Productive Infection of Human Breast Cancer Cell Lines with Human Cytomegalovirus (HCMV)
Abstract
:1. Introduction
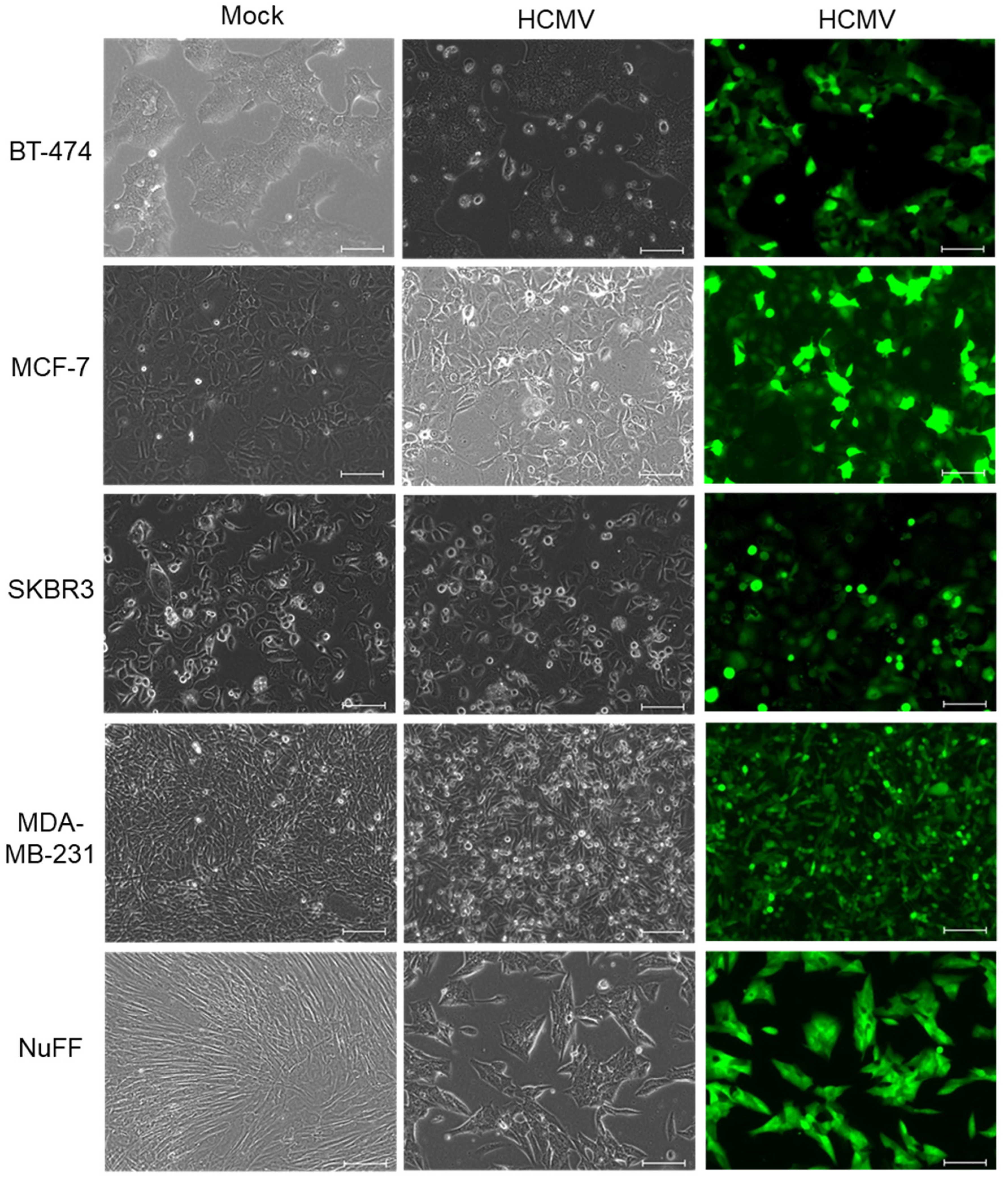
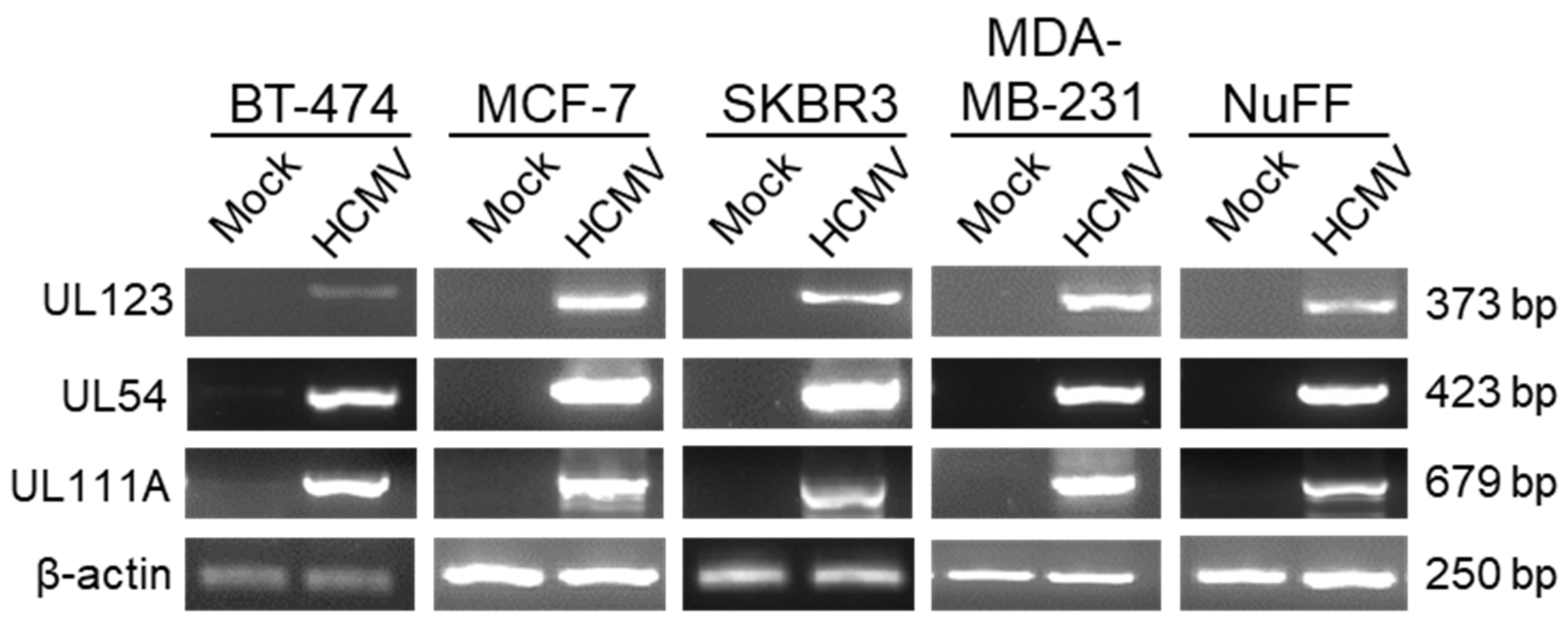
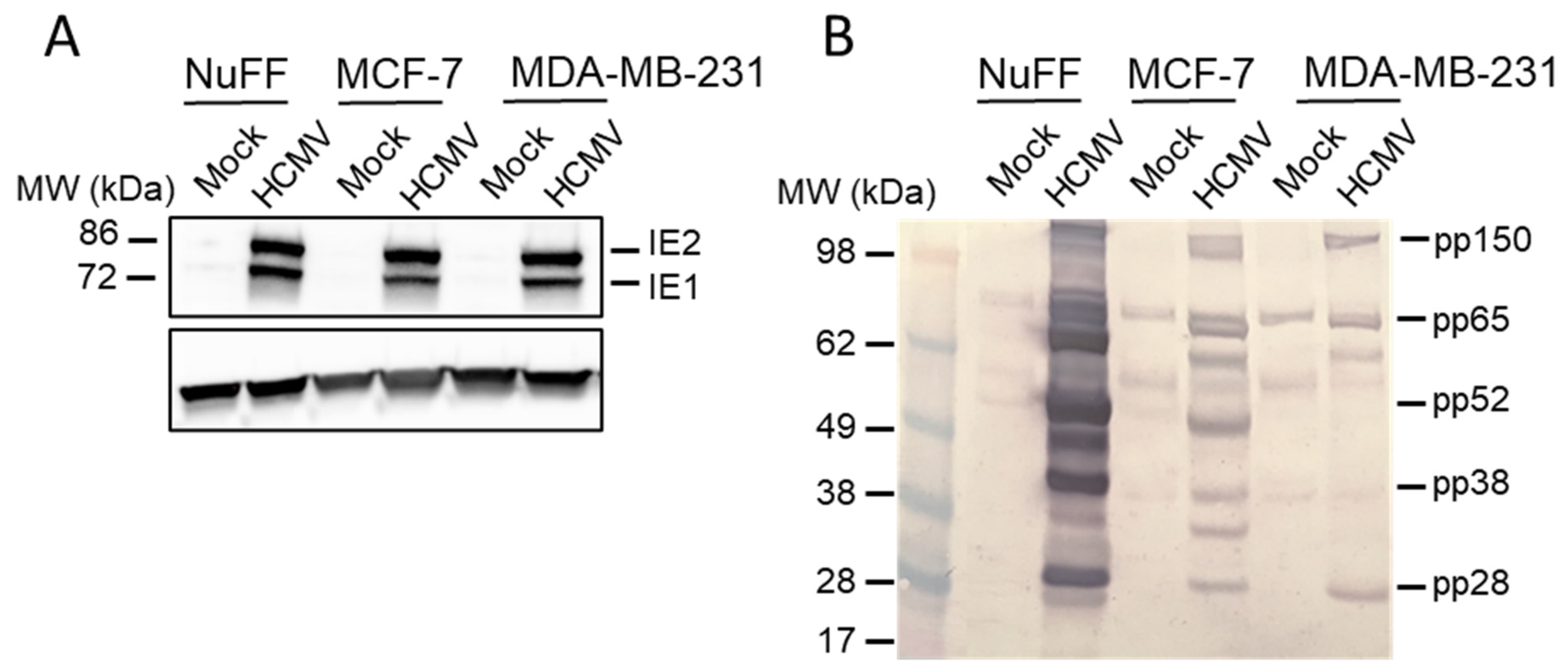
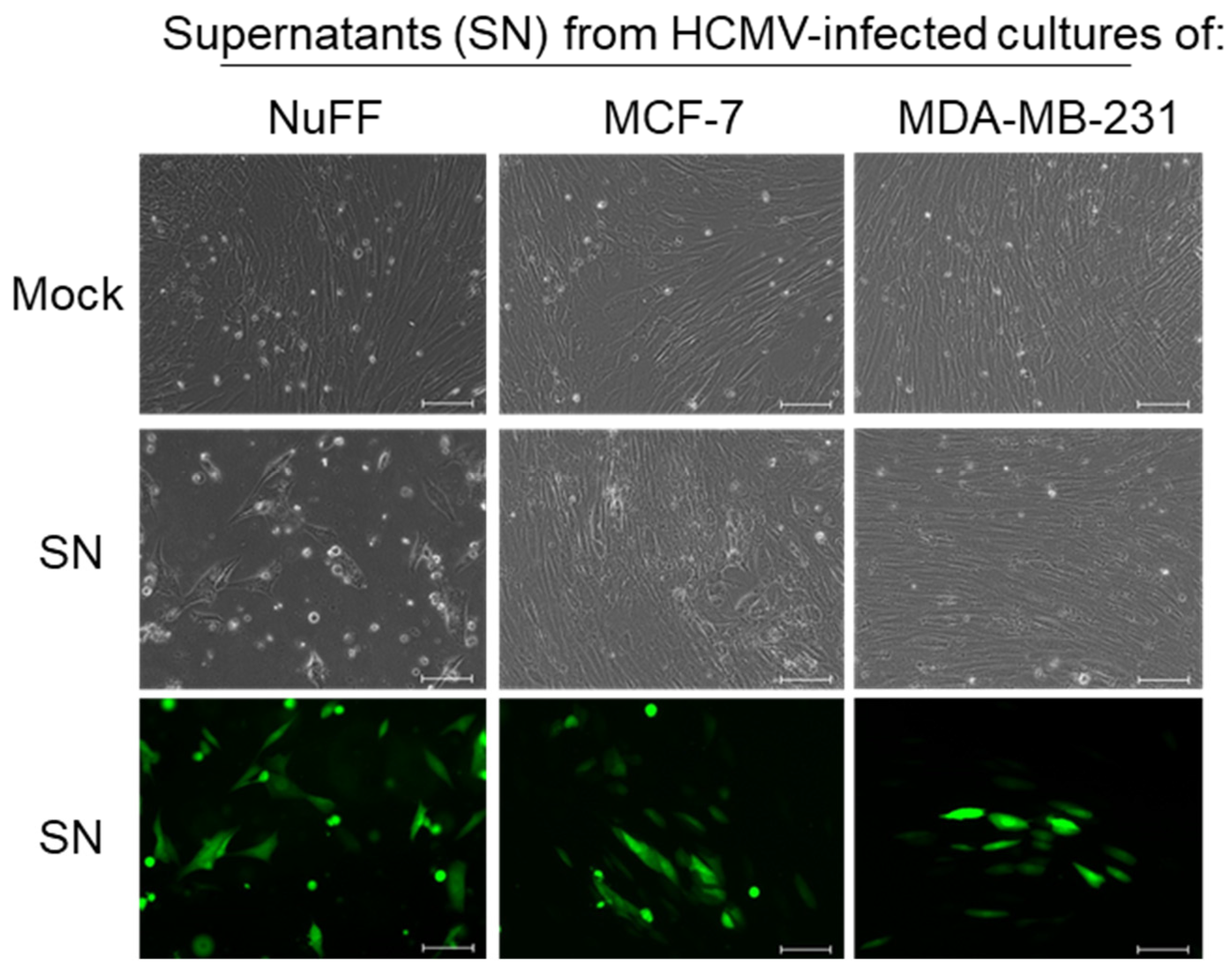
2. Results
3. Discussion
4. Materials and Methods
4.1. Cells and Viruses
4.2. Reverse Transcription Polymerase Chain Reaction (RT-PCR)
4.3. Immunoblotting
4.4. Virus Infectivity Assay
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alibek, K.; Kakpenova, A.; Mussabekova, A.; Sypabekova, M.; Karatayeva, N. Role of viruses in the development of breast cancer. Infect. Agents Cancer 2013, 8, 32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- White, M.K.; Pagano, J.S.; Khalili, K. Viruses and Human Cancers: A Long Road of Discovery of Molecular Paradigms. Clin. Microbiol. Rev. 2014, 27, 463–481. [Google Scholar] [CrossRef] [Green Version]
- Richardson, A. Is breast cancer caused by late exposure to a common virus? Med. Hypotheses 1997, 48, 491–497. [Google Scholar] [CrossRef]
- Buehring, G.C.; Shen, H.M.; Jensen, H.M.; Choi, K.Y.; Sun, D.; Nuovo, G. Bovine Leukemia Virus DNA in Human Breast Tissue. Emerg. Infect. Dis. 2014, 20, 772–782. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buehring, G.C.; Shen, H.M.; Jensen, H.M.; Jin, D.L.; Hudes, M.; Block, G. Exposure to Bovine Leukemia Virus Is Associated with Breast Cancer: A Case-Control Study. PLoS ONE 2015, 10, e0134304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melana, S.M.; Nepomnaschy, I.; Hasa, J.; Djougarian, A.; Djougarian, A.; Holland, J.F.; Pogo, B.G. Detection of human mammary tumor virus proteins in human breast cancer cells. J. Virol. Methods 2010, 163, 157–161. [Google Scholar] [CrossRef] [PubMed]
- Tsai, J.-H.; Hsu, C.-S.; Tsai, C.-H.; Su, J.-M.; Liu, Y.-T.; Cheng, M.-H.; Wei, J.C.-C.; Chen, F.-L.; Yang, C.-C. Relationship between viral factors, axillary lymph node status and survival in breast cancer. J. Cancer Res. Clin. Oncol. 2007, 133, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Joshi, D.; Quadri, M.; Gangane, N.; Joshi, R.; Gangane, N. Association of Epstein Barr Virus Infection (EBV) with Breast Cancer in Rural Indian Women. PLoS ONE 2009, 4, e8180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fawzy, S.; Sallam, M.; Awad, N.M. Detection of Epstein–Barr virus in breast carcinoma in Egyptian women. Clin. Biochem. 2008, 41, 486–492. [Google Scholar] [CrossRef]
- Hachana, M.; Amara, K.; Ziadi, S.; Romdhane, E.; Gacem, R.B.; Trimeche, M. Investigation of Epstein-Barr virus in breast carcinomas in Tunisia. Pathol. Res. Pract. 2011, 207, 695–700. [Google Scholar] [CrossRef] [PubMed]
- Harkins, L.E.; Matlaf, L.A.; Soroceanu, L.; Klemm, K.; Britt, W.J.; Wang, W.; Bland, K.I.; Cobbs, C.S. Detection of human cytomegalovirus in normal and neoplastic breast epithelium. Herpesviridae 2010, 1, 8. [Google Scholar] [CrossRef] [Green Version]
- Taher, C.; De Boniface, J.; Mohammad, A.-A.; Religa, P.; Hartman, J.; Yaiw, K.-C.; Frisell, J.; Rahbar, A.; Söderberg-Naucler, C. High Prevalence of Human Cytomegalovirus Proteins and Nucleic Acids in Primary Breast Cancer and Metastatic Sentinel Lymph Nodes. PLoS ONE 2013, 8, e56795. [Google Scholar] [CrossRef]
- Herbein, G.; Kumar, A. The oncogenic potential of human cytomegalovirus and breast cancer. Front. Oncol. 2014, 4, 230. [Google Scholar] [CrossRef] [PubMed]
- Taher, C.; Frisk, G.; Fuentes, S.; Religa, P.; Costa, H.; Assinger, A.; Vetvik, K.K.; Bukholm, I.R.; Yaiw, K.C.; Smedby, K.E.; et al. High prevalence of human cytomegalovirus in brain metastases of patients with primary breast and colorectal cancers. Transl. Oncol. 2014, 7, 732–740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richardson, A.K.; Currie, M.J.; Robinson, B.A.; Morrin, H.; Phung, Y.; Pearson, J.F.; Anderson, T.P.; Potter, J.D.; Walker, L.C. Cytomegalovirus and Epstein-Barr virus in breast cancer. PLoS ONE 2015, 10, e0118989. [Google Scholar] [CrossRef] [Green Version]
- Richardson, A.K.; Cox, B.; McCredie, M.R.; Dite, G.S.; Chang, J.H.; Gertig, D.M.; Southey, M.C.; Giles, G.G.; Hopper, J.L. Cytomegalovirus, Epstein-Barr virus and risk of breast cancer before age 40 years: A case-control study. Br. J. Cancer 2004, 90, 2149–2152. [Google Scholar] [CrossRef]
- Yang, Z.; Tang, X.; Meng, G.; Benesch, M.G.K.; Mackova, M.; Belon, A.P.; Serrano-Lomelin, J.; Goping, I.S.; Brindley, D.N.; Hemmings, D.G. Latent Cytomegalovirus Infection in Female Mice Increases Breast Cancer Metastasis. Cancers 2019, 11, 447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohamed, H.T.; El-Shinawi, M.; Nouh, M.A.; Bashtar, A.-R.; Elsayed, E.T.; Schneider, R.J.; Mohamed, M.M. Inflammatory Breast Cancer: High Incidence of Detection of Mixed Human Cytomegalovirus Genotypes Associated with Disease Pathogenesis. Front. Oncol. 2014, 4, 246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soroceanu, L.; Cobbs, C.S. Is HCMV a tumor promoter? Virus Res. 2011, 157, 193–203. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michaelis, M.; Doerr, H.W.; Cinatl, J. The story of human cytomegalovirus and cancer: Increasing evidence and open questions. Neoplasia 2009, 11, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Sanchez, V.; Spector, D.H. Subversion of Cell Cycle Regulatory Pathways. Curr. Topics Microbiol. Immunol. 2008, 325, 243–262. [Google Scholar] [CrossRef]
- Asanuma, H.; Numazaki, K.; Nagata, N.; Hotsubo, T.; Horino, K.; Chiba, S. Role of Milk Whey in the Transmission of Human Cytomegalovirus Infection by Breast Milk. Microbiol. Immunol. 1996, 40, 201–204. [Google Scholar] [CrossRef]
- Vochem, M.; Hamprecht, K.; Jahn, G.; Speer, C.P. Transmission of cytomegalovirus to preterm infants through breast milk. Pediatr. Infect. Dis. J. 1998, 17, 53–58. [Google Scholar] [CrossRef]
- Cox, B.; Richardson, A.; Graham, P.; E Gislefoss, R.; Jellum, E.; Rollag, H. Breast cancer, cytomegalovirus and Epstein–Barr virus: A nested case–control study. Br. J. Cancer 2010, 102, 1665–1669. [Google Scholar] [CrossRef] [Green Version]
- Costa, H.; Touma, J.; Davoudi, B.; Benard, M.; Sauer, T.; Geisler, J.; Vetvik, K.; Rahbar, A.; Soderberg-Naucler, C. Human cytomegalovirus infection is correlated with enhanced cyclooxygenase-2 and 5-lipoxygenase protein expression in breast cancer. J. Cancer Res. Clin. Oncol. 2019, 145, 2083–2095. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oberstein, A.; Shenk, T. Cellular responses to human cytomegalovirus infection: Induction of a mesenchymal-to-epithelial transition (MET) phenotype. Proc. Natl. Acad. Sci. USA 2017, 114, E8244–E8253. [Google Scholar] [CrossRef] [Green Version]
- Nogalski, M.T.; Shenk, T. HSATII RNA is induced via a noncanonical ATM-regulated DNA damage response pathway and promotes tumor cell proliferation and movement. Proc. Natl. Acad. Sci. USA 2020, 117, 31891–31901. [Google Scholar] [CrossRef] [PubMed]
- Landini, M.P.; Rossier, E.; Schmitz, H. Antibodies to human cytomegalovirus structural polypeptides during primary infection. J. Virol. Methods 1988, 22, 309–317. [Google Scholar] [CrossRef]
- Landini, M.P.; Guan, M.X.; Jahn, G.; Lindenmaier, W.; Mach, M.; Ripalti, A.; Necker, A.; Lazzarotto, T.; Plachter, B. Large-scale screening of human sera with cytomegalovirus recombinant antigens. J. Clin. Microbiol. 1990, 28, 1375–1379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vornhagen, R.; Plachter, B.; Hinderer, W.; The, T.H.; Van Zanten, J.; Matter, L.; Schmidt, C.A.; Sonneborn, H.H.; Jahn, G. Early serodiagnosis of acute human cytomegalovirus infection by enzyme-linked immunosorbent assay using recombinant antigens. J. Clin. Microbiol. 1994, 32, 981–986. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamprecht, K.; Maschmann, J.; Vochem, M.; Dietz, K.; Speer, C.P.; Jahn, G. Epidemiology of transmission of cytomegalovirus from mother to preterm infant by breastfeeding. Lancet 2001, 357, 513–518. [Google Scholar] [CrossRef]
- Reddehase, M.J.; Lemmermann, N.A.W. Cellular reservoirs of latent cytomegaloviruses. Med. Microbiol. Immunol. 2019, 208, 391–403. [Google Scholar] [CrossRef] [PubMed]
- Sinzger, C.; Digel, M.; Jahn, G. Cytomegalovirus Cell Tropism. Curr. Topics Microbiol. Immunol. 2008, 325, 63–83. [Google Scholar] [CrossRef]
- Rahbar, A.; Touma, J.; Costa, H.; Davoudi, B.; Bukholm, I.R.; Sauer, T.; Vetvik, K.; Geisler, J.; Söderberg-Naucler, C. Low Expression of Estrogen Receptor-α and Progesterone Receptor in Human Breast Cancer Tissues Is Associated With High-Grade Human Cytomegalovirus Protein Expression. Clin. Breast Cancer 2017, 17, 526–535.e1. [Google Scholar] [CrossRef] [Green Version]
- Kumar, A.; Tripathy, M.K.; Pasquereau, S.; Al Moussawi, F.; Abbas, W.; Coquard, L.; Khan, K.A.; Russo, L.; Algros, M.-P.; Valmary-Degano, S.; et al. The Human Cytomegalovirus Strain DB Activates Oncogenic Pathways in Mammary Epithelial Cells. EBioMedicine 2018, 30, 167–183. [Google Scholar] [CrossRef] [Green Version]
- Sinzger, C.; Hahn, G.; Digel, M.; Katona, R.; Sampaio, K.L.; Messerle, M.; Hengel, H.; Koszinowski, U.; Brune, W.; Adler, B. Cloning and sequencing of a highly productive, endotheliotropic virus strain derived from human cytomegalovirus TB40/E. J. Gen. Virol. 2008, 89, 359–368. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, C.M.; Shenk, T. Human Cytomegalovirus pUS27 G Protein-Coupled Receptor Homologue Is Required for Efficient Spread by the Extracellular Route but Not for Direct Cell-to-Cell Spread. J. Virol. 2011, 85, 3700–3707. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Young, V.P.; Mariano, M.C.; Tu, C.C.; Allaire, K.M.; Avdic, S.; Slobedman, B.; Spencer, J.V. Modulation of the Host Environment by Human Cytomegalovirus with Viral Interleukin 10 in Peripheral Blood. J. Infect. Dis. 2017, 215, 874–882. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Branch, K.M.; Garcia, E.C.; Chen, Y.M.; McGregor, M.; Min, M.; Prosser, R.; Whitney, N.; Spencer, J.V. Productive Infection of Human Breast Cancer Cell Lines with Human Cytomegalovirus (HCMV). Pathogens 2021, 10, 641. https://doi.org/10.3390/pathogens10060641
Branch KM, Garcia EC, Chen YM, McGregor M, Min M, Prosser R, Whitney N, Spencer JV. Productive Infection of Human Breast Cancer Cell Lines with Human Cytomegalovirus (HCMV). Pathogens. 2021; 10(6):641. https://doi.org/10.3390/pathogens10060641
Chicago/Turabian StyleBranch, Kaitlin M., Erica C. Garcia, Yin Maggie Chen, Matthew McGregor, Mikayla Min, Rachel Prosser, Natalia Whitney, and Juliet V. Spencer. 2021. "Productive Infection of Human Breast Cancer Cell Lines with Human Cytomegalovirus (HCMV)" Pathogens 10, no. 6: 641. https://doi.org/10.3390/pathogens10060641





