.; Svanborg, C.; Miethke, T. A comparative analysis of the mechanism of toll-like receptor-disruption by tir-containing protein C from uropathogenic Escherichia coli. Pathogens 2016, 5, 25. [Google Scholar] [CrossRef] [PubMed]
- Salcedo, S.P.; Marchesini, M.I.; Degos, C.; Terwagne, M.; von Bargen, K.; Lepidi, H.; Herrmann, C.K.; Lacerda, T.L.S.; Imbert, P.R.; Pierre, P.; et al. BtpB, a novel Brucella TIR-containing effector protein with immune modulatory functions. Front. Cell. Infect. Microbiol. 2013, 3, 28. [Google Scholar] [CrossRef] [PubMed]
- Waldhuber, A.; Puthia, M.; Wieser, A.; Cirl, C.; Durr, S.; Neumann-Pfeifer, S.; Albrecht, S.; Rommler, F.; Muller, T.; Zheng, Y.; et al. Uropathogenic Escherichia coli strain CFT073 disrupts NLRP3 inflammasome activation. J. Clin. Investig. 2016, 126, 2425–2436. [Google Scholar] [CrossRef] [PubMed]
- Hering, N.A.; Richter, J.F.; Fromm, A.; Wieser, A.; Hartmann, S.; Gunzel, D.; Bucker, R.; Fromm, M.; Schulzke, J.D.; Troeger, H. TcpC protein from E. coli Nissle improves epithelial barrier function involving PKCzeta and ERK1/2 signaling in HT-29/B6 cells. Mucosal Immunol. 2014, 7, 369–378. [Google Scholar] [CrossRef]
- Alvarez, C.S.; Badia, J.; Bosch, M.; Gimenez, R.; Baldoma, L. Outer membrane vesicles and soluble factors released by probiotic Escherichia coli nissle 1917 and commensal ECOR63 enhance barrier function by regulating expression of tight junction proteins in intestinal epithelial cells. Front. Microbiol. 2016, 7, 1981. [Google Scholar] [CrossRef]
- Schubert, S.; Norenberg, D.; Clermont, O.; Magistro, G.; Wieser, A.; Romann, E.; Hoffmann, C.; Weinert, K.; Denamur, E. Prevalence and phylogenetic history of the TcpC virulence determinant in Escherichia coli. Int. J. Med. Microbiol. 2010, 300, 429–434. [Google Scholar] [CrossRef] [PubMed]
- Snyder, G.A.; Deredge, D.; Waldhuber, A.; Fresquez, T.; Wilkins, D.Z.; Smith, P.T.; Durr, S.; Cirl, C.; Jiang, J.; Jennings, W.; et al. Crystal structures of the Toll/Interleukin-1 receptor (TIR) domains from the Brucella protein TcpB and host adaptor TIRAP reveal mechanisms of molecular mimicry. J. Biol. Chem. 2014, 289, 669–679. [Google Scholar] [CrossRef]
- Pasi, S.; Kant, R.; Surolia, A. Toll/interleukin-1 receptor domain derived from TcpC (TIR-TcpC) ameliorates experimental autoimmune arthritis by down-modulating Th17 cell response. J. Biol. Chem. 2016, 291, 12358–12369. [Google Scholar] [CrossRef]
- Essuman, K.; Summers, D.W.; Sasaki, Y.; Mao, X.; Yim, A.K.Y.; di Antonio, A.; Milbrandt, J. TIR domain proteins are an ancient family of NAD(+)-consuming enzymes. Curr. Biol. 2018, 28, 421–430. [Google Scholar] [CrossRef]
- Welch, R.A.; Burland, V.; Plunkett, G., 3rd; Redford, P.; Roesch, P.; Rasko, D.; Buckles, E.L.; Liou, S.R.; Boutin, A.; Hackett, J.; et al. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 2002, 99, 17020–17024. [Google Scholar] [CrossRef]
- Mann, R.; Mediati, D.G.; Duggin, I.G.; Harry, E.J.; Bottomley, A.L. Metabolic adaptations of uropathogenic E. coli in the urinary tract. Front. Cell. Infect. Microbiol. 2017, 7, 241. [Google Scholar] [CrossRef] [PubMed]
- Fontenot, C.R.; Tasnim, H.; Valdes, K.A.; Popescu, C.V.; Ding, H. Ferric uptake regulator (Fur) reversibly binds a [2Fe-2S] cluster to sense intracellular iron homeostasis in Escherichia coli. J. Biol. Chem. 2020, 295, 15454–15463. [Google Scholar] [CrossRef]
- Yadav, M.; Zhang, J.; Fischer, H.; Huang, W.; Lutay, N.; Cirl, C.; Lum, J.; Miethke, T.; Svanborg, C. Inhibition of TIR domain signaling by TcpC: MyD88-dependent and independent effects on Escherichia coli virulence. PLoS Pathog. 2010, 6, e1001120. [Google Scholar] [CrossRef] [PubMed]
- Cassiano, M.H.A.; Silva-Rocha, R. Benchmarking bacterial promoter prediction tools: Potentialities and limitations. mSystems 2020, 5. [Google Scholar] [CrossRef] [PubMed]
- Mendoza-Vargas, A.; Olvera, L.; Olvera, M.; Grande, R.; Vega-Alvarado, L.; Taboada, B.; Jimenez-Jacinto, V.; Salgado, H.; Juarez, K.; Contreras-Moreira, B.; et al. Genome-wide identification of transcription start sites, promoters and transcription factor binding sites in E. coli. PLoS ONE 2009, 4, e7526. [Google Scholar] [CrossRef]
- Coronas-Serna, J.M.; Louche, A.; Rodriguez-Escudero, M.; Roussin, M.; Imbert, P.R.C.; Rodriguez-Escudero, I.; Terradot, L.; Molina, M.; Gorvel, J.P.; Cid, V.J.; et al. The TIR-domain containing effectors BtpA and BtpB from Brucella abortus impact NAD metabolism. PLoS Pathog. 2020, 16, e1007979. [Google Scholar] [CrossRef] [PubMed]
- Sriskanda, V.; Shuman, S. Conserved residues in domain Ia are required for the reaction of Escherichia coli DNA ligase with NAD+. J. Biol. Chem. 2002, 277, 9695–9700. [Google Scholar] [CrossRef]
- Justice, S.S.; Hunstad, D.A.; Seed, P.C.; Hultgren, S.J. Filamentation by Escherichia coli subverts innate defenses during urinary tract infection. Proc. Natl. Acad. Sci. USA 2006, 103, 19884–19889. [Google Scholar] [CrossRef]
- Justice, S.S.; Hunstad, D.A.; Cegelski, L.; Hultgren, S.J. Morphological plasticity as a bacterial survival strategy. Nat. Rev. Microbiol. 2008, 6, 162–168. [Google Scholar] [CrossRef]
- Justice, S.S.; Hung, C.; Theriot, J.A.; Fletcher, D.A.; Anderson, G.G.; Footer, M.J.; Hultgren, S.J. Differentiation and developmental pathways of uropathogenic Escherichia coli in urinary tract pathogenesis. Proc. Natl. Acad. Sci. USA 2004, 101, 1333–1338. [Google Scholar] [CrossRef]
- Andersen, T.E.; Khandige, S.; Madelung, M.; Brewer, J.; Kolmos, H.J.; Moller-Jensen, J. Escherichia coli uropathogenesis in vitro: Invasion, cellular escape, and secondary infection analyzed in a human bladder cell infection model. Infect. Immun. 2012, 80, 1858–1867. [Google Scholar] [CrossRef]
- Rosen, D.A.; Hooton, T.M.; Stamm, W.E.; Humphrey, P.A.; Hultgren, S.J. Detection of intracellular bacterial communities in human urinary tract infection. PLoS Med. 2007, 4, e329. [Google Scholar] [CrossRef]
- Meyer, C.; Hoffmann, C.; Haas, R.; Schubert, S. The role of the galU gene of uropathogenic Escherichia coli in modulating macrophage TNF-alpha response. Int. J. Med. Microbiol. 2015, 305, 893–901. [Google Scholar] [CrossRef]
- Hagan, E.C.; Lloyd, A.L.; Rasko, D.A.; Faerber, G.J.; Mobley, H.L. Escherichia coli global gene expression in urine from women with urinary tract infection. PLoS Pathog. 2010, 6, e1001187. [Google Scholar] [CrossRef]
- Subashchandrabose, S.; Hazen, T.H.; Brumbaugh, A.R.; Himpsl, S.D.; Smith, S.N.; Ernst, R.D.; Rasko, D.A.; Mobley, H.L. Host-specific induction of Escherichia coli fitness genes during human urinary tract infection. Proc. Natl. Acad. Sci. USA 2014, 111, 18327–18332. [Google Scholar] [CrossRef] [PubMed]
- Mobley, H.L.; Green, D.M.; Trifillis, A.L.; Johnson, D.E.; Chippendale, G.R.; Lockatell, C.V.; Jones, B.D.; Warren, J.W. Pyelonephritogenic Escherichia coli and killing of cultured human renal proximal tubular epithelial cells: Role of hemolysin in some strains. Infect. Immun. 1990, 58, 1281–1289. [Google Scholar] [CrossRef] [PubMed]
- Datsenko, K.A.; Wanner, B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 2000, 97, 6640–6645. [Google Scholar] [CrossRef]
- Zaslaver, A.; Bren, A.; Ronen, M.; Itzkovitz, S.; Kikoin, I.; Shavit, S.; Liebermeister, W.; Surette, M.G.; Alon, U. A comprehensive library of fluorescent transcriptional reporters for Escherichia coli. Nat. Methods 2006, 3, 623–628. [Google Scholar] [CrossRef] [PubMed]
- Amann, E.; Ochs, B.; Abel, K.J. Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene 1988, 69, 301–315. [Google Scholar] [CrossRef]
- Sommer, E.; Koler, M.; Frank, V.; Sourjik, V.; Vaknin, A. The sensory histidine kinases TorS and EvgS tend to form clusters in Escherichia coli cells. PLoS ONE 2013, 8, e77708. [Google Scholar] [CrossRef] [PubMed]











| Bacterial Host | Construct | Plasmid Back Bone | Resist-Ance | Plasmid Name | Function |
|---|---|---|---|---|---|
| CFT073 | 5′UTR-c2397-c2398 | pACYC-184 | cm | pTcpC [1] | contains a DNA fragment starting at 535 bp 5′ of the start codon of c2397 and ends at the stop codon of c2398. |
| CFT073 | tcpC-full-length (bp 1–924)-Strep-tag | pASK-IBA5plus | amp † | p(Strep-tag tcpC 1-924):AMP | non-leaky, ATc-inducible expression of Strep tag-TcpC |
| CFT073 | tcpC-full-length (bp 1–924) Strep-tag, E244A | pASK-IBA5plus | amp † | p(Strep-tag E244A tcpC 1-924):AMP | non-leaky, ATc-inducible expression of Strep tag-E244A mutated TcpC |
| CFT073 | tcpC-full-length(bp 1–924)-eYFP | pASK-IBA3plus | amp | p(tcpC 1-924):eYFP:Strep-tag:AMP | non-leaky, ATc-inducible expression of TcpC-YFP-Strep-tag |
| CFT073 | Pc2397:gfpmut2 | pUA66 [38] | kan ‡ | pPc2397:gfpmut2:KAN | P1 reporter |
| CFT073 | Pc2398:gfpmut2 | pUA66 | kan | pPc2398:gfpmut2:KAN | P2 reporter |
| CFT073 | Pc2397–Pc2398:gfpmut2 | pUA66 | kan | p(Pc2397-Pc2398):gfpmut2:KAN | P1+P2 reporter |
| CFT073 | - | pTrc99A [39] | amp | pAMP | negative control |
| CFT073 | tcpC-fragment (bp 1–150)-eYFP | pDK112 [40] | amp | p(tcpC 1–150): eYFP:AMP | overexpression of YFP-labeled tcpC bp 1–150 |
| CFT073 | tcpC-fragment (bp 1–507)-eYFP | pDK112 | amp | p(tcpC 1–507): eYFP:AMP | overexpression of YFP-labeled tcpC bp 1–507 |
| CFT073 | tcpC-fragment (bp 127–507)-eYFP | pDK112 | amp | p(tcpC 127–507): eYFP:AMP | overexpression of YFP-labeled tcpC bp 127–507 |
| CFT073 | tcpC-fragment (bp 508–924)-eYFP | pDK112 | amp | p(tcpC 508–924): eYFP:AMP | overexpression of YFP-labeled tcpC bp 508–924 |
| CFT073 | tcpC-fragment (bp 127–924)-eYFP | pDK112 | amp | p(tcpC 127–924): eYFP:AMP | overexpression of YFP-labeled tcpC bp 127–924 |
| CFT073 | tcpC-full-length (bp 1–924)-eYFP | pDK112 | amp | p(tcpC 1–924): eYFP:AMP | overexpression of YFP-labeled tcpC bp 1–924 |
| Designation | Sequence |
|---|---|
| pASK-IBA5 16_tcpC_BsaI_fw | 5′-ATGGTAggtctcAGCGCCGTGATAGCATATGAAA-ACATAG-3′ |
| pASK-IBA5 16_tcpC_BsaI_rev | 5′-ATGGTAggtctcATATCATCTTCTCCTGTATGCT-ATTTC-3′ |
| pASK-IBA3_tcpC_BsaI_fw | 5′-ATGGTAggtctcAAATGGTGATAGCATATGAAAA-CATAG-3′ |
| pASK-IBA3_eYFP_BsaI_rev | 5′-ATGGTAggtctcAGCGCTCTTGTACAGCTCGTCC-ATGCCG-3′ |
| c2397 5’UTR_XhoI_fw | 5′-TACTATctcgagCACCTCTTGCTGTTTATACG-3′ |
| c2397 5’UTR_BamHI_rev | 5′-ATAGTAggatccGCCATTAAAAATATAATCTC-3′ |
| c2398 5’UTR_XhoI_fw | 5′-TACTATctcgagGCAGGAGTCTATGGTAACG-3′ |
| c2398 5’UTR_BamHI_rev | 5′-ATAGTAggatccCATATGCTATCACATTTTGAG-3′ |
| tcpC bp 1_NcoI_fw | 5′-TACTATccatgGTGATAGCATATGAAAACATAG-3′ |
| tcpC bp 150_BamHI_rev | 5′-ATAGTAggatccCTCTTTGGTTTTTAGGTGCTG-3′ |
| tcpC bp 924_BamHI_rev | 5′-ATAGTAggatccTCTTCTCCTGTATGCTATTTCA-G-3′ |
| tcpC bp 508_NcoI_fw | 5′-TACTATccatggACTATGATTTTTTCATATCC-3′ |
| tcpC bp 507_BamHI_rev | 5′-ATAGTAggatccCGTATTATTGTTATCTTGC-3′ |
| tcpC bp 127_NcoI_fw | 5′-TACTATccatggGGAAACAGCACCTAAAAACC-3′ |
| c2397fw | 5‘-ATGGCGATTTTTCATCTG-3‘ |
| c2398fw | 5‘-GTGATAGCATATGAAAACATAG-3‘ |
| c2398rev | 5‘-CTTCTCCTGTATGCTATTTC-3‘ |
| GSP1 | 5‘-CTTTGCCTCAACCTCCTT-3‘ |
| GSP2 | 5‘-CTCCCATCTATAATCGTGGAT-3‘ |
| GSP3 | 5‘-GCTTGGTGAATGTTTTGG-3‘ |
| AAP (Abridged Anchor Primer) | 5‘-GGCCACGCGTCGACTAGTACGGGIIGGGIIGGGIIG-3‘ |
| AUAP (Abridged Universal Amplification Primer) | 5‘-GGCCACGCGTCGACTAGTAC-3‘ |
| Construct | Forward Primer | Reverse Primer |
|---|---|---|
| Strep tag-tcpC-full-length (bp 1–924) | pASK-IBA5 16_tcpC_BsaI_fw | pASK-IBA5 16_tcpC_BsaI_rev |
| tcpC-full-length (bp 1–924)-eYFP | pASK-IBA3_tcpC_BsaI_fw | pASK-IBA3_eYFP_BsaI_rev |
| Pc2397:gfpmut2 | c2397 5’UTR_XhoI_fw | c2397 5’UTR_BamHI_rev |
| Pc2398:gfpmut2 | c2398 5’UTR_XhoI_fw | c2398 5’UTR_BamHI_rev |
| Pc2397–Pc2398:gfpmut2 | c2397 5’UTR_XhoI_fw | c2398 5’UTR_BamHI_rev |
| tcpC-fragment (bp 1–150)-eYFP | tcpC bp 1_NcoI_fw | tcpC bp 150_BamHI_rev |
| tcpC-fragment (bp 1–507)-eYFP | tcpC bp 1_NcoI_fw | tcpC bp 507_BamHI_rev |
| tcpC-fragment (bp 127–507)-eYFP | tcpC bp 127_NcoI_fw | tcpC bp 507_BamHI_rev |
| tcpC-fragment (bp 508–924)-eYFP | tcpC bp 508_NcoI_fw | tcpC bp 924_BamHI_rev |
| tcpC-fragment (bp 127–924)-eYFP | tcpC bp 127_NcoI_fw | tcpC bp 924_BamHI_rev |
| tcpC-full-length (bp 1–924)-eYFP | tcpC bp 1_NcoI_fw | tcpC bp 924_BamHI_rev |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ittensohn, J.; Hemberger, J.; Griffiths, H.; Keller, M.; Albrecht, S.; Miethke, T. Regulation of Expression of the TIR-Containing Protein C Gene of the Uropathogenic Escherichia coli Strain CFT073. Pathogens 2021, 10, 549. https://doi.org/10.3390/pathogens10050549
Ittensohn J, Hemberger J, Griffiths H, Keller M, Albrecht S, Miethke T. Regulation of Expression of the TIR-Containing Protein C Gene of the Uropathogenic Escherichia coli Strain CFT073. Pathogens. 2021; 10(5):549. https://doi.org/10.3390/pathogens10050549
Chicago/Turabian StyleIttensohn, Julia, Jacqueline Hemberger, Hannah Griffiths, Maren Keller, Simone Albrecht, and Thomas Miethke. 2021. "Regulation of Expression of the TIR-Containing Protein C Gene of the Uropathogenic Escherichia coli Strain CFT073" Pathogens 10, no. 5: 549. https://doi.org/10.3390/pathogens10050549
APA StyleIttensohn, J., Hemberger, J., Griffiths, H., Keller, M., Albrecht, S., & Miethke, T. (2021). Regulation of Expression of the TIR-Containing Protein C Gene of the Uropathogenic Escherichia coli Strain CFT073. Pathogens, 10(5), 549. https://doi.org/10.3390/pathogens10050549







