First Report of Blood Fluke Pathogens with Potential Risk for Emerging Yellowtail Kingfish (Seriola lalandi) Aquaculture on the Chilean Coast, with Descriptions of Two New Species of Paradeontacylix (Aporocotylidae)
Abstract
:1. Introduction
2. Results
2.1. Morphometric Analysis
2.2. Molecular Analysis
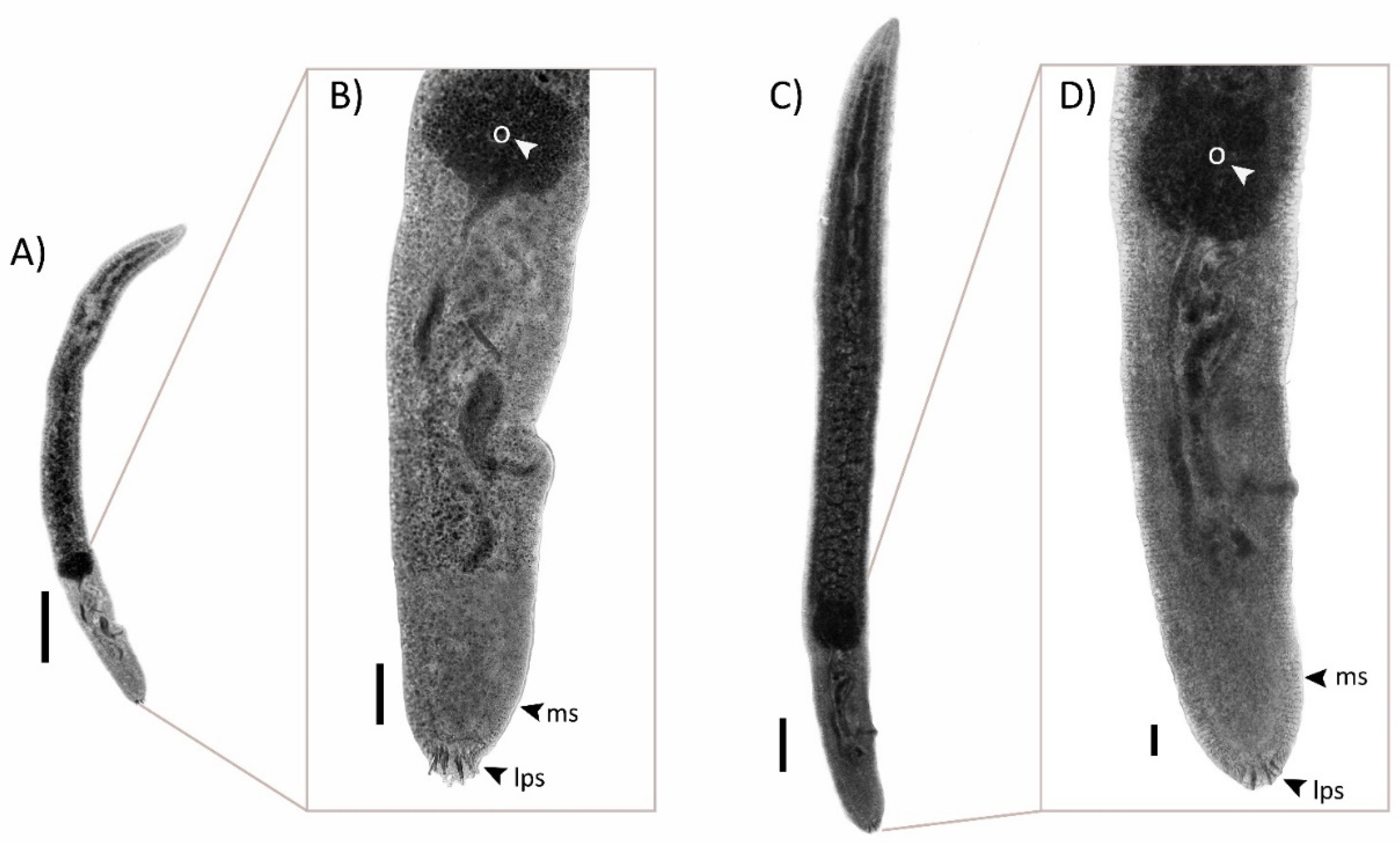
2.3. Morphologic Descriptions
2.3.1. Paradeontacylix humboldti n. sp.
Description
Remarks
2.3.2. Paradeontacylix olivai n. sp.
Description
Remarks
3. Discussion
4. Material and Methods
4.1. Sample Collection
4.2. Morphological Description and Morphometrical Analyses
4.3. Molecular Analysis
4.3.1. DNA Extraction and Amplification
4.3.2. Phylogenetic Reconstruction
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Ethics Statements
References
- Rohde, K. Marine Parasitology; Csiro Publishing: Melbourne, Australia; CAB International: Melbourne, Australia, 2005; p. 592. [Google Scholar]
- Holzer, A.S.; Montero, F.E.; Repullés, A.; Nolan, M.J.; Sitja-Bobadilla, A.; Alvarez-Pellitero, P.; Zarza, C.; Raga, J.A. Cardicola aurata sp. n. (Digenea: Sanguinicolidae) from Mediterranean Sparus aurata L. (Teleostei: Sparidae) and its unexpected phylogenetic relationship with Paradeontacylix McIntosh, 1934. Parasitol. Int. 2008, 57, 472–482. [Google Scholar] [CrossRef] [Green Version]
- Power, C.; Nowak, B.F.; Cribb, T.H.; Bott, N.J. Bloody flukes: A review of aporocotylids as parasites of cultured marine fishes. Int. J. Parasitol. 2020, 50, 743–753. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, K.; Egusa, S. Two new species of Paradeontacylix Mcintosh, 1934 (trematoda: Sanguinicolidae) from the vascular system of a cultured marine fish, Seriola purpurascens. Fish Pathol. 1986, 21, 15–19. [Google Scholar] [CrossRef]
- Crespo, S.; Grau, A.; Padros, F. Sanguinocoliasis in the cultured amberjack Seriola dumerili Risso, from the spanish Mediterranean area. Bull. Eur. Ass. Fish Pathol. 1992, 12, 157. [Google Scholar]
- Shirakashi, S.; Tani, K.; Ishimaru, K.; Shin, S.P.; Honryo, T.; Uchida, H.; Ogawa, K. Discovery of intermediate hosts for two species of blood flukes Cardicola orientalis and Cardicola forsteri (Trematoda: Aporocotylidae) infecting Pacific bluefin tuna in Japan. Parasitol. Int. 2016, 65, 128–136. [Google Scholar] [CrossRef] [Green Version]
- Sugihara, Y.; Yamada, T.; Ichimaru, T.; Matsukura, K.; Kanai, K. Detection of bluefin tuna blood flukes (Cardicola spp.) from wild juvenile Pacific bluefin tuna Thunnus orientalis caught for aquaculture. Aquaculture 2016, 452, 9–11. [Google Scholar] [CrossRef]
- Ogawa, K.; Hattori, K.; Hatai, K.; Kubota, S. Histopathology of cultured marine fish, Seriola Purpurascens (Carangidae) infected with Paradeontacylix spp. (Trematoda: Sanguinicolidae) in its vascular system. Fish Pathol. 1989, 24, 75–81. [Google Scholar] [CrossRef]
- Kirk, R.S.; Lewis, J.W. Histopathology of Sanguinicola inermis infection in carp, Cyprinus carpio. J. Helminthol. 1998, 72, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Padrós, F.; Zarza, C.; Crespo, S. Histopathology of cultured sea bream Sparus aurata infected with sanguinicolid trematodes. Dis. Aquat. Organ. 2001, 44, 47–52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paperna, I.; Dzikowski, R. Chapter 10 Digenea (Phylum Platyhelminthes). In Fish Diseases and Disorders, Volume 1: Protozoan and Metazoan Infections; Woo, P.T.K., Ed.; CABI: Wallingford, UK; University of Guelph: Guelph, ON, Canada, 2006; p. 800. [Google Scholar]
- Ogawa, K.; Fukudome, M. Mass mortality caused by blood fluke (Paradeontacylix) among Amberjack (Seriola dumeili) imported to Japan. Fish Pathol. 1994, 29, 265–269. [Google Scholar] [CrossRef]
- Crespo, S.; Grau, A.; Padros, F. The intensive culture of 0-group amberjack in the western Mediterranean is compromised by disease problems. Aquac. Int. J. Eur. Aquac. Soc. 1994, 2, 262–265. [Google Scholar] [CrossRef]
- McIntosh, A. A new blood trematode, Paradeontacylix sanguinicoloides n.g., n.sp., from Seriola lalandi with a key to the species of the family aporocotylidae. Parasitology 1934, 26, 463–467. [Google Scholar] [CrossRef]
- Repullés-Albelda, A.; Montero, F.E.; Holzer, A.S.; Ogawa, K.; Hutson, K.S.; Raga, J.A. Speciation of the Paradeontacylix spp. (Sanguinicolidae) of Seriola dumerili. Two new species of the genus Paradeontacylix from the Mediterranean. Parasitol. Int. 2008, 57, 405–414. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, K.; Akiyama, K.; Grabner, D. Paradeontacylix buri n. sp. (Trematoda: Aporocotylidae) from Seriola quinqueradiata cultured in Japan with a description of unidentified Paradeontacylix sp. from S. lalandi. Fish Pathol. 2015, 50, 183–191. [Google Scholar] [CrossRef] [Green Version]
- Hutson, K.S.; Whittington, I.D. Paradeontacylix godfreyi n. sp. (Digenea: Sanguinicolidae) from the heart of wild Seriola lalandi (Perciformes: Carangidae) in southern Australia. Zootaxa 2006, 1151, 55. [Google Scholar] [CrossRef] [Green Version]
- Martinez-Takeshita, N.; Purcell, C.M.; Chabot, C.L.; Craig, M.T.; Paterson, C.N.; Hyde, J.R.; Allen, L.G. A tale of three tails: Cryptic speciation in a globally distributed marine fish of the genus Seriola. Copeia 2015, 103, 357–368. [Google Scholar] [CrossRef]
- Layman, E.M. Parasitic worms from the fishes of Peter The Great Bay. Bull. Pac. Sci. Fish. Res. Stn. 1930, 3, 1–120. [Google Scholar]
- Lakshmi, T.T.; Madhavi, R. Paradeontacylix megalaspium n. sp. (Digenea: Sanguinicolidae) from the carangid fish, Megalaspis cordyla of Bay of Bengal. Zootaxa 2007, 1512, 65–68. [Google Scholar] [CrossRef]
- Hutson, K.S.; Ernst, I.; Whittington, I.D. Risk assessment for metazoan parasites of yellowtail kingfish Seriola lalandi (Perciformes: Carangidae) in South Australian sea-cage aquaculture. Aquaculture 2007, 271, 85–99. [Google Scholar] [CrossRef]
- Bullard, S.A.; Overstreet, R.M. Potential pathological effects of blood flukes (Digenea: Sanguinicolidae) on Pen-reared Marine Fishes. Fac. Publ. Harold W. Manter Lab. Parasitol. 2002, 414, 10–25. [Google Scholar]
- Ogawa, K.; Yokoyama, H. Parasitic diseases of cultured marine fish in Japan. Fish Pathol. 1998, 33, 303–309. [Google Scholar] [CrossRef] [Green Version]
- Ogawa, K. Diseases of cultured marine fishes caused by Platyhelminthes (Monogenea, Digenea, Cestoda). Parasitology 2015, 142, 178–195. [Google Scholar] [CrossRef] [PubMed]
- Orellana, J.; Waller, U.; Wecker, B. Culture of yellowtail kingfish (Seriola lalandi) in a marine recirculating aquaculture system (RAS) with artificial seawater. Aquac. Eng. 2014, 58, 20–28. [Google Scholar] [CrossRef]
- Fernández, G.; Cichero, D.; Patel, A.; Martínez, V. Estructura genética de poblaciones chilenas de Seriola lalandi para la diversificación de la acuicultura nacional en el norte de Chile. Lat. Am. J. Aquat. Res. 2015, 43, 374–379. [Google Scholar] [CrossRef]
- Bravo, S.; Hurtado, C.F.; Silva, M.T. Coinfection of Caligus lalandei and Benedenia seriolae on the yellowtail kingfish Seriola lalandi farmed in a net cage in northern Chile. Lat. Am. J. Aquat. Res. 2017, 45, 852–857. [Google Scholar] [CrossRef]
- Sepúlveda, F.A.; González, M.T. Patterns of genetic variation and life history traits of Zeuxapta seriolae infesting Seriola lalandi across the coastal and oceanic areas in the southeastern Pacific Ocean: Potential implications for aquaculture. Parasit. Vectors 2015, 8, 282. [Google Scholar] [CrossRef] [Green Version]
- Sepúlveda, F.A.; González, M.T. Molecular and morphological analyses reveal that the pathogen Benedenia seriolae (Monogenea: Capsalidae) is a complex species: Implications for yellowtail Seriola spp. aquaculture. Aquaculture 2014, 418–419, 94–100. [Google Scholar] [CrossRef]
- Baeza, J.A.; Sepúlveda, F.A.; González, M.T. The complete mitochondrial genome and description of a new cryptic species of Benedenia Diesing, 1858 (Monogenea: Capsalidae), a major pathogen infecting the yellowtail kingfish Seriola lalandi Valenciennes in the South-East Pacific. Parasit. Vectors 2019, 12, 490. [Google Scholar] [CrossRef]
- Sepúlveda, F.A.; González, M.T. DNA barcoding evidence for the first recorded transmission of Neobenedenia sp. from wild fish species to Seriola lalandi cultured in an open recirculating system on the Coast of Northern Chile. Aquaculture 2019, 501, 239–246. [Google Scholar] [CrossRef]
- Pečnikar, Ž.F.; Buzan, E.V. 20 years since the introduction of DNA barcoding: From theory to application. J. Appl. Genet. 2014, 55, 43–52. [Google Scholar] [CrossRef]
- Nolan, M.J.; Cribb, T.H. Two new blood flukes (Digenea: Sanguinicolidae) from Epinephelinae (Perciformes: Serranidae) of the Pacific Ocean. Parasitol. Int. 2004, 53, 327–335. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Ponce de León, G.; Hernández-Mena, D.I. Testing the higher-level phylogenetic classification of Digenea (Platyhelminthes, Trematoda) based on nuclear rDNA sequences before entering the age of the ‘next-generation’ tree of life. J. Helminthol. 2019, 93, 260–276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morgan, J.A.T.; Blair, D. Relative merits of nuclear ribosomal internal transcribed spacers and mitochondrial CO1 and ND1 genes for distinguishing among Echinostoma species (Trematoda). Parasitology 1998, 116, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Callejón, R.; Cutillas, C.; Nadler, S.A. Nuclear and mitochondrial genes for inferring Trichuris phylogeny. Parasitol. Res. 2015, 114, 4591–4599. [Google Scholar] [CrossRef]
- Nugroho, E.; Taniguchi, N.; Kato, K.; Miyashita, S. Genetic difference among seed populations of greater amberjack used in aquaculture farm of Japan. Suisanzoshoku 2000, 48, 665–674. [Google Scholar] [CrossRef]
- Nugroho, E.; Ferrell, D.J.; Smith, P.; Taniguchi, N. Genetic divergence of kingfish from Japan, Australia and New Zealand inferred by microsatellite DNA and mitochondrial DNA control region markers. Fish. Sci. 2001, 67, 843–850. [Google Scholar] [CrossRef] [Green Version]
- Šegvić-Bubić, T.; Marrone, F.; Grubišić, L.; Izquierdo-Gomez, D.; Katavić, I.; Arculeo, M.; Lo Brutto, S. Two seas, two lineages: How genetic diversity is structured in Atlantic and Mediterranean greater amberjack Seriola dumerili Risso, 1810 (Perciformes, Carangidae). Fish. Res. 2016, 179, 271–279. [Google Scholar] [CrossRef]
- Premachandra, H.K.A.; Lafarga-De la Cruz, F.; Takeuchi, Y.; Miller, A.; Fielder, S.; O’Connor, W.; Frère, C.H.; Nguyen, N.H.; Bar, I.; Knibb, W. Genomic DNA variation confirmed Seriola lalandi comprises three different populations in the Pacific, but with recent divergence. Sci. Rep. 2017, 7, 9386. [Google Scholar] [CrossRef] [Green Version]
- Slatkin, M. Gene flow and the geographic structure of natural populations. Science 1987, 236, 787–792. [Google Scholar] [CrossRef] [PubMed]
- Nadler, S.A. Microevolution and the genetic structure of parasite populations. J. Parasitol. 1995, 81, 395. [Google Scholar] [CrossRef]
- Sepúlveda, F.A.; González, M.T. Spatio-temporal patterns of genetic variations in populations of yellowtail kingfish Seriola lalandi from the south-eastern Pacific Ocean and potential implications for its fishery management. J. Fish Biol. 2017, 90, 249–264. [Google Scholar] [CrossRef] [PubMed]
- Miller, P.A.; Fitch, A.J.; Gardner, M.; Hutson, K.S.; Mair, G. Genetic population structure of Yellowtail Kingfish (Seriola lalandi) in temperate Australasian waters inferred from microsatellite markers and mitochondrial DNA. Aquaculture 2011, 319, 328–336. [Google Scholar] [CrossRef]
- Cribb, T.H.; Adlard, R.D.; Hayward, C.J.; Bott, N.J.; Ellis, D.; Evans, D.; Nowak, B.F. The life cycle of Cardicola forsteri (Trematoda: Aporocotylidae), a pathogen of ranched southern bluefin tuna, Thunnus maccoyi. Int. J. Parasitol. 2011, 41, 861–870. [Google Scholar] [CrossRef] [PubMed]
- Shirakashi, S.; Ogawa, K. Blood fluke infections in marine cultured fish. Fish Pathol. 2016, 51, 92–98. [Google Scholar] [CrossRef] [Green Version]
- Ogawa, K. Marine parasitology with special reference to Japanese fisheries and mariculture. Vet. Parasitol. 1996, 64, 95–105. [Google Scholar] [CrossRef]
- Montero, F.E.; Kostadinova, A.; Raga, J.A. Development and habitat selection of a new sanguinicolid parasite of cultured greater amberjack, Seriola dumerili, in the Mediterranean. Aquaculture 2009, 288, 132–139. [Google Scholar] [CrossRef]
- Bush, A.O.; Lafferty, K.D.; Lotz, J.M.; Shostak, A.W. Parasitology meets ecology on its own terms: Margolis et al. revisited. J. Parasitol. 1997, 83, 575. [Google Scholar] [CrossRef] [PubMed]
- Abràmoff, M.D.; Magalhães, P.J.; Ram, S.J. Image processing with imageJ. Biophotonics Int. 2004, 11, 36–41. [Google Scholar] [CrossRef]
- Quinn, G.P.; Keough, M.J. Chapter 17: Principal components and correspondence analysis. In Experimental Design and Data Analysis for Biologists; Quinn, G.P., Keough, M.J., Eds.; Cambridge University Press: Cambridge, UK, 2002; pp. 444–472. [Google Scholar]
- Miller, S.A.; Dykes, D.D.; Polesky, H.F. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988, 16, 1215. [Google Scholar] [CrossRef] [Green Version]
- Chisholm, L.A.; Whittington, I.D.; Morgan, J.A.T.; Adlard, R.D. The Calicotyle conundrum: Do molecules reveal more than morphology? Syst. Parasitol. 2001, 49, 81–87. [Google Scholar] [CrossRef]
- Leung, T.L.; Donald, K.M.; Keeney, D.B.; Koehler, A.V.; Peoples, R.C.; Poulin, R. Trematode parasites of Otago Harbour (New Zealand) soft-sediment intertidal ecosystems: Life cycles, ecological roles and DNA barcodes. N. Z. J. Mar. Freshw. Res. 2009, 43, 857–865. [Google Scholar] [CrossRef] [Green Version]
- Filatov, D.A. PROSEQ: A software for preparation and evolutionary analysis of DNA sequence data sets. Mol. Ecol. Notes 2002, 2, 621–624. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Posada, D. jModelTest: Phylogenetic model averaging. Mol. Biol. Evol. 2008, 25, 1253–1256. [Google Scholar] [CrossRef] [PubMed]
- Massidon, W.P.; Maddison, D.R. Mesquite: A modular System for Evolutionary Analysis. Version 2.75. Available online: https://ci.nii.ac.jp/naid/20001328793 (accessed on 15 January 2021).
- Huelsenbeck, J.P.; Ronquist, F. MrBAYES: Bayesian inference for phylogeny trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarization in bayesian phylogenetics using tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef] [Green Version]
- Trifinopoulos, J.; Nguyen, L.T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef] [Green Version]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [Green Version]




| Species | N | Host | Country | Access Number | Author(s) | |
|---|---|---|---|---|---|---|
| 28S | cox1 | |||||
| P. balearicus | 2/1 | Seriola dumerili | Spain | AM489594 | AM489604 | Repullés-Albelda et al., 2008 |
| P. buri | -/1 | S. quinqueradiata | Japan | AB904154 | - | Ogawa et al., 2015 |
| P. godfreyi | 2/1 | S. lalandi | Australia | AM489597 | AM489607 | Repullés-Albelda et al., 2008 |
| P. grandispinus | 2/1 | S. dumerili | Japan | AM489596 | AM489606 | Repullés-Albelda et al., 2008 |
| P. humboldti | 2/3 | S. lalandi | Chile | MW599287 | MW598468 | This study |
| P. ibericus | 2/1 | S. dumerili | Spain | AM489593 | AM489603 | Repullés-Albelda et al., 2008 |
| P. kampachi | 2/1 | S. dumerili | Japan | AM489595 | AM489605 | Repullés-Albelda et al., 2008 |
| P. olivai | 8/5 | S. lalandi | Chile | MW599288 | MW598468-70 | This study |
| P. sanguinicoloides | 2/- | S. dorsalis | Florida | - | - | McIntosh, 1934 |
| Cardicola forsteri | - | Thunnus thynnus | Spain | EF653388 | KP988302 | Aiken et al., 2007/Palacios-Abella et al., 2015 |
| Cardicola opisthorchis | - | Thunnus thynnus | Spain | KP217052 | KP988305 | Unpublished/Palacios-Abella et al., 2015 |
| Psettarium nolani | - | Arothron hispidus | Australia | MG709043 | - | Yong et al., 2018 |
| Psettarium sinensis | - | Fugu rubripes | China | EU368853 | - | Unpublished |
| Plethorchis acanthus | - | Mugil cephalus | Australia | AY222178 | - | Olson et al., 2003 |
| Species | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |
|---|---|---|---|---|---|---|---|---|---|
| 1 | P. humboldti | 10.26 | 10.50 | - | 11.69 | 16.47 | 11.93 | 13.84 | |
| 2 | P. olivai | 1.25 | 10.74 | - | 12.17 | 15.51 | 11.22 | 11.93 | |
| 3 | P. grandispinus | 1.02 | 1.02 | - | 11.69 | 13.84 | 6.20 | 11.46 | |
| 4 | P. buri | 2.39 | 2.39 | 2.05 | - | - | - | - | |
| 5 | P. ibericus | 1.59 | 1.59 | 1.25 | 1.94 | 15.99 | 11.46 | 6.921 | |
| 6 | P. godfreyi | 1.48 | 1.48 | 1.14 | 2.05 | 0.79 | 13.6 | 15.04 | |
| 7 | P. balearicus | 1.02 | 1.02 | 0.23 | 2.05 | 1.25 | 1.14 | 11.46 | |
| 8 | P. kampachi | 1.48 | 1.48 | 1.14 | 1.82 | 0.23 | 0.68 | 1.14 |
| Body Measurement | Paradeontacylix humboldti n. sp. | Paradeontacylix olivai n. sp. |
|---|---|---|
| Body length | 1858–2353 (2105, n = 2) | 3567–5214 (4099, n = 8) |
| Body maximum width | 155–179 (167, n = 2) | 204–369 (324, n = 8) |
| Marginal tegumental spine rows | 286–436 (361, n = 4) | 478–650 (563, n = 6) |
| Marginal tegumental spine length | 4 | 5–8 (6, n = 7) |
| Marginal tegumental spine width | 1 | 1–2 (1.6, n = 7) |
| Marginal tegumental spine per row | [4–9] | [3–14] |
| No. of large posterior tegumental spines [No. spines × No. rows] | ([(2–3) × 4]–[(2–3) × 4]) | [(3–5) × 4] |
| Large posterior tegumental spines length | 15–22 (19, n = 18) | 15–39 (24, n = 30) |
| Large posterior tegumental spines width (maximum wide at spine base) | 3–5 (4, n = 18) | 4–7 (5, n = 30) |
| No. of medium posterior tegumental spines [No. spines × No. rows] | [3 × 3] | [(3–4) × 3] |
| Medium posterior tegumental spines length | 7–13 (9, n = 12) | 6–17 (11, n = 16) |
| Medium posterior tegumental spines width | 1–2 (2, n = 12) | 2–4 (3, n = 18) |
| Mouth opening from anterior end | 28 (n = 2) | 8–22 (16, n = 7) |
| Oesophagus length | 533–660 (597, n = 2) | 496–1268 (1040, n = 7) |
| Oesophagus (percentage of body length) | 28–29% (n = 2) | 14–30% (25%, n = 7) |
| Oesophagus’s glands cell from anterior end length | 342–451 (396, n = 2) | 706–962 (814, n = 6) |
| Intestine anterior caeca length | 47–48 (48, n = 4) | 60–126 (103, n = 14) |
| Intestine posterior caeca length | 688–878 (783, n = 4) | 1622–2436 (1848, n = 12) |
| Testes number | 63–69 (66, n = 2) | 41–45 (43, n = 7) |
| Testes length (average) | 26–28 (27, n = 2) | 47–72 (53, n = 5) |
| Testes width (average) | 20–22 (21, n = 2) | 74–108 (92, n = 5) |
| Testicular field length | 613–766 (689, n = 2) | 1165–1862 (1491, n = 8) |
| Testicular field (percentage of body length) | 33% (n = 2) | 33–40% (36%, n = 8) |
| Vesicula seminalis length | 86–112 (99, n = 2) | 187–268 (234, n = 6) |
| Vesicula seminalis wide | 11–16 (13.5, n = 2) | 23–43 (31, n = 6) |
| Cirrus sac length | 86–114 (100, n = 2) | 184–283 (226, n = 7) |
| Cirrus sac width | 30–49 (39.6, n = 2) | 60–94 (76, n = 7) |
| Cirrus length | - | 47–65 (55, n = 3) |
| Cirrus width | 24–34 (29, n = 2) | 37–60 (45.27, n = 3) |
| Ovary length | 99–124 (111, n = 2) | 174–342 (244, n = 8) |
| Ovary width | 99–124 (111, n = 2) | 115–306 (211, n = 8) |
| Ovary from its posterior end to posterior end of body | 481–597 (539, n = 2) | 778–1100 (911, n = 8) |
| Ovary from its posterior end of body end (percentage of body length) | 25–26% (25.5%, n = 2) | 21–23% (22%, n = 8) |
| Oviduct length | 163–174 (169, n = 2) | 183–336 (262, n = 7) |
| Oviducal seminal receptacle length | 81–93 (87, n = 2) | 174–254 (214, n = 7) |
| Oviducal seminal receptacle width | 19–21 (20, n = 2) | 34–54 (44, n = 7) |
| Oötype length | 20–22 (21, n = 2) | 42–66 (57, n = 6) |
| Oötype width | 14–18 (16, n = 2) | 33–75 (50, n = 6) |
| Oötype from posterior end of body | 202–236 (219, n = 2) | 330–443 (361, n = 6) |
| Female genital pore from sinistral body margin | 28–31 (30, n = 2) | 102–174 (143, n = 5) |
| Distance between male and female genital pores | 145–208 (177, n = 2) | 167–204 (190, n = 5) |
| Female genital pore from posterior end of body | 409–548 (478, n = 2) | 616–753 (664, n = 5) |
| Eggs length | 18–21 (20, n = 20) | 26–37 (31, n = 15) |
| Eggs width | 13–16 (15, n = 20) | 28–28 (24, n = 15) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sepúlveda, F.A.; Ñacari, L.A.; González, M.T. First Report of Blood Fluke Pathogens with Potential Risk for Emerging Yellowtail Kingfish (Seriola lalandi) Aquaculture on the Chilean Coast, with Descriptions of Two New Species of Paradeontacylix (Aporocotylidae). Pathogens 2021, 10, 849. https://doi.org/10.3390/pathogens10070849
Sepúlveda FA, Ñacari LA, González MT. First Report of Blood Fluke Pathogens with Potential Risk for Emerging Yellowtail Kingfish (Seriola lalandi) Aquaculture on the Chilean Coast, with Descriptions of Two New Species of Paradeontacylix (Aporocotylidae). Pathogens. 2021; 10(7):849. https://doi.org/10.3390/pathogens10070849
Chicago/Turabian StyleSepúlveda, Fabiola A., Luis A. Ñacari, and Maria Teresa González. 2021. "First Report of Blood Fluke Pathogens with Potential Risk for Emerging Yellowtail Kingfish (Seriola lalandi) Aquaculture on the Chilean Coast, with Descriptions of Two New Species of Paradeontacylix (Aporocotylidae)" Pathogens 10, no. 7: 849. https://doi.org/10.3390/pathogens10070849
APA StyleSepúlveda, F. A., Ñacari, L. A., & González, M. T. (2021). First Report of Blood Fluke Pathogens with Potential Risk for Emerging Yellowtail Kingfish (Seriola lalandi) Aquaculture on the Chilean Coast, with Descriptions of Two New Species of Paradeontacylix (Aporocotylidae). Pathogens, 10(7), 849. https://doi.org/10.3390/pathogens10070849





