Immunogenicity of Adjuvanted Psoralen-Inactivated SARS-CoV-2 Vaccines and SARS-CoV-2 Spike Protein DNA Vaccines in BALB/c Mice
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Preparation and Inactivation of SARS-CoV-2
2.3. Purification and Characterization of SARS-CoV-2 PsIV
2.4. DNA Vaccines
2.5. Immunogenicity Assessment of SARS-CoV-2 Vaccine Candidates in Mice
2.6. SARS-CoV-2 Microneutralization Assay
2.7. T-Cell Assays
2.7.1. Extraction of Splenocytes from Mouse Spleen
2.7.2. Cytokine ELISPOT
2.7.3. ELISA Endpoint Determination for Total IgG, IgG1 and IgG2a
2.8. Data Analysis
3. Results
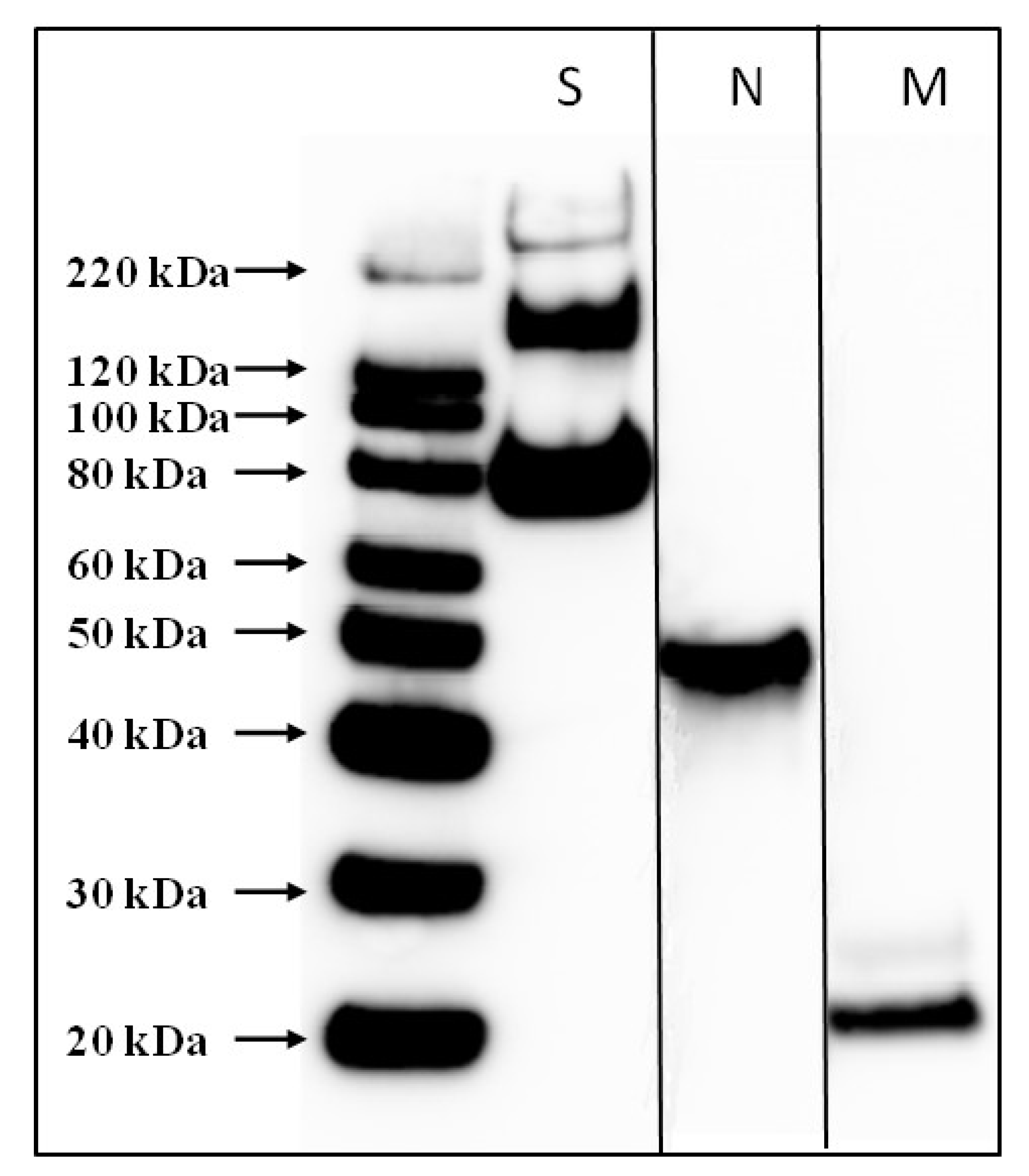
3.1. Preparation and Purification of Inactivated SARS-CoV-2 Vaccine
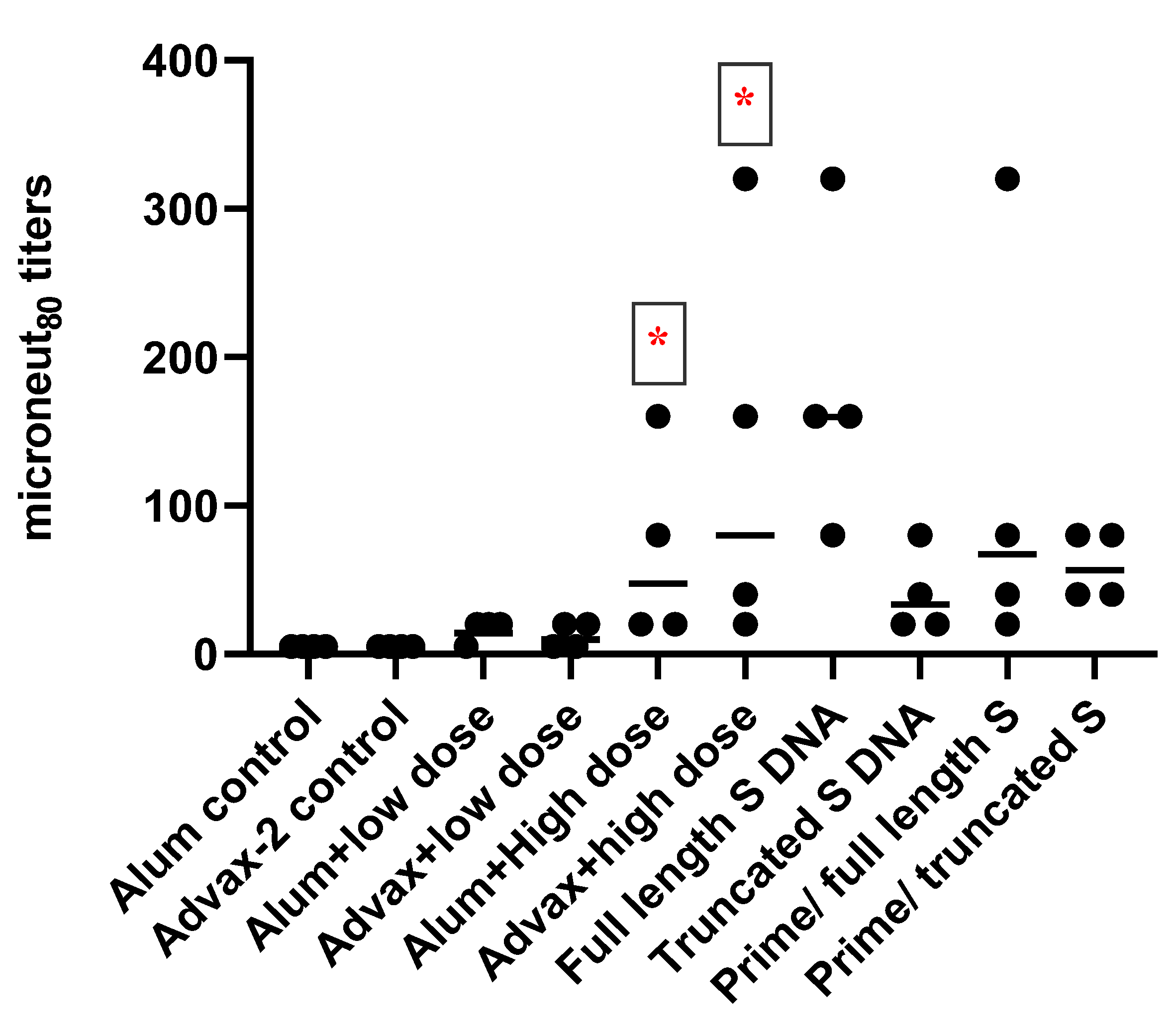
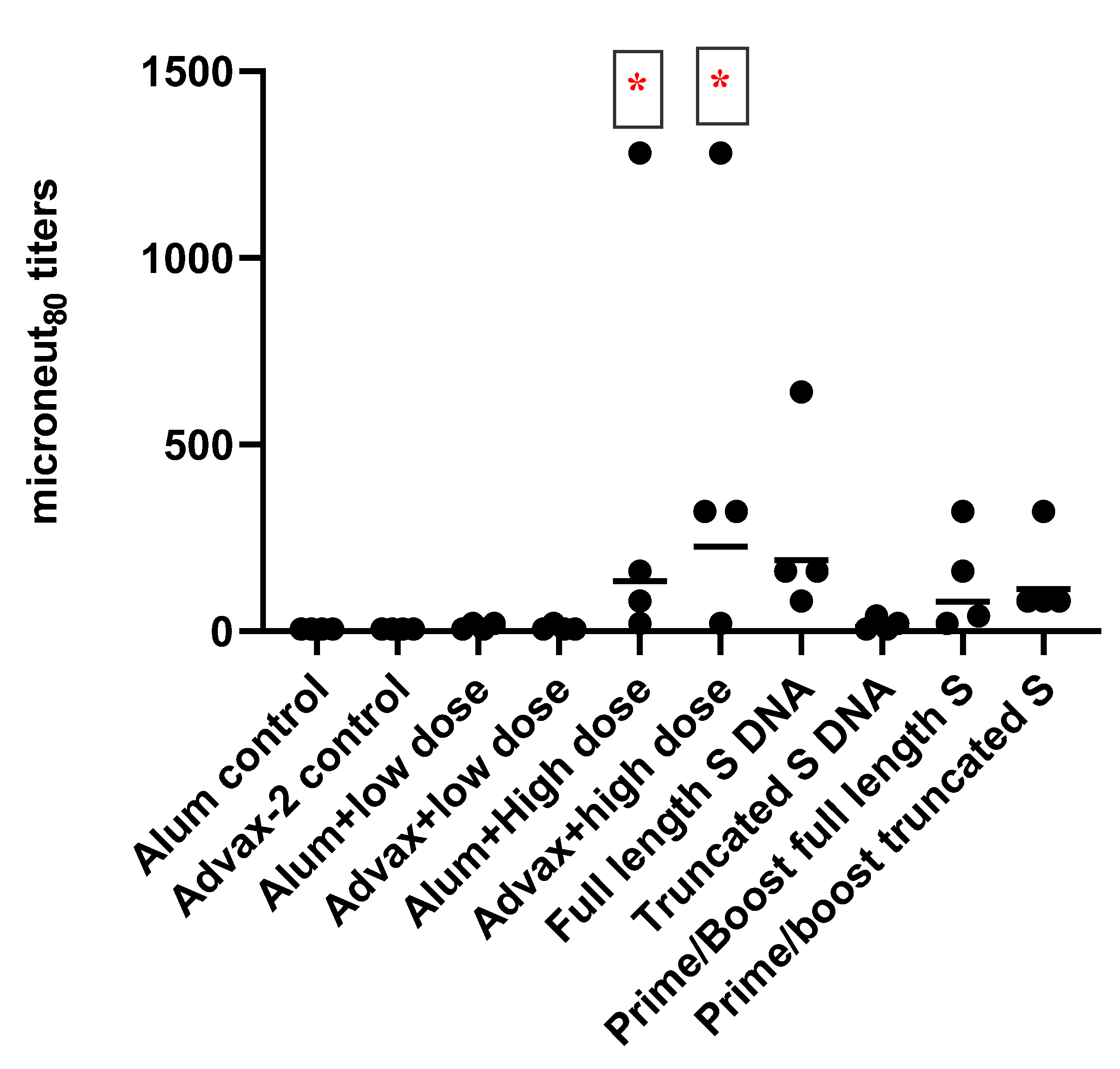
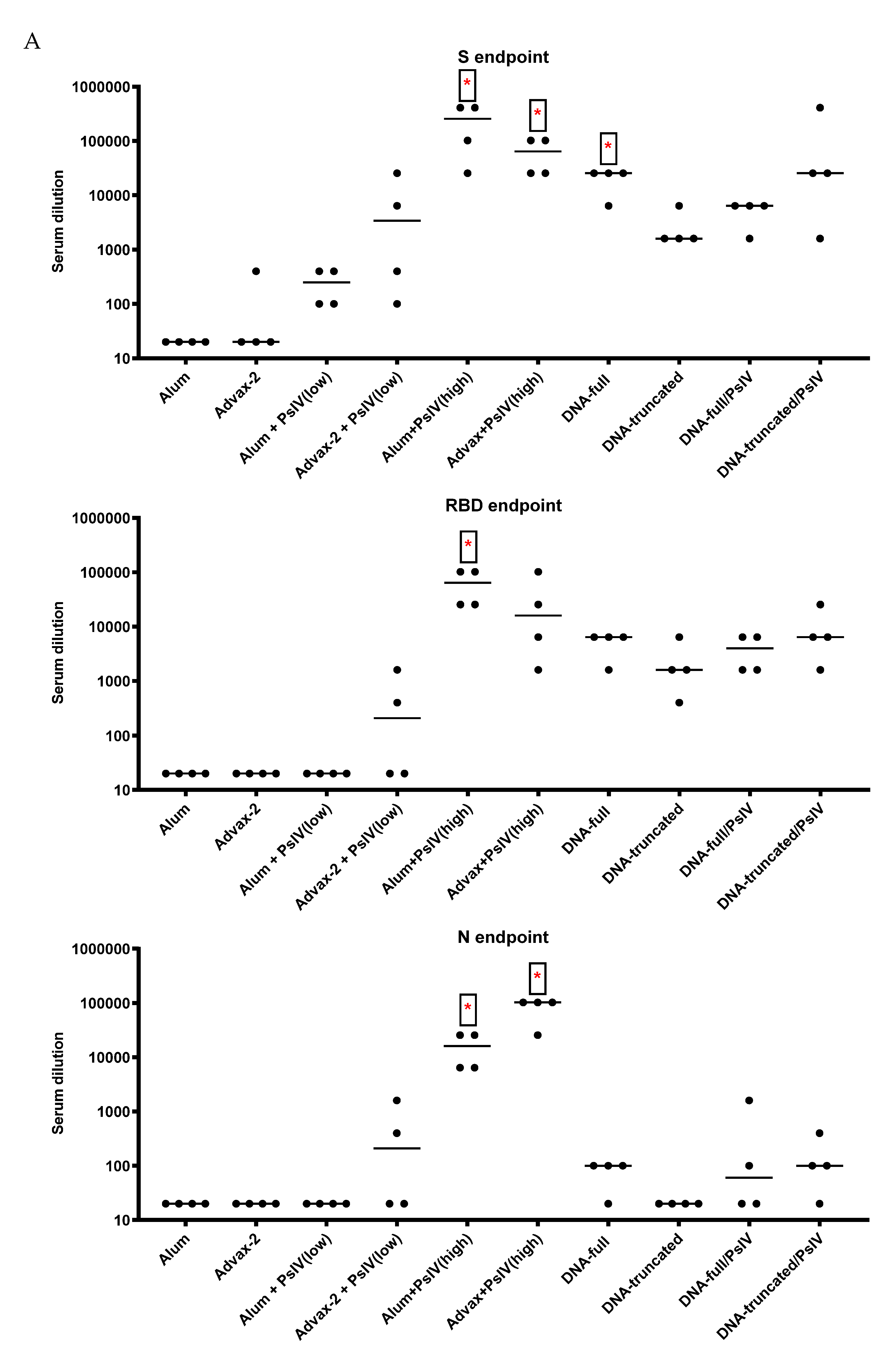
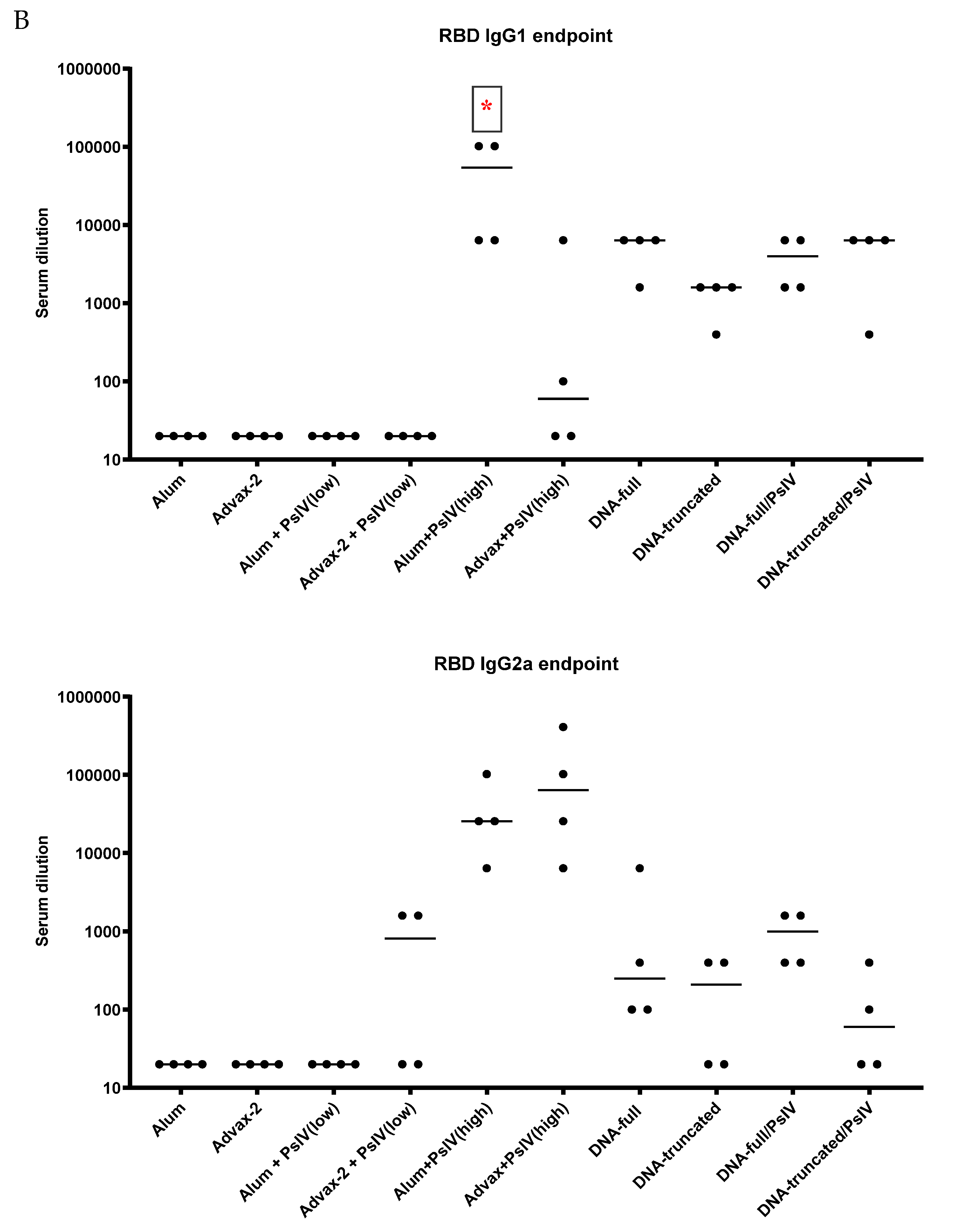
3.2. Antibody Responses to SARS-CoV-2 PsIV in Mice
Total IgG and IgG Subclasses
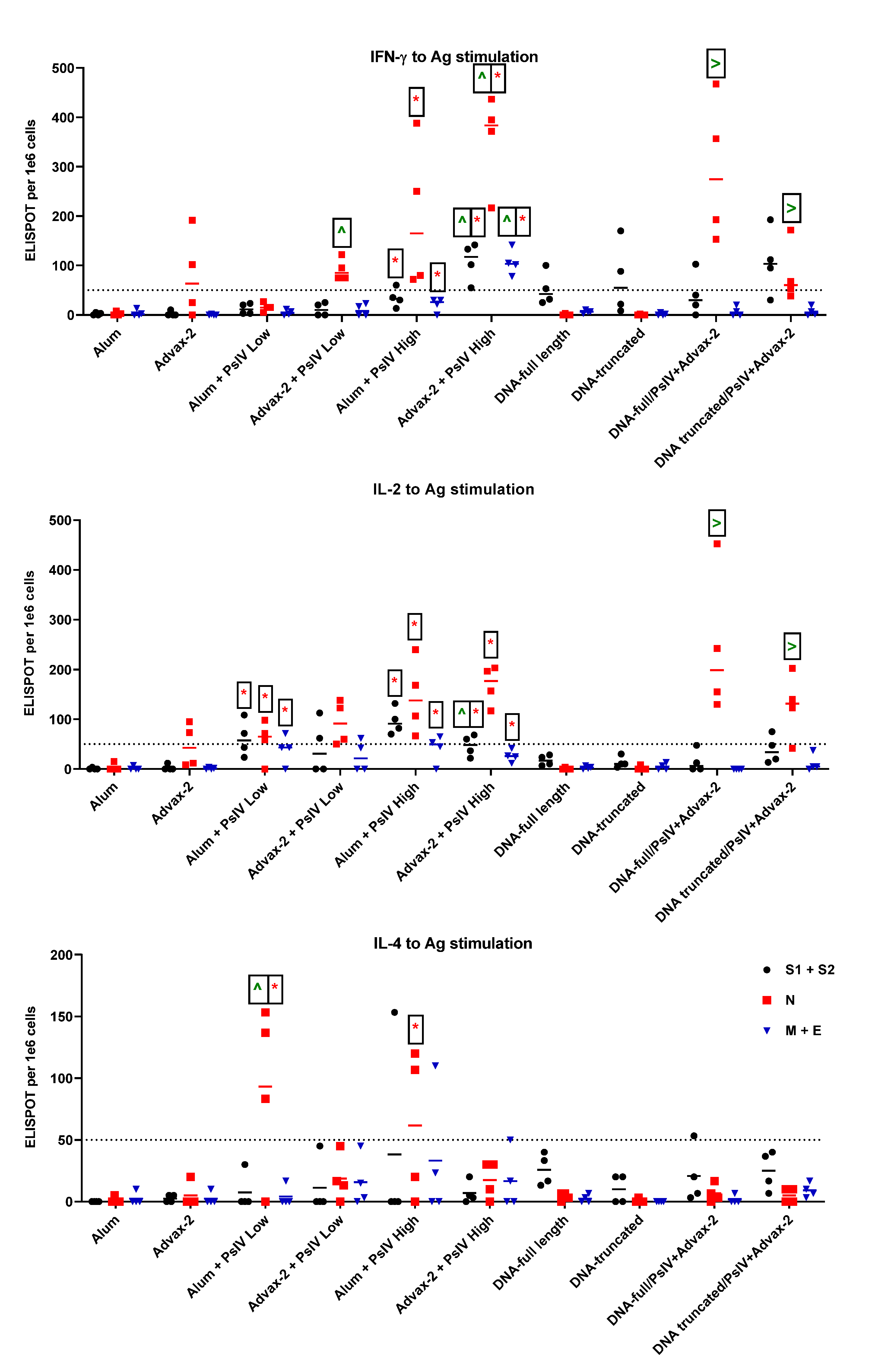
3.3. T-Cell Responses to SARS-CoV-2 PsIV in Mice
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
Disclaimer
References
- Cascella, M.; Rajnik, M.; Cuomo, A.; Dulebohn, S.C.; Di Napoli, R. Features, Evaluation and Treatment Coronavirus (COVID-19); StatPearls: Treasure Island, FL, USA, 2020. [Google Scholar]
- Walls, A.C.; Park, Y.J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020. [Google Scholar] [CrossRef] [PubMed]
- WHO Coronavirus (COVID-19) Dashboard. Available online: https://covid19.who.int/ (accessed on 15 October 2020).
- Zhao, J.; Zhao, S.; Ou, J.; Zhang, J.; Lan, W.; Guan, W.; Wu, X.; Yan, Y.; Zhao, W.; Wu, J.; et al. COVID-19: Coronavirus Vaccine Development Updates. Front. Immunol. 2020, 11, 602256. [Google Scholar] [CrossRef] [PubMed]
- Forni, G.; Mantovani, A.; Covid-19 Commission of Accademia Nazionale dei Lincei, R. COVID-19 vaccines: Where we stand and challenges ahead. Cell Death Differ. 2021, 28, 626–639. [Google Scholar] [CrossRef]
- Chung, J.Y.; Thone, M.N.; Kwon, Y.J. COVID-19 vaccines: The status and perspectives in delivery points of view. Adv. Drug Deliv. Rev. 2021, 170, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhang, Y.; Huang, B.; Deng, W.; Quan, Y.; Wang, W.; Xu, W.; Zhao, Y.; Li, N.; Zhang, J.; et al. Development of an Inactivated Vaccine Candidate, BBIBP-CorV, with Potent Protection against SARS-CoV-2. Cell 2020, 182, 713–721.e9. [Google Scholar] [CrossRef]
- Gao, Q.; Bao, L.; Mao, H.; Wang, L.; Xu, K.; Yang, M.; Li, Y.; Zhu, L.; Wang, N.; Lv, Z.; et al. Development of an inactivated vaccine candidate for SARS-CoV-2. Science 2020, 369, 77–81. [Google Scholar] [CrossRef] [PubMed]
- Spaninger, E.; Bren, U. Carcinogenesis of beta-Propiolactone: A Computational Study. Chem. Res. Toxicol. 2020, 33, 769–781. [Google Scholar] [CrossRef]
- Bhattacharyya, A.; Gupta, B.; Singh, A.; Sah, K.; Gupta, V. Probing natural substitute for formalin: Comparing honey, sugar, and jaggery syrup as fixatives. Natl. J. Maxillofac. Surg. 2018, 9, 14–21. [Google Scholar] [CrossRef]
- Raviprakash, K.; Sun, P.; Raviv, Y.; Luke, T.; Martin, N.; Kochel, T. Dengue virus photo-inactivated in presence of 1,5-iodonaphthylazide (INA) or AMT, a psoralen compound (4’-aminomethyl-trioxsalen) is highly immunogenic in mice. Hum. Vaccines Immunother. 2013, 9, 2336–2341. [Google Scholar] [CrossRef]
- Sundaram, A.K.; Ewing, D.; Blevins, M.; Liang, Z.; Sink, S.; Lassan, J.; Raviprakash, K.; Defang, G.; Williams, M.; Porter, K.R.; et al. Comparison of purified psoralen-inactivated and formalin-inactivated dengue vaccines in mice and nonhuman primates. Vaccine 2020, 38, 3313–3320. [Google Scholar] [CrossRef]
- Wollowitz, S. Fundamentals of the psoralen-based Helinx technology for inactivation of infectious pathogens and leukocytes in platelets and plasma. Semin. Hematol. 2001, 38, 4–11. [Google Scholar] [CrossRef]
- Hanson, C.V.; Riggs, J.L.; Lennette, E.H. Photochemical inactivation of DNA and RNA viruses by psoralen derivatives. J. Gen. Virol. 1978, 40, 345–358. [Google Scholar] [CrossRef]
- Hanson, C.V. Photochemical inactivation of viruses with psoralens: An overview. Blood Cells 1992, 18, 7–25. [Google Scholar] [PubMed]
- Raviprakash, K.; Porter, K.R.; Kochel, T.J.; Ewing, D.; Simmons, M.; Phillips, I.; Murphy, G.S.; Weiss, W.R.; Hayes, C.G. Dengue virus type 1 DNA vaccine induces protective immune responses in rhesus macaques. J. Gen. Virol. 2000, 81, 1659–1667. [Google Scholar] [CrossRef] [PubMed]
- Raviprakash, K.; Kochel, T.J.; Ewing, D.; Simmons, M.; Phillips, I.; Hayes, C.G.; Porter, K.R. Immunogenicity of dengue virus type 1 DNA vaccines expressing truncated and full length envelope protein. Vaccine 2000, 18, 2426–2434. [Google Scholar] [CrossRef]
- Frey, A.; Di Canzio, J.; Zurakowski, D. A statistically defined endpoint titer determination method for immunoassays. J. Immunol. Methods 1998, 221, 35–41. [Google Scholar] [CrossRef]
- Hayashi, M.; Aoshi, T.; Haseda, Y.; Kobiyama, K.; Wijaya, E.; Nakatsu, N.; Igarashi, Y.; Standley, D.M.; Yamada, H.; Honda-Okubo, Y.; et al. Advax, a Delta Inulin Microparticle, Potentiates In-built Adjuvant Property of Co-administered Vaccines. EBioMedicine 2017, 15, 127–136. [Google Scholar] [CrossRef] [PubMed]
- Nolan, T.; Richmond, P.C.; Formica, N.T.; Hoschler, K.; Skeljo, M.V.; Stoney, T.; McVernon, J.; Hartel, G.; Sawlwin, D.C.; Bennet, J.; et al. Safety and immunogenicity of a prototype adjuvanted inactivated split-virus influenza A (H5N1) vaccine in infants and children. Vaccine 2008, 26, 6383–6391. [Google Scholar] [CrossRef] [PubMed]
- Petrovsky, N.; Larena, M.; Siddharthan, V.; Prow, N.A.; Hall, R.A.; Lobigs, M.; Morrey, J. An inactivated cell culture Japanese encephalitis vaccine (JE-ADVAX) formulated with delta inulin adjuvant provides robust heterologous protection against West Nile encephalitis via cross-protective memory B cells and neutralizing antibody. J. Virol. 2013, 87, 10324–10333. [Google Scholar] [CrossRef]
- Gordon, D.L.; Sajkov, D.; Honda-Okubo, Y.; Wilks, S.H.; Aban, M.; Barr, I.G.; Petrovsky, N. Human Phase 1 trial of low-dose inactivated seasonal influenza vaccine formulated with Advax delta inulin adjuvant. Vaccine 2016, 34, 3780–3786. [Google Scholar] [CrossRef]
- Wong, T.M.; Petrovsky, N.; Bissel, S.J.; Wiley, C.A.; Ross, T.M. Delta inulin-derived adjuvants that elicit Th1 phenotype following vaccination reduces respiratory syncytial virus lung titers without a reduction in lung immunopathology. Hum. Vaccines Immunother. 2016, 12, 2096–2105. [Google Scholar] [CrossRef] [PubMed]
- Xia, S.; Zhang, Y.; Wang, Y.; Wang, H.; Yang, Y.; Gao, G.F.; Tan, W.; Wu, G.; Xu, M.; Lou, Z.; et al. Safety and immunogenicity of an inactivated SARS-CoV-2 vaccine, BBIBP-CorV: A randomised, double-blind, placebo-controlled, phase 1/2 trial. Lancet Infect. Dis. 2020. [Google Scholar] [CrossRef]
- Williams, M.; Ewing, D.; Blevins, M.; Sun, P.; Sundaram, A.K.; Raviprakash, K.S.; Porter, K.R.; Sanders, J.W. Enhanced immunogenicity and protective efficacy of a tetravalent dengue DNA vaccine using electroporation and intradermal delivery. Vaccine 2019, 37, 4444–4453. [Google Scholar] [CrossRef] [PubMed]
- Grifoni, A.; Weiskopf, D.; Ramirez, S.I.; Mateus, J.; Dan, J.M.; Moderbacher, C.R.; Rawlings, S.A.; Sutherland, A.; Premkumar, L.; Jadi, R.S.; et al. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell 2020, 181, 1489–1501. [Google Scholar] [CrossRef] [PubMed]

| Groups | Adjuvant and Vaccine | No. of Animals | Dose |
|---|---|---|---|
| 1 | Alum and PBS | 4 | N/A |
| 2 | Advax-2 and PBS | 4 | N/A |
| 3 | Alum and SARS-CoV-2 PsIV (low dose) | 4 | 105 particles of SARS-Cov-2 PsIV |
| 4 | Advax-2 and SARS-CoV-2 PsIV (low dose) | 4 | 105 particles of SARS-Cov-2 PsIV |
| 5 | Alum and SARS-CoV-2 PsIV (high dose) | 4 | 107 particles of SARS-Cov-2 PsIV |
| 6 | Advax-2 and SARS-CoV-2 PsIV (high dose) | 4 | 107 particles of SARS-Cov-2 PsIV |
| 7 | DNA vaccine encoding full-length SARS-CoV-2 spike protein | 4 | 50 µg of DNA |
| 8 | DNA vaccine encoding truncated SARS-CoV-2 spike protein | 4 | 50 µg of DNA |
| 9 | Prime/Boost (2 doses of DNA; full-length S protein and PsIV) | 4 | 50 µg of DNA (2 doses) and 107 particles of SARS-Cov-2 PsIV |
| 10 | Prime/Boost (2 doses of DNA; truncated S protein and PsIV) | 4 | 50 µg of DNA (2 doses) 107 particles of SARS-Cov-2 PsIV |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sundaram, A.K.; Ewing, D.; Liang, Z.; Jani, V.; Cheng, Y.; Sun, P.; Raviprakash, K.; Wu, S.-J.; Petrovsky, N.; Defang, G.; et al. Immunogenicity of Adjuvanted Psoralen-Inactivated SARS-CoV-2 Vaccines and SARS-CoV-2 Spike Protein DNA Vaccines in BALB/c Mice. Pathogens 2021, 10, 626. https://doi.org/10.3390/pathogens10050626
Sundaram AK, Ewing D, Liang Z, Jani V, Cheng Y, Sun P, Raviprakash K, Wu S-J, Petrovsky N, Defang G, et al. Immunogenicity of Adjuvanted Psoralen-Inactivated SARS-CoV-2 Vaccines and SARS-CoV-2 Spike Protein DNA Vaccines in BALB/c Mice. Pathogens. 2021; 10(5):626. https://doi.org/10.3390/pathogens10050626
Chicago/Turabian StyleSundaram, Appavu K., Daniel Ewing, Zhaodong Liang, Vihasi Jani, Ying Cheng, Peifang Sun, Kanakatte Raviprakash, Shuenn-Jue Wu, Nikolai Petrovsky, Gabriel Defang, and et al. 2021. "Immunogenicity of Adjuvanted Psoralen-Inactivated SARS-CoV-2 Vaccines and SARS-CoV-2 Spike Protein DNA Vaccines in BALB/c Mice" Pathogens 10, no. 5: 626. https://doi.org/10.3390/pathogens10050626
APA StyleSundaram, A. K., Ewing, D., Liang, Z., Jani, V., Cheng, Y., Sun, P., Raviprakash, K., Wu, S.-J., Petrovsky, N., Defang, G., Williams, M., & Porter, K. R. (2021). Immunogenicity of Adjuvanted Psoralen-Inactivated SARS-CoV-2 Vaccines and SARS-CoV-2 Spike Protein DNA Vaccines in BALB/c Mice. Pathogens, 10(5), 626. https://doi.org/10.3390/pathogens10050626






