Digital Biosurveillance for Zoonotic Disease Detection in Kenya
Abstract
:1. Introduction
Definitions
- Disease report: Open-source digital media reports including news articles, governmental and international bulletins, and other online disease warning/surveillance system notifications that report a disease occurrence, harvested through Biofeeds.
- Disease event: A single or group of epidemiologically related disease occurrences at a given location and time that is characterized by one or more common epidemiological indicators.
- Official disease event: A disease event whose occurrence was confirmed by one or more national and/or international health agencies, e.g., WHO or the World Organization for Animal Health (OIE), in the form of an official release report.
- Epidemiological indicators: The epidemiological information present in disease reports used to identify and describe a disease event.
- Disease event taxonomy: A tree-based structure of epidemiological indicators used to systematically identify and describe disease event information present in disease reports.
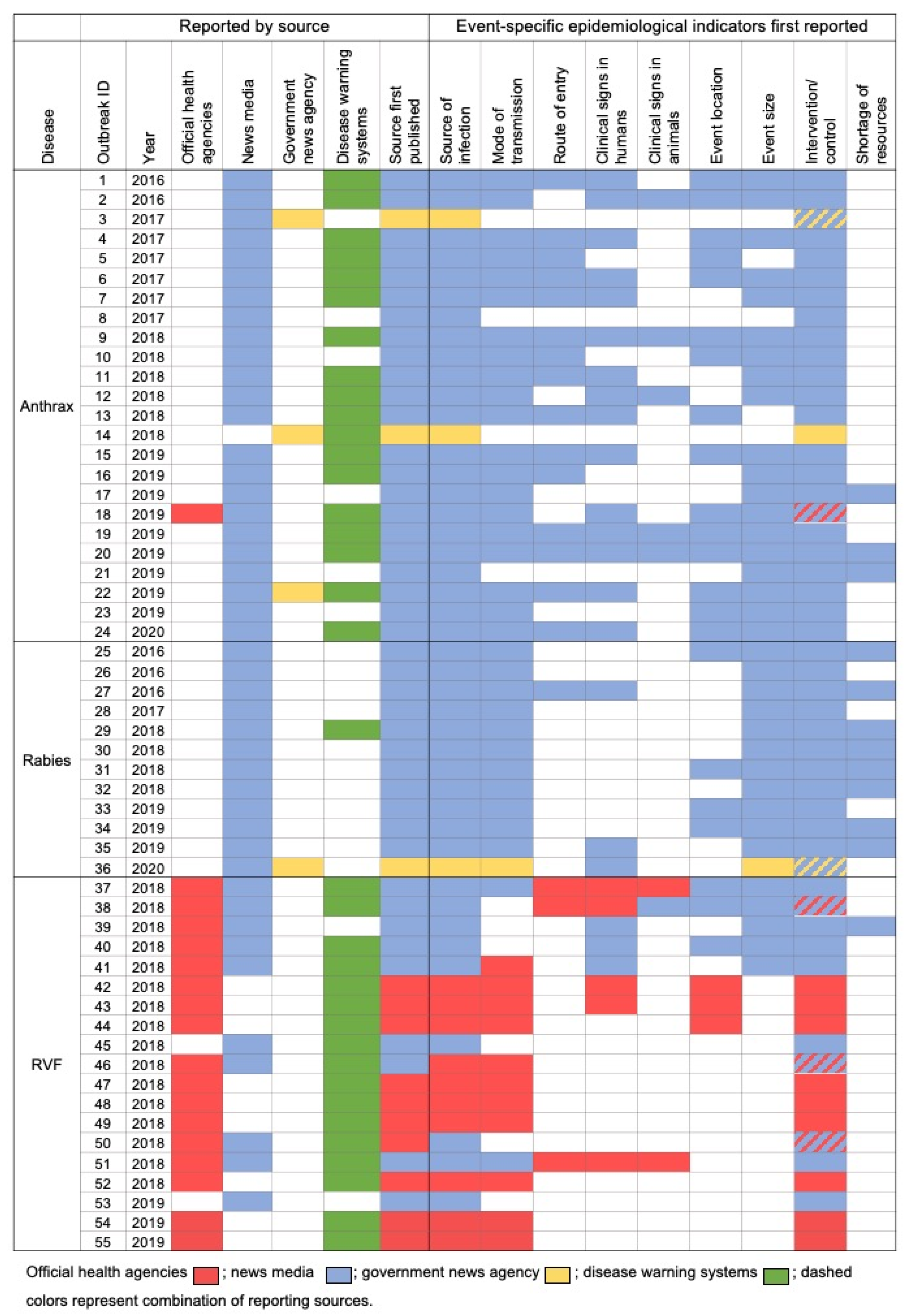
2. Results
3. Discussion
4. Methodology
4.1. Data Collection and Handling
4.2. Disease Event Taxonomy Construction and Report Tagging
4.3. Variable Construction and Data Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Murray, J.; Cohen, A.L. Infectious Disease Surveillance. In International Encyclopedia of Public Health; Elsevier Inc.: Amsterdam, The Netherlands, 2016; pp. 222–229. ISBN 9780128037089. [Google Scholar]
- Milinovich, G.J.; Williams, G.M.; Clements, A.C.A.; Hu, W. Internet-Based Surveillance Systems for Monitoring Emerging Infectious Diseases. Lancet Infect. Dis. 2014, 14, 160–168. [Google Scholar] [CrossRef]
- O’Shea, J.Ç. Digital disease detection: A systematic review of event-based internet biosurveillance systems. Int. J. Med. Inform. 2017, 101, 15–22. [Google Scholar] [CrossRef]
- Munyua, P.; Bitek, A.; Osoro, E.; Pieracci, E.G.; Muema, J.; Mwatondo, A.; Kungu, M.; Nanyingi, M.; Gharpure, R.; Njenga, K.; et al. Prioritization of Zoonotic Diseases in Kenya, 2015. PLoS ONE 2016, 11, e0161576. [Google Scholar] [CrossRef]
- Morse, S.S. Public Health Surveillance and Infectious Disease Detection. Biosecur. Bioterror. 2012. [Google Scholar] [CrossRef] [PubMed]
- Keller, M.; Blench, M.; Tolentino, H.; Freifeld, C.C.; Mandl, K.D.; Mawudeku, A.; Eysenbach, G.; Brownstein, J.S. Use of Unstructured Event-Based Reports for Global Infectious Disease Surveillance. Emerg. Infect. Dis. 2009, 15, 689–695. [Google Scholar] [CrossRef]
- Milinovich, G.J.; Magalhães, R.J.S.; Hu, W. Role of Big Data in the Early Detection of Ebola and Other Emerging Infectious Diseases. Lancet Glob. Health 2015, 3, e20–e21. [Google Scholar] [CrossRef] [Green Version]
- Madoff, L.C. ProMED-Mail: An Early Warning System for Emerging Diseases. Clin. Infect. Dis. 2004, 39, 227–232. [Google Scholar] [CrossRef] [Green Version]
- Freifeld, C.C.; Mandl, K.D.; Reis, B.Y.; Brownstein, J.S. HealthMap: Global Infectious Disease Monitoring through Automated Classification and Visualization of Internet Media Reports. J. Am. Med. Inform. Assoc. 2008, 15, 150–157. [Google Scholar] [CrossRef] [PubMed]
- Dion, M.; AbdelMalik, P.; Mawudeku, A. Big Data and the Global Public Health Intelligence Network (GPHIN). Can. Commun. Dis. Rep. 2015, 41, 209–214. [Google Scholar] [CrossRef]
- Karo, B.; Haskew, C.; Khan, A.S.; Polonsky, J.A.; Mazhar, M.K.A.; Buddha, N. World Health Organization Early Warning, Alert and Response System in the Rohingya Crisis, Bangladesh, 2017–2018. Emerg. Infect. Dis. 2018, 24, 2074–2076. [Google Scholar] [CrossRef]
- Guy, S.; Ratzki-Leewing, A.; Bahati, R.; Gwadry-Sridhar, F. Social Media: A Systematic Review to Understand the Evidence and Application in Infodemiology. In Lecture Notes of the Institute for Computer Sciences, Social-Informatics and Telecommunications Engineering; Springer: Berlin/Heidelberg, Germany, 2011; pp. 1–8. [Google Scholar]
- Grein, T.W.; Kamara, K.B.O.; Rodier, G.; Plant, A.J.; Bovier, P.; Ryan, M.J.; Ohyama, T.; Heymann, D.L. Rumors of Disease in the Global Village: Outbreak Verification. Emerg. Infect. Dis. 2000, 6, 97–102. [Google Scholar] [CrossRef] [PubMed]
- Heymann, D.L.; Rodier, G.R. Hot Spots in a Wired World: WHO Surveillance of Emerging and Re-Emerging Infectious Diseases. Lancet Infect. Dis. 2001, 1, 345–353. [Google Scholar] [CrossRef]
- Baker, H.; Grady, A.; Schwantes, C.; Iarocci, E.; Campbell, R.; Calapristi, G.; Dowson, S.; Hart, M.; Charles, L.E.; Quitugua, T. NBIC Biofeeds: Deploying a New, Digital Tool for Open Source Biosurveillance across Federal Agencies. Online J. Public Health Inform. 2018. [Google Scholar] [CrossRef] [Green Version]
- Falzon, L.C.; Alumasa, L.; Amanya, F.; Kang’ethe, E.; Kariuki, S.; Momanyi, K.; Muinde, P.; Murungi, M.K.; Njoroge, S.M.; Ogendo, A.; et al. One Health in Action: Operational Aspects of an Integrated Surveillance System for Zoonoses in Western Kenya. Front. Vet. Sci. 2019, 6, 252. [Google Scholar] [CrossRef] [Green Version]
- Munyua, P.M.; Njenga, M.K.; Osoro, E.M.; Onyango, C.O.; Bitek, A.O.; Mwatondo, A.; Muturi, M.K.; Musee, N.; Bigogo, G.; Otiang, E.; et al. Successes and Challenges of the One Health Approach in Kenya over the Last Decade. BMC Public Health 2019, 19. [Google Scholar] [CrossRef]
- OIE—World Organisation for Animal Health. OIE-Listed Diseases 2021. Available online: https://www.oie.int/en/animal-health-in-the-world/oie-listed-diseases-2021/ (accessed on 19 February 2021).
- WHO. WHO|Other Neglected Zoonotic Diseases. 2013. Available online: https://www.who.int/neglected_diseases/zoonoses/en/ (accessed on 15 February 2021).
- Hassan, A.; Muturi, M.; Mwatondo, A.; Omolo, J.; Bett, B.; Gikundi, S.; Konongoi, L.; Ofula, V.; Makayotto, L.; Kasiti, J.; et al. Epidemiological Investigation of a Rift Valley Fever Outbreak in Humans and Livestock in Kenya, 2018. Am. J. Trop. Med. Hyg. 2020, 103, 1649–1655. [Google Scholar] [CrossRef]
- Kitala, P.M.; McDermott, J.J.; Kyule, M.N.; Gathuma, J.M. Community-Based Active Surveillance for Rabies in Machakos District, Kenya. Prev. Vet. Med. 2000, 44, 73–85. [Google Scholar] [CrossRef]
- Bitek, A.O.; Osoro, E.; Munyua, P.M.; Nanyingi, M.; Muthiani, Y.; Kiambi, S.; Muturi, M.; Mwatondo, A.; Muriithi, R.; Cleaveland, S.; et al. A Hundred Years of Rabies in Kenya and the Strategy for Eliminating Dog-Mediated Rabies by 2030. AAS Open Res. 2019, 1, 23. [Google Scholar] [CrossRef]
- Muturi, M.; Gachohi, J.; Mwatondo, A.; Lekolool, I.; Gakuya, F.; Bett, A.; Osoro, E.; Bitek, A.; Thumbi, S.M.; Munyua, P.; et al. Recurrent Anthrax Outbreaks in Humans, Livestock, and Wildlife in the Same Locality. Am. J. Trop. Med. Hyg. 2018, 99, 833–839. [Google Scholar] [CrossRef] [Green Version]
- Bukachi, S.A.; Wandibba, S.; Nyamongo, I.K. The Socio-Economic Burden of Human African Trypanosomiasis and the Coping Strategies of Households in the South Western Kenya Foci. PLoS Negl. Trop. Dis. 2017, 11, e0006002. [Google Scholar] [CrossRef] [Green Version]
- Mableson, H.E.; Okello, A.; Picozzi, K.; Welburn, S.C. Neglected Zoonotic Diseases-The Long and Winding Road to Advocacy. PLoS Negl. Trop. Dis. 2014, 8, e2800. [Google Scholar] [CrossRef] [PubMed]
- Schwind, J.S.; Norman, S.A.; Karmacharya, D.; Wolking, D.J.; Dixit, S.M.; Rajbhandari, R.M.; Mekaru, S.R.; Brownstein, J.S. Online Surveillance of Media Health Event Reporting in Nepal: Digital Disease Detection from a One Health Perspective. BMC Int. Health Hum. Rights 2017, 17, 26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- OIE—World Organisation for Animal Health. Terrestrial Code Online Access. Available online: https://www.oie.int/en/what-we-do/standards/codes-and-manuals/terrestrial-code-online-access/ (accessed on 20 May 2021).
| Outbreak ID | Disease | Number of Days News Media Reported before Official Reports |
|---|---|---|
| 18 | Anthrax | 3 |
| 37 | RVF | 2 |
| 38 | RVF | 2 |
| 39 | RVF | 8 |
| 40 | RVF | 1 |
| 41 | RVF | 5 |
| 46 | RVF | 14 |
| 50 | RVF | −4 |
| 51 | RVF | 7 |
| Disease Events (n = Total Events) | Primary Affected Group Reported | ||
|---|---|---|---|
| Human (Official/Total) | Livestock (Official/Total) | Wildlife (Official/Total) | |
| Anthrax (n = 24) | 0% (0/20) | 0% (0/3) | 100% (1/1) |
| Rabies (n = 12) | 0% (0/10) | 0% (0/2) | 0% (0/0) |
| RVF (n = 19) | 80% (4/5) | 93% (13/14) | 0% (0/0) |
| Parameters | Category | Not Reported | Reported | Odds Ratio | LCL | UCL | p-Value |
|---|---|---|---|---|---|---|---|
| Disease | Anthrax | 23 | 1 | Reference | <0.001 | ||
| Rabies | 12 | 0 | 0.00 | ** | ** | ||
| RVF | 2 | 17 | 195.50 | 24.01 | 4756.43 | ||
| Government news agency reporting | Not Reported | 33 | 18 | Reference | 0.291 | ||
| Reported | 4 | 0 | 0.00 | ** | ** | ||
| News media reporting | Not Reported | 1 | 9 | Reference | 0.030 | ||
| Reported | 36 | 9 | 0.03 | 0.00 | 0.17 | ||
| Surveillance system reporting | Not Reported | 18 | 1 | Reference | 0.004 | ||
| Reported | 19 | 17 | 16.11 | 2.84 | 305.22 | ||
| Location of the event (Province) | Central | 6 | 2 | Reference | 0.015 | ||
| Coast | 1 | 2 | 6.00 | 0.38 | 182.92 | ||
| Eastern | 11 | 5 | 1.36 | 0.21 | 11.60 | ||
| Nairobi | 1 | 0 | 0.00 | ** | ** | ||
| Northeastern | 0 | 5 | ** | 0.00 | ** | ||
| Nyanza | 6 | 2 | 1.00 | 0.09 | 10.76 | ||
| Rift Valley | 12 | 2 | 0.50 | 0.05 | 5.01 | ||
| Primary affected group | Humans | 31 | 4 | Reference | <0.001 | ||
| Livestock | 6 | 13 | 16.79 | 4.41 | 78.71 | ||
| Wildlife | 0 | 1 | ** | 0.00 | ** | ||
| Number of reports | Less than two | 17 | 4 | Reference | 0.224 | ||
| Three to six | 8 | 7 | 3.72 | 0.87 | 17.98 | ||
| Seven and more | 12 | 7 | 2.48 | 0.61 | 11.32 |
| Epidemiological Indicator | Sub-Categories |
|---|---|
| Disease | Anthrax, Rift Valley fever, rabies, brucellosis, trypanosomiasis |
| Species affected | Humans: children, adult, elderly Domesticated/Livestock: bovine, sheep, goat, poultry, camel, donkey, horse, dog, cat Wildlife: buffalo, rhinoceros, gazelle, giraffe, warthog, waterbuck |
| Source of infection | Humans to humans, animals (domestic/wildlife) to humans, environment to humans, animals to animals, environment to animals |
| Mode of transmission | Direct contact: infected animals, infected carcass/meat, infected animal byproducts, infected birthing fluids/placenta, animal bite Indirect contact: soil, water, air, mechanical vector, biological vector |
| Route of entry | Ingestion, inhalation, cutaneous/contact |
| Clinical presentation | General/non-specific, pulmonary, gastrointestinal, cutaneous, neurological, musculoskeletal, circulatory, reproductive, behavioral |
| Event location | Hospital/clinic, home residence, small farm, production farm, national park, slaughterhouse |
| Event size | Isolated case, single cluster (within village/town/city), multiple clusters/regional, multiple regions (counties/provinces/states), multiple countries, epidemic, pandemic |
| Intervention/control | Hospitalization, home isolation, movement control, passive surveillance, active surveillance, area containment/closure, travel ban, quarantine, culling, disposal, disinfection, vaccination, warning/advisory, monitoring, recall/market removal, meat inspection, vector control |
| Shortage of resources | Diagnostic facilities, vaccines, drugs/medication, ventilators, personal protective equipment, health care workers, misdiagnosis/medical error, delayed/no medical attention |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Keshavamurthy, R.; Thumbi, S.M.; Charles, L.E. Digital Biosurveillance for Zoonotic Disease Detection in Kenya. Pathogens 2021, 10, 783. https://doi.org/10.3390/pathogens10070783
Keshavamurthy R, Thumbi SM, Charles LE. Digital Biosurveillance for Zoonotic Disease Detection in Kenya. Pathogens. 2021; 10(7):783. https://doi.org/10.3390/pathogens10070783
Chicago/Turabian StyleKeshavamurthy, Ravikiran, Samuel M. Thumbi, and Lauren E. Charles. 2021. "Digital Biosurveillance for Zoonotic Disease Detection in Kenya" Pathogens 10, no. 7: 783. https://doi.org/10.3390/pathogens10070783
APA StyleKeshavamurthy, R., Thumbi, S. M., & Charles, L. E. (2021). Digital Biosurveillance for Zoonotic Disease Detection in Kenya. Pathogens, 10(7), 783. https://doi.org/10.3390/pathogens10070783







