Temperature Variation and Host Immunity Regulate Viral Persistence in a Salmonid Host
Abstract
:1. Introduction
2. Materials and Methods
2.1. Virus and Cell Culture
2.2. Viral Replication in Cell Culture at Different Temperatures
2.3. Steelhead Survival after Virus Exposure at Different Temperatures
2.4. Viral Decay in Steelhead Exposed to Virus at Three Temperatures
2.5. Individual Steelhead Tissues Harboring Long-Term Persistent IHNV RNA
2.6. Steelhead Specific Immune Response to IHNV
3. Results
3.1. Temperature-Dependent Virus Replication in Cell Culture
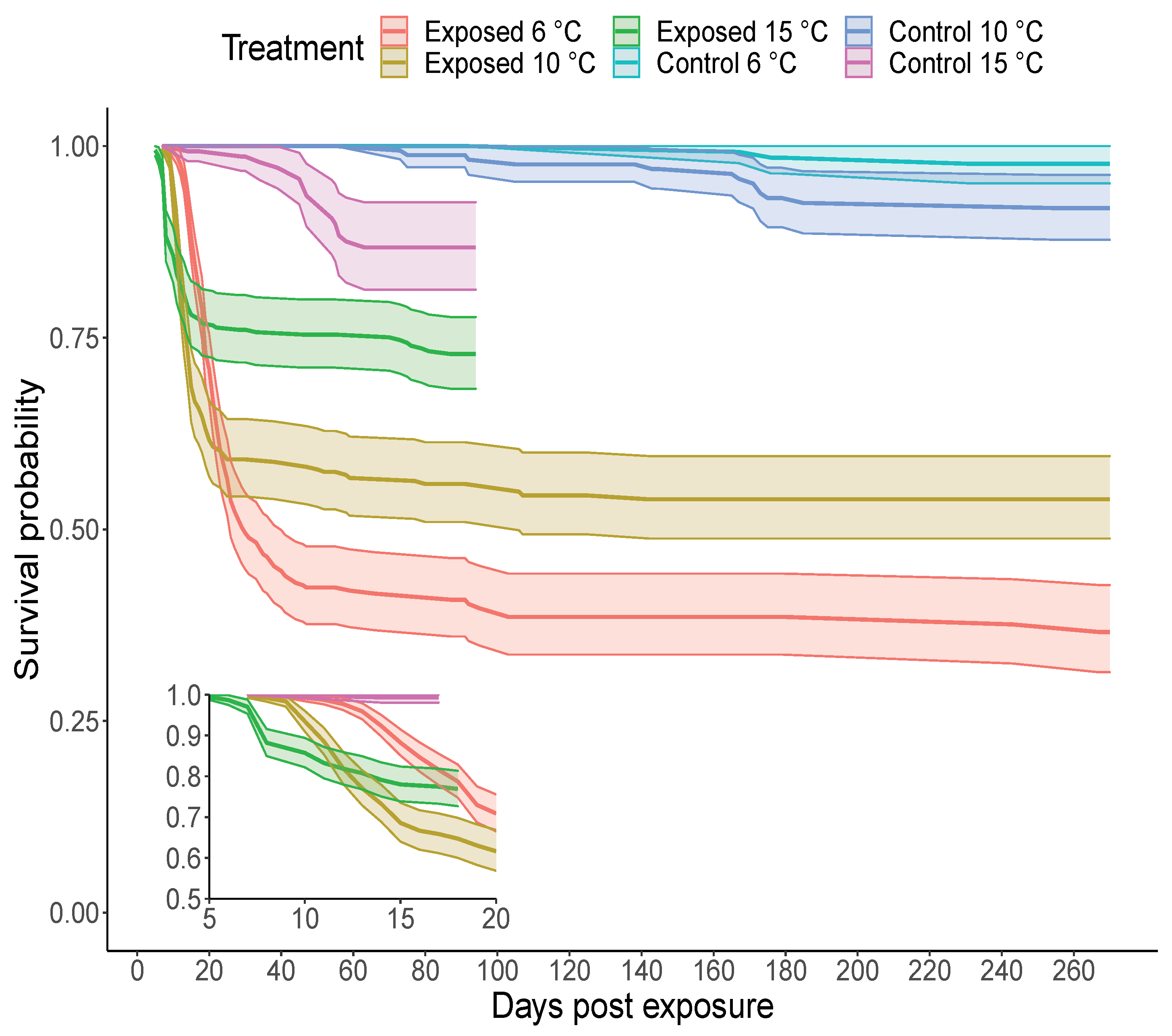
3.2. Survival of Steelhead Following Virus Exposure at Three Temperatures
3.3. Viral Decay in Steelhead Exposed to Virus at Three Temperatures
3.4. Specific Steelhead Tissues Harboring IHNV RNA
3.5. Steelhead Specific Immune Response to IHNV
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| MDPI | Multidisciplinary Digital Publishing Institute |
| DOAJ | Directory of open access journals |
| TLA | Three letter acronym |
| LD | Linear dichroism |
References
- Elderd, B.D.; Rehill, B.J.; Haynes, K.J.; Dwyer, G. Induced plant defenses, host–pathogen interactions, and forest insect outbreaks. Proc. Natl. Acad. Sci. USA 2013, 110, 14978–14983. [Google Scholar] [CrossRef] [Green Version]
- Kyle, C.H.; Liu, J.; Gallagher, M.E.; Dukic, V.; Dwyer, G. Stochasticity and Infectious Disease Dynamics: Density and Weather Effects on a Fungal Insect Pathogen. Am. Nat. 2020, 195, 504–523. [Google Scholar] [CrossRef]
- Hall, S.R.; Becker, C.R.; Duffy, M.A.; Cáceres, C.E. Variation in resource acquisition and use among host clones creates key epidemiological trade-offs. Am. Nat. 2010, 176, 557–565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boots, M. The evolution of resistance to a parasite is determined by resources. Am. Nat. 2011, 178, 214–220. [Google Scholar] [CrossRef] [PubMed]
- Hedrick, R. Relationships of the host, pathogen, and environment: Implications for diseases of cultured and wild fish populations. J. Aquat. Anim. Health 1998, 10, 107–111. [Google Scholar] [CrossRef]
- Marcos-López, M.; Gale, P.; Oidtmann, B.; Peeler, E. Assessing the impact of climate change on disease emergence in freshwater fish in the United Kingdom. Transbound. Emerg. Dis. 2010, 57, 293–304. [Google Scholar] [CrossRef]
- Okamura, B.; Feist, S.W. Emerging diseases in freshwater systems. Freshw. Biol. 2011, 56, 627–637. [Google Scholar] [CrossRef]
- Kurath, G. Molecular epidemiology and evolution of Novirhabdoviruses. In Molecular Taxonomy, Evolution, Genomics, Ecology, Cytopathology, and Control; Dietzgen, R., Kuzmin, I., Eds.; Caister Academic Press: Norfolk, UK, 2012; pp. 89–116. [Google Scholar]
- Bootland, L.; Leong, J.C. Infectious hematopoietic necrosis virus. In Fish Diseases and Disorders. Volume 3; Woo, P.T.K., Bruno, D.W., Eds.; CAB International: Wallingford, UK, 2011; pp. 66–109. [Google Scholar]
- Kurath, G.; Garver, K.A.; Troyer, R.M.; Emmenegger, E.J.; Einer-Jensen, K.; Anderson, E.D. Phylogeography of infectious haematopoietic necrosis virus in North America. J. Gen. Virol. 2003, 84, 803–814. [Google Scholar] [CrossRef]
- LaPatra, S.; Lauda, K.; Jones, G.; Walker, S.; Shewmaker, B.; Morton, A. Characterization of IHNV isolates associated with neurotropism. Vet. Res. 1995, 26, 433–437. [Google Scholar]
- Engelking, H.M.; Leong, J. Glycoprotein from infectious hematopoietic necrosis virus (IHNV) induces protective immunity against five IHNV types. J. Aquat. Anim. Health 1989, 1, 291–300. [Google Scholar] [CrossRef]
- Engelking, H.M.; Harry, J.B.; Leong, J.A.C. Comparison of representative strains of infectious hematopoietic necrosis virus by serological neutralization and cross-protection assays. Appl. Environ. Microbiol. 1991, 57, 1372–1378. [Google Scholar] [CrossRef] [Green Version]
- Purcell, M.K.; Laing, K.J.; Winton, J.R. Immunity to fish rhabdoviruses. Viruses 2012, 4, 140–166. [Google Scholar] [CrossRef] [Green Version]
- Müller, A.; Sutherland, B.J.; Koop, B.F.; Johnson, S.C.; Garver, K.A. Infectious hematopoietic necrosis virus (IHNV) persistence in Sockeye Salmon: Influence on brain transcriptome and subsequent response to the viral mimic poly (I:C). BMC Genom. 2015, 16, 634. [Google Scholar] [CrossRef] [PubMed]
- Dixon, P.; Paley, R.; Alegria-Moran, R.; Oidtmann, B. Epidemiological characteristics of infectious hematopoietic necrosis virus (IHNV): A review. Vet. Res. 2016, 47, 63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hershberger, P.K.; van der Leeuw, B.K.; Gregg, J.L.; Grady, C.A.; Lujan, K.M.; Gutenberger, S.K.; Purcell, M.K.; Woodson, J.C.; Winton, J.R.; Parsley, M.J. Amplification and transport of an endemic fish disease by an introduced species. Biol. Invasions 2010, 12, 3665–3675. [Google Scholar] [CrossRef]
- Neukirch, M. Demonstration of persistent viral haemorrhagic septicaemia (VHS) virus in rainbow trout after experimental waterborne infection. J. Vet. Med. Ser. B 1986, 33, 471–476. [Google Scholar] [CrossRef]
- Amend, D.F. Control of infectious hematopoietic necrosis virus disease by elevating the water temperature. J. Fish. Board Can. 1970, 27, 265–270. [Google Scholar] [CrossRef]
- Hetrick, F.M.; Knittel, M.D.; Fryer, J. Increased susceptibility of rainbow trout to infectious hematopoietic necrosis virus after exposure to copper. Appl. Environ. Microbiol. 1979, 37, 198–201. [Google Scholar] [CrossRef] [Green Version]
- Garver, K.A.; Batts, W.N.; Kurath, G. Virulence comparisons of infectious hematopoietic necrosis virus U and M genogroups in sockeye salmon and rainbow trout. J. Aquat. Anim. Health 2006, 18, 232–243. [Google Scholar] [CrossRef] [PubMed]
- Amend, D.F. Detection and transmission of infectious hematopoietic necrosis virus in rainbow trout. J. Wildl. Dis. 1975, 11, 471–478. [Google Scholar] [CrossRef] [PubMed]
- Drolet, B.S.; Chiou, P.P.; Heidel, J.; Leong, J. Detection of truncated virus particles in a persistent RNA virus infection in vivo. J. Virol. 1995, 69, 2140–2147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, C.H.; Dummer, D.M.; Chiou, P.P.; Leong, J.A.C. Truncated particles produced in fish surviving infectious hematopoietic necrosis virus infection: Mediators of persistence? J. Virol. 1999, 73, 843–849. [Google Scholar] [CrossRef] [Green Version]
- St-Hilaire, S.; Ribble, C.; Traxler, G.; Davies, T.; Kent, M.L. Evidence for a carrier state of infectious hematopoietic necrosis virus in chinook salmon Oncorhynchus tshawytscha. Dis. Aquat. Org. 2001, 46, 173–179. [Google Scholar] [CrossRef]
- Foott, J.S.; Free, D.; McDowell, T.; Arkush, K.D.; Hedrick, R.P. Infectious hematopoietic necrosis virus transmission and disease among juvenile Chinook salmon exposed in culture compared to environmentally relevant conditions. San Fr. Estuary Watershed Sci. 2006, 4. [Google Scholar] [CrossRef] [Green Version]
- Amos, K.H.; Hopper, K.A.; Levander, L. Absence of infectious hematopoietic necrosis virus in adult sockeye salmon. J. Aquat. Anim. Health 1989, 1, 281–283. [Google Scholar] [CrossRef]
- LaPatra, S.; Batts, W.; Overturf, K.; Jones, G.; Shewmaker, W.; Winton, J. Negligible risk associated with the movement of processed rainbow trout, Oncorhynchus mykiss (Walbaum), from an infectious haematopoietic necrosis virus (IHNV) endemic area. J. Fish Dis. 2001, 24, 399–408. [Google Scholar] [CrossRef]
- Garver, K.A.; Mahony, A.A.; Stucchi, D.; Richard, J.; Van Woensel, C.; Foreman, M. Estimation of parameters influencing waterborne transmission of infectious hematopoietic necrosis virus (IHNV) in Atlantic salmon (Salmo salar). PLoS ONE 2013, 8, e82296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Breyta, R.; Jones, A.; Stewart, B.; Brunson, R.; Thomas, J.; Kerwin, J.; Bertolini, J.; Mumford, S.; Patterson, C.; Kurath, G. Emergence of MD type infectious hematopoietic necrosis virus in Washington State coastal steelhead trout. Dis. Aquat. Org. 2013, 104, 179–195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Batts, W.; Winton, J. Enhanced detection of infectious hematopoietic necrosis virus and other fish viruses by pretreatment of cell monolayers with polyethylene glycol. J. Aquat. Anim. Health 1989, 1, 284–290. [Google Scholar] [CrossRef]
- Burnham, K.; Anderson, D. Model Selection and Multimodel Inference: A Practical Information-Theoretic Approach; Springer: New York, NY, USA, 2003. [Google Scholar]
- Schemper, M.; Wakounig, S.; Heinze, G. The estimation of average hazard ratios by weighted Cox regression. Stat. Med. 2009, 28, 2473–2489. [Google Scholar] [CrossRef]
- Therneau, T.M.; Grambsch, P.M. Modeling Survival Data: Extending the Cox Model; Springer: New York, NY, USA, 2000. [Google Scholar]
- Dunkler, D.; Ploner, M.; Schemper, M.; Heinze, G. Weighted Cox Regression Using the R Package coxphw. J. Stat. Softw. 2018, 84, 1–26. [Google Scholar] [CrossRef] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018. [Google Scholar]
- Purcell, M.K.; Thompson, R.L.; Garver, K.A.; Hawley, L.M.; Batts, W.N.; Sprague, L.; Sampson, C.; Winton, J.R. Universal reverse-transcriptase real-time PCR for infectious hematopoietic necrosis virus (IHNV). Dis. Aquat. Org. 2013, 106, 103–115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pinheiro, J.; Bates, D.; DebRoy, S.; Sarkar, D.; R Core Team. NLME: Linear and Nonlinear Mixed Effects Models, 2019. R Package Version 3.1-141. Available online: https://CRAN.R-project.org/package=nlme (accessed on 14 June 2021).
- LaPatra, S.; Turner, T.; Lauda, K.; Jones, G.; Walker, S. Characterization of the humoral response of rainbow trout to infectious hematopoietic necrosis virus. J. Aquat. Anim. Health 1993, 5, 165–171. [Google Scholar] [CrossRef]
- Hart, L.M.; MacKenzie, A.; Purcell, M.K.; Powers, R.L.; Hershberger, P.K. Optimization of a Plaque Neutralization Test (PNT) to Identify the Exposure History of Pacific Herring to Viral Hemorrhagic Septicemia Virus (VHSV). J. Aquat. Anim. Health 2017, 29, 74–82. [Google Scholar] [CrossRef] [PubMed]
- Stewart, A.; Hablützel, P.I.; Brown, M.; Watson, H.V.; Parker-Norman, S.; Tober, A.V.; Thomason, A.G.; Friberg, I.M.; Cable, J.; Jackson, J.A. Half the story: Thermal effects on within-host infectious disease progression in a warming climate. Glob. Chang. Biol. 2018, 24, 371–386. [Google Scholar] [CrossRef] [PubMed]
- Harvell, C.D.; Mitchell, C.E.; Ward, J.R.; Altizer, S.; Dobson, A.P.; Ostfeld, R.S.; Samuel, M.D. Climate warming and disease risks for terrestrial and marine biota. Science 2002, 296, 2158–2162. [Google Scholar] [CrossRef] [Green Version]
- Mulcahy, D.; Pascho, R.; Jenes, C. Comparison of in vitro growth characteristics of ten isolates of infectious haematopoietic necrosis virus. J. Gen. Virol. 1984, 65, 2199–2207. [Google Scholar] [CrossRef]
- Amend, D.F.; Smith, L. Pathophysiology of infectious hematopoietic necrosis virus disease in rainbow trout (Salmo gairdneri): Early changes in blood and aspects of the immune response after injection of IHN virus. J. Fish. Board Can. 1974, 31, 1371–1378. [Google Scholar] [CrossRef]
- Troyer, R.M.; LaPatra, S.E.; Kurath, G. Genetic analyses reveal unusually high diversity of infectious haematopoietic necrosis virus in rainbow trout aquaculture. J. Gen. Virol. 2000, 81, 2823–2832. [Google Scholar] [CrossRef] [PubMed]
- Garver, K.A.; Troyer, R.M.; Kurath, G. Two distinct phylogenetic clades of infectious hematopoietic necrosis virus overlap within the Columbia River basin. Dis. Aquat. Org. 2003, 55, 187–203. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.H.; Yusuff, S.; Vakharia, V.N.; Evensen, Ø. Interchange of L polymerase protein between two strains of viral hemorrhagic septicemia virus (VHSV) genotype IV alters temperature sensitivities in vitro. Virus Res. 2015, 195, 203–206. [Google Scholar] [CrossRef]
- Holt, R.; Amandi, A.; Rohovec, J.; Fryer, J. Relation of water temperature to bacterial cold-water disease in coho salmon, chinook salmon, and rainbow trout. J. Aquat. Anim. Health 1989, 1, 94–101. [Google Scholar] [CrossRef]
- Purcell, M.K.; Powers, R.; Evered, J.; Kerwin, J.; Meyers, T.R.; Stewart, B.; Winton, J. Molecular testing of adult Pacific salmon and trout (Oncorhynchus spp.) for several RNA viruses demonstrates widespread distribution of piscine orthoreovirus in Alaska and Washington. J. Fish Dis. 2018, 41, 347–355. [Google Scholar] [CrossRef] [PubMed]
- Rozas-Serri, M.; Lobos, C.; Correa, R.; Ildefonso, R.; Vásquez, J.; Muñoz, A.; Maldonado, L.; Jaramillo, V.; Coñuecar, D.; Oyarzún, C.; et al. Atlantic salmon pre-smolt survivors of Renibacterium salmoninarum infection show inhibited cell-mediated adaptive immune response and a higher risk of death during the late stage of infection at lower water temperatures. Front. Immunol. 2020, 11, 1378. [Google Scholar] [CrossRef]
- Hershberger, P.; Rhodes, L.; Kurath, G.; Winton, J. Infectious diseases of fishes in the Salish Sea. Fisheries 2013, 38, 402–409. [Google Scholar] [CrossRef]
- Evelyn, T. An improved growth medium for the kidney disease bacterium and some notes on using the medium. Bull. Off. Int. Epizoot. 1977, 87, 511–513. [Google Scholar]
- Ray, R.A.; Holt, R.A.; Bartholomew, J.L. Relationship between temperature and Ceratomyxa shasta–induced mortality in Klamath River salmonids. J. Parasitol. 2012, 98, 520–526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arkush, K.D.; Mendonca, H.L.; McBride, A.M.; Yun, S.; McDowell, T.S.; Hedrick, R.P. Effects of temperature on infectivity and of commercial freezing on survival of the North American strain of viral hemorrhagic septicemia virus (VHSV). Dis. Aquat. Org. 2006, 69, 145–151. [Google Scholar] [CrossRef]
- Purcell, M.K.; Kurath, G.; Garver, K.A.; Herwig, R.P.; Winton, J.R. Quantitative expression profiling of immune response genes in rainbow trout following infectious haematopoietic necrosis virus (IHNV) infection or DNA vaccination. Fish Shellfish Immunol. 2004, 17, 447–462. [Google Scholar] [CrossRef]
- Penaranda, M.M.D.; Purcell, M.K.; Kurath, G. Differential virulence mechanisms of infectious hematopoietic necrosis virus in rainbow trout (Oncorhynchus mykiss) include host entry and virus replication kinetics. J. Gen. Virol. 2009, 90, 2172–2182. [Google Scholar] [CrossRef] [PubMed]
- Lorenzen, E.; Einer-Jensen, K.; Rasmussen, J.S.; Kjær, T.E.; Collet, B.; Secombes, C.; Lorenzen, N. The protective mechanisms induced by a fish rhabdovirus DNA vaccine depend on temperature. Vaccine 2009, 27, 3870–3880. [Google Scholar] [CrossRef] [PubMed]
- Le Morvan, C.; Troutaud, D.; Deschaux, P. Differential effects of temperature on specific and nonspecific immune defences in fish. J. Exp. Biol. 1998, 201, 165–168. [Google Scholar] [CrossRef] [PubMed]
- Lorenzen, N.; Lapatra, S.E. Immunity to rhabdoviruses in rainbow trout: The antibody response. Fish Shellfish Immunol. 1999, 9, 345–360. [Google Scholar] [CrossRef]
- Press, C.M.; Evensen, Ø. The morphology of the immune system in teleost fishes. Fish Shellfish Immunol. 1999, 9, 309–318. [Google Scholar] [CrossRef] [Green Version]
- Randall, R.E.; Griffin, D.E. Within host RNA virus persistence: Mechanisms and consequences. Curr. Opin. Virol. 2017, 23, 35–42. [Google Scholar] [CrossRef] [Green Version]
- Hershberger, P.K.; MacKenzie, A.; Gregg, J.L.; Wilmot, M.D.; Powers, R.L.; Purcell, M.K. Long-term shedding from fully convalesced individuals indicates that Pacific herring are a reservoir for viral hemorrhagic septicemia virus. Dis. Aquat. Org. 2021, 144, 245–252. [Google Scholar] [CrossRef]
- Purcell, M.K.; Powers, R. Survival, Viral Load and Neutralizing Antibodies in Steelhead Trout and Cell Cultures Exposed to Infectious Hematopoietic Necrosis Virus (IHNV) at 3 Temperatures: U.S. Geological Survey Data Release. 2021. Available online: https://doi.org/10.5066/P9T4PH4Z (accessed on 14 June 2021).



| Temperature | Final Viral Load (L) | Rate of Decay (VL/Day) | Half-Time (dpe) |
|---|---|---|---|
| 6 | 1.98 (1.33, 2.62) | 0.026 (0.008, 0.043) | 59 (35, 189) |
| 10 | 2.14 (1.75, 2.52) | 0.087 (0.041, 0.134) | 12 (7, 30) |
| 15 | 1.59 (1.15, 2.03) | 0.242 (−0.232, 0.716) | 3 (NA) |
| Days Post Exposure | ||||
|---|---|---|---|---|
| Temperature | Dilution | 31 | 55 | 91 |
| Negative | 1.0 (15/15) | 0.73 (22/30) | 0.87 (13/15) | |
| Small | - | 0.13 (4/30) | 0.13 (2/15) | |
| 6 | Intermediate | - | 0.10 (3/30) | - |
| Large | - | 0.03 (1/30) | - | |
| Negative | 1.0 (15/15) | 0.33 (10/30) | 0.73(11/15) | |
| Small | - | 0.20 (6/30) | 0.20 (3/15) | |
| 10 | Intermediate | - | 0.26 (8/30) | 0.07 (1/15) |
| Large | - | 0.20 (6/30) | - | |
| Negative | 0.78 (14/18) | 0.38 (11/29) | 0.61 (17/28) | |
| Small | 0.22 (4/18) | 0.38 (11/29) | 0.14 (4/28) | |
| 15 | Intermediate | - | 0.10 (3/29) | 0.10 (3/28) |
| Large | - | 0.14 (4/29) | 0.14 (4/28) | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Páez, D.J.; Powers, R.L.; Jia, P.; Ballesteros, N.; Kurath, G.; Naish, K.A.; Purcell, M.K. Temperature Variation and Host Immunity Regulate Viral Persistence in a Salmonid Host. Pathogens 2021, 10, 855. https://doi.org/10.3390/pathogens10070855
Páez DJ, Powers RL, Jia P, Ballesteros N, Kurath G, Naish KA, Purcell MK. Temperature Variation and Host Immunity Regulate Viral Persistence in a Salmonid Host. Pathogens. 2021; 10(7):855. https://doi.org/10.3390/pathogens10070855
Chicago/Turabian StylePáez, David J., Rachel L. Powers, Peng Jia, Natalia Ballesteros, Gael Kurath, Kerry A. Naish, and Maureen K. Purcell. 2021. "Temperature Variation and Host Immunity Regulate Viral Persistence in a Salmonid Host" Pathogens 10, no. 7: 855. https://doi.org/10.3390/pathogens10070855
APA StylePáez, D. J., Powers, R. L., Jia, P., Ballesteros, N., Kurath, G., Naish, K. A., & Purcell, M. K. (2021). Temperature Variation and Host Immunity Regulate Viral Persistence in a Salmonid Host. Pathogens, 10(7), 855. https://doi.org/10.3390/pathogens10070855






