Abstract
Streptococcus suis is a pathogen that causes invasive infections in humans and pigs. In this study, 448 S. suis isolates recovered from human infections in Thailand were characterized with regard to their antimicrobial susceptibility and antimicrobial resistance genes, including, for non-penicillin-susceptible isolates, sequence analyses of five genes encoding penicillin-binding proteins (pbp1a, pbp1b, pbp2a, pbp2b, and pbp2x). All 448 isolates were susceptible to cefepime and ceftriaxone, whereas 99.6%, 91.7%, and 72.9% of the isolates were susceptible to levofloxacin, penicillin, and chloramphenicol, respectively. Almost all isolates were resistant to tetracycline (98.2%), clindamycin (94%), erythromycin (92.4%), and azithromycin (82.6%). Genes tet(O) and ermB were the predominant resistance genes detected among macrolide- and tetracycline-resistant isolates. A total of 37 out of 448 isolates (8.2%) showed intermediately resistance to penicillin. Most of these isolates (59.5%) belonged to serotype 2-ST233. Comparison of the predicted translated sequences of five PBP proteins of a penicillin-susceptible isolate (strain P1/7) to the respective PBP sequences of ten non-penicillin-susceptible isolates revealed multiple amino acid substitutions. Isolates of CC221/234 showed highly variable amino acid substitutions in all PBP proteins. An ST104 isolate had a higher number of amino acid substitutions in PBP2X. Isolates belonging to CC233/379 had numerous substitutions in PBP2B and PBP2X. ST25 isolates exhibited fewer amino acid substitutions than isolates of other STs in all five PBPs. The antimicrobial resistance of S. suis is increasing worldwide; therefore, restrictions on antimicrobial use, continuous control, and the surveillance of this bacterium throughout the pork supply chain are crucial for ensuring public health and must be a priority concern.
1. Introduction
Streptococcus suis, an important zoonotic pathogen, causes invasive infections in humans in close contact with infected pigs or contaminated pork-derived products [1]. The number of reported human cases, especially in Southeast Asian countries, has dramatically increased in the past few years [1,2]. Among the 29 serotypes of S. suis, serotype 2 is the most common cause of human infections, although human cases due to serotypes 4, 5, 9, 14, 16, 21, 24, and 31 have also been reported [1,3,4,5,6,7,8,9].
Although some have raised the alarm about the risks posed by increasing antimicrobial resistance in S. suis, including acquisition/transmission and the organism serving as a reservoir for resistance genes, S. suis has not yet been broadly acknowledged as a concern by the medical community [10,11]. One reason for this is that antimicrobial resistance data for S. suis continue to be scarce. Furthermore, most available antimicrobial resistance data have been generated by investigating S. suis isolates recovered from pigs [11,12,13,14,15,16,17], whereas those for human S. suis isolates are minimal [16,18,19,20]. The mass use of antibiotics in the livestock industry in many countries, especially in pigs for growth promotion, routine prophylaxis, or treatment for controlling the spread of infection, has potentially led to antimicrobial resistance in the microbial communities of food animal production under such conditions [2].
Here, we report the antimicrobial susceptibility and resistance gene profiles of 448 S. suis isolates of different serotypes and genetic backgrounds recovered from human infections in Thailand. We also sequenced the genomes of non-penicillin-susceptible isolates to begin to reveal the molecular reasons for decreased penicillin susceptibility in S. suis.
2. Results
2.1. Antimicrobial Susceptibility
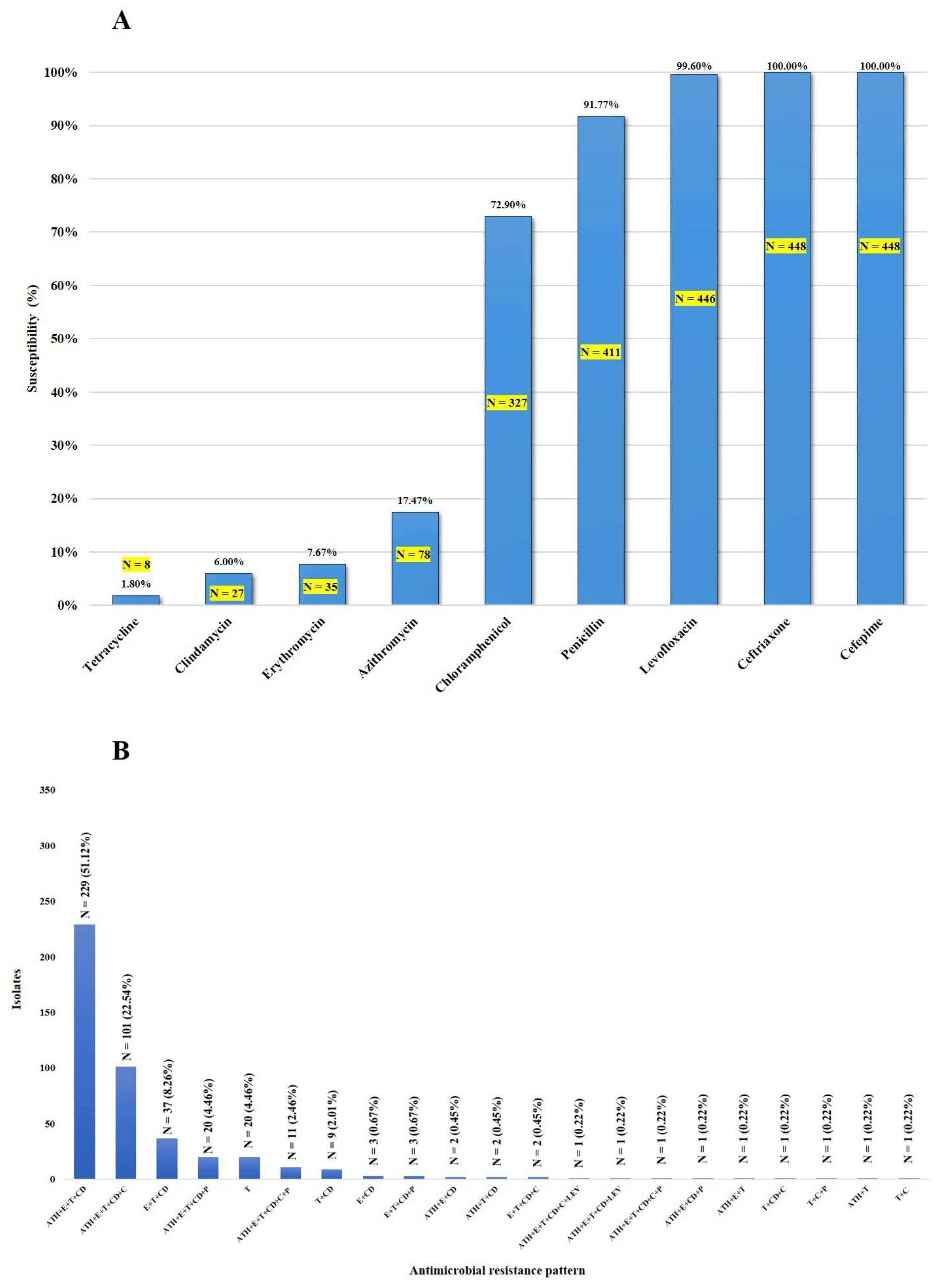
As shown in Figure 1A, all 448 isolates (one isolate/patient) were susceptible to cefepime and ceftriaxone, whereas 99.6%, 91.7%, and 72.9% were susceptible to levofloxacin, penicillin, and chloramphenicol, respectively. Thirty-seven isolates (8.3%) showed intermediate resistance to penicillin (Table 1). Among these non-penicillin-susceptible isolates, serotype 2-ST233 was predominant (22/37; 59.5%). The vast majority of the isolates were resistant to tetracycline (98.2%), clindamycin (94%), erythromycin (92.4%), and azithromycin (82.6%). As shown in Figure 1B and Table S1, the macrolide-lincosamide–tetracycline (azithromycin (ATH)-erythromycin (E)-clindamycin (CD)-tetracycline-T) resistance pattern was the most frequently found (51.1%), followed by macrolide-lincosamide-tetracycline-phenicol (azithromycin (ATH)-erythromycin (E)-clindamycin (CD)-tetracycline (T)-chloramphenicol (C)) (22.5%). Of the 448 isolates, 410 (91.5%) were considered to be multidrug-resistant (MDR), as shown in Figure 1B. This study demonstrates that some S. suis isolates exhibit non-susceptibility to five antimicrobial categories (at least one agent in each category), either macrolide-lincosamide-tetracycline-chloramphenicol-penicillin (N = 10) or macrolide-lincosamide-tetracycline-chloramphenicol-levofloxacin (N = 1), as shown in Figure 1B. All 37 non-penicillin-susceptible isolates exhibited the MDR phenotype (Table S1).
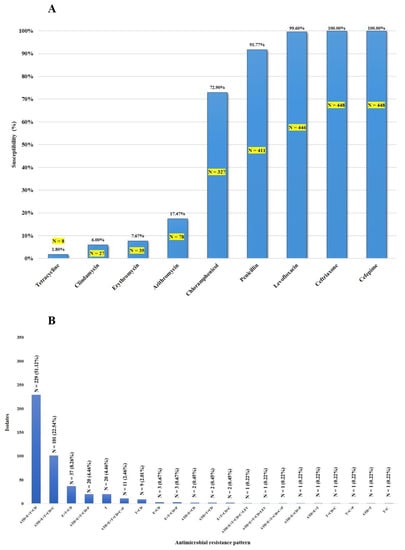
Figure 1.
Susceptibility of S. suis isolates to antimicrobial agents (A) and the distribution of antimicrobial resistance profiles of S. suis isolates (B).

Table 1.
Minimal inhibitory concentrations of penicillin against intermediately resistant S. suis isolates.
Information about the antibiotic treatments of patients was available for 445 isolates (Table S2). Indeed, 95.7% of cases were treated with ceftriaxone and 4.3% were treated with a combination of penicillin and ceftriaxone. According to antimicrobial susceptibility results, all of these isolates were susceptible to ceftriaxone and cefepime that belonged to third-generation cephalosporins.
2.2. Macrolide- and Tetracycline-Resistance Genes
Table 2 shows that ermB was the most commonly detected macrolide-resistance gene among the isolates (N = 384; 85.7%). None of the isolates carried mef (A/E) and msrD. However, 64 isolates (14.3%) were phenotypically resistant to macrolides, although ermB, mef (A/E), or msrD were not detected, suggesting that the macrolide resistance exhibited by these isolates is due to other mechanisms. In the case of tetracycline-resistance genes, gene tet (O) was the most commonly identified (N = 409, 91.3%). A total of 26 isolates (5.8%) carried tet (O) + tet (W). Other tet genes detected were tet (M), tet (M)+tet (L), and tet (W). Nine isolates (2%) had a tetracycline-resistant phenotype, but we did not detect tet, tcr3, otrB, or otrC genes by PCR in these organisms. Among the macrolide- and tetracycline-resistance genes detected in 410 MDR S. suis isolates, ermB+tet (O) was predominant, found in 337 (82.2%) isolates (Table S1). Only tet (O) was found in 39 (9.5%) isolates, and ermB + tet (O) + tet (W) was detected in 24 (5.9%) isolates. The remaining MDR S. suis isolates exhibited ermB for 6 (1.5%) isolates, tet (L) + tet (M) for 2 (0.5%) isolates, and one isolate each for tet (O) + tet (W) and only tet (W), as shown in Table S1.

Table 2.
Distribution of macrolide- and tetracycline-resistance genes in S. suis.
2.3. Whole-Genome Analysis of Non-penicillin-Susceptible S. suis Isolates
Of the 37 non-penicillin-susceptible isolates, 10 were selected for whole-genome sequence analysis, including 4 ST233 isolates and 1 isolate each of ST25, ST104, ST105, ST221, ST234, and ST379. Resfinder and CARD were used to identify antimicrobial resistance genes in these isolates (Table 3). Genes ermB and tet (O) were found in all 10 isolates. Additional macrolide resistance genes included gene Inu (A), detected in ST105 (serotype 14; ID32494), whereas genes Inu (B) and Isa (E) were found in ST104 (serotype 2; ID27715). In addition, this ST104 also carried aminoglycoside-resistance genes aac (6′)-aph (2″) and aph (3′)-III (Table 3).

Table 3.
Whole-genome analysis of 10 non-penicillin-susceptible S. suis isolates isolated from humans.
Analysis of plasmid replicon types with PlasmidFinder identified a rep21-type replicon only in the ST105 isolate. However, PLACNETw analysis revealed that MOBP and MOPT replicon types were frequently detected in our isolates (N = 8 in both cases). Interestingly, isolates in CC104 and CC233/379 had the same MOB replicon types, whereas ST221 and ST234 exhibited MOBP and MOBV. ST25 carried plasmid replicon types MOBV and MOBT. ST235 (serotype 5) had three MOB replicon types (MOBP, MOBV, and MOBT). It was apparent that MOB type plasmids were specific to ST or CC. A summary of the plasmid replicon types is shown in Table 3.
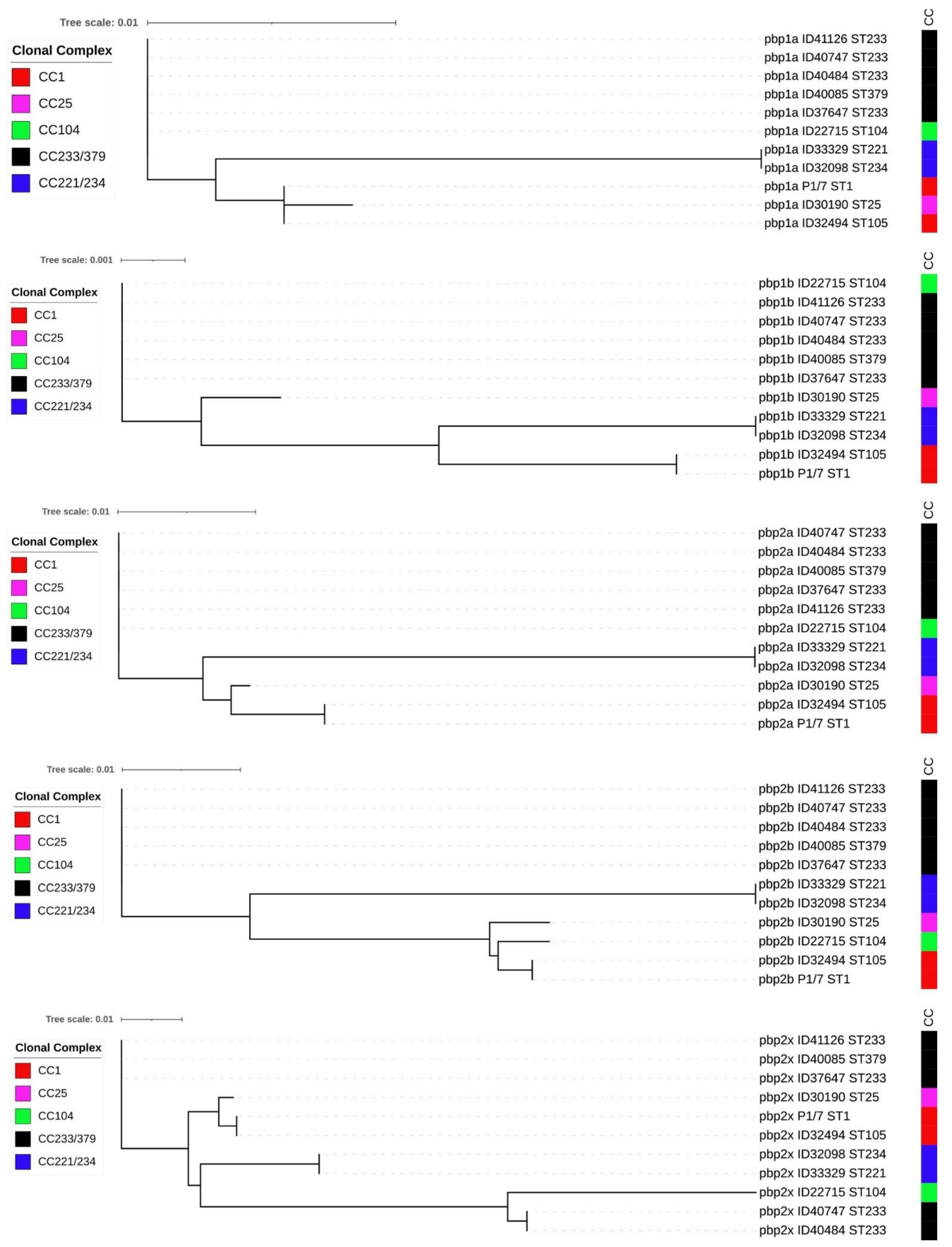
Analysis of five pbp genes (pbp1a, pbp1b, pbp2a, pbp2b, and pbp2x) of ten penicillin-non-susceptible isolates compared to S. suis P1/7 (penicillin-susceptible isolate) revealed multiple nucleotide substitutions impacting primary protein sequences throughout all pbp genes (Table 3 and Supplementary Files 1–5). In contrast with other isolates, strains ID33329 (ST221) and ID32098 (ST234) of CC221/234 had highly variable numbers of amino acid substitutions. Isolate ID27715 (ST104) had higher rates of amino acid substitutions in the predicted translated sequence of pbp2x than in other pbp gene-encoded proteins. Isolates in CC233/379 had high amino acid substitutions in the predicted pbp2b and pbp2x gene products. Isolate ID30190 (ST25) exhibited few amino acid substitutions in the translated sequences of all five pbp genes. In contrast, we did not identify substitutions in an ST105 isolate (ID32494), although it displayed intermediate resistance.
In addition, insertions of amino acids were detected between amino acid positions 708 and 709 in PBP1A of isolate ID32098 (ST234) and ID33329 (ST221), and between positions 433 and 434 of PBP2B for isolate ID30190 (ST25) (Supplementary Files 1 and 4). We also detected deletions of amino acid in the PBP2B of seven isolates at position 433 and two isolates at position 672 (Supplementary File 4). Phylogenetic trees were constructed using the PBP amino acid sequences of these non-penicillin-susceptible isolates (Figure 2). Five of these phylogenetic trees showed different clustering of the isolates, which were concordant with STs. ST104 (CC104), ST233 (CC233/379), and ST379 (CC233/379) isolates clustered together in the phylogenetic trees of PBP1A, PBP1B, PBP2A, and PBP2B, although they belong to different clonal complex. However, the PBP2X tree revealed that some isolates of ST233 and ST379 clustered separately from ST104 and ST233 isolates (Figure 2).
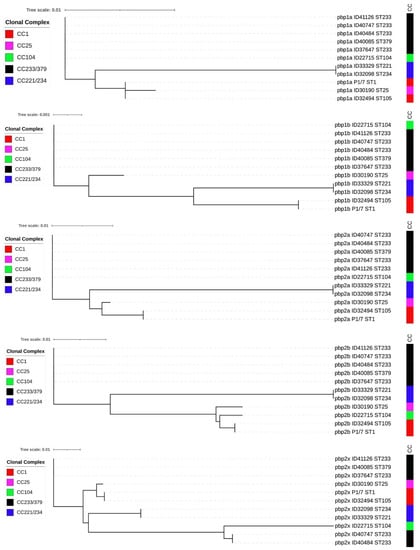
Figure 2.
Phylogenetic tree of 5 PBP amino acid sequences of 10 non-penicillin-susceptible S. suis isolates and penicillin-susceptible S. suis P1/7.
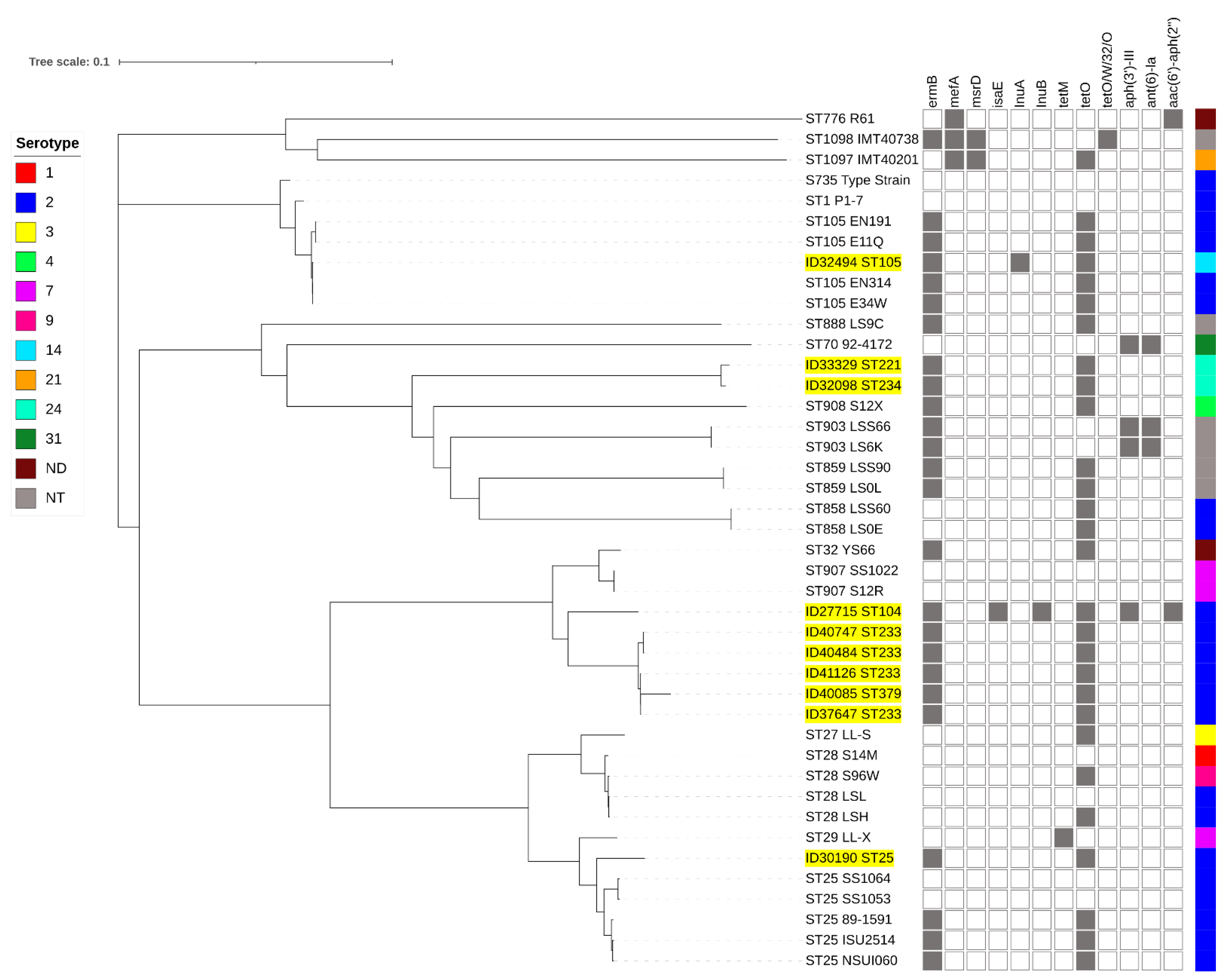
Full-genome SNP-based phylogenetic analysis showed that the penicillin-non-susceptible isolates were distributed throughout the phylogenetic tree (Figure 3). However, they were clustered accordingly to their STs or CCs. Isolates of STs 104 (ID27715), 233 (ID37647, ID40484, ID40747, and ID41126), and 379 (ID40085) clustered together, whereas ID30190-ST25 clustered with other ST25 isolates. ID32494 (ST105) clustered with CC1 isolates. The serotype 14-ST105 (ID32494) isolate was very closely related to serotype 2-ST105 isolates (E34W, EN314, E11Q, and EN191) originating from Vietnam. ST221 (ID33329) and ST234 (ID32098) isolates were very closely related to ST903, 908, 858, and 859 isolates.
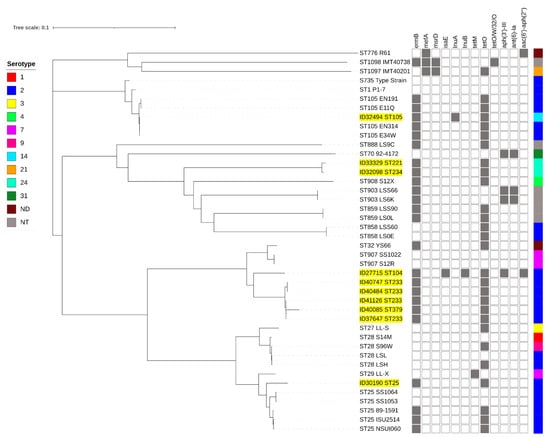
Figure 3.
Whole-genome phylogeny analysis of S. suis generated by CSI phylogeny and visualized with an interactive life tool tree. S. suis strains used in this study are highlighted in yellow. Presentation of the antimicrobial-resistance genes in each S. suis strain is shown by filled squares. ND, serotype is not determined; NT, non-typable.
3. Discussion
S. suis infections have a significant economic impact on the swine industry and are also a public health concern, with a multitude of reported human infections in Southeast Asian countries [1,11]. Other than the loss of lives, and frequent sequalae, a study in Thailand recently showed that S. suis human infections are responsible for an estimated USD 11.3 million loss in productivity-adjusted life years in the gross domestic product, which equates to USD 36,033 lost per person [21]. In addition to the economic burden, this bacterium is one of the main reasons for antimicrobial use in swine farms for routine prophylaxis or metaphylaxis, and a reservoir of antimicrobial-resistant genes [2,11]. Antimicrobial resistance is a major health problem; data suggest an increasing and alarming contribution of S. suis to this global threat [2].
Worldwide antimicrobial resistance data available for S. suis indicate that most isolates recovered from both humans and pigs have high resistance to tetracycline and moderate to high resistance to macrolides, e.g., erythromycin [12,13,14,15,16,17,19,22]. Our study confirms those observations. In addition, our data show that clindamycin (lincosamide) resistance was very common (94%) among S. suis human isolates characterized here. Other studies have shown a wide range of rates of resistance to clindamycin in S. suis isolates from pigs (38.5–98.7%) and humans (23.8–81.5%) [12,14,15,16,17,18,23]. On the other hand, a study from The Netherlands showed very low levels of S. suis resistance to clindamycin (0.5%) [24]. This may be due to differences in the usage of antimicrobials between Asia and Europe: there is highly extensive use in Asian countries but low levels of use in European countries [25,26].
Tetracycline-resistance gene presence in S. suis is variable. Genes tet (B), tet (L), tet (M), tet (O), tet (S), tet (W), tet (40), tet (O/32/O), and tet (O/W/32/O) have been reported [12,14,17,19,27,28]. Of these tet genes, tet(O), encoding the ribosome protection protein, is the most frequently described gene in S. suis isolates from pigs and humans worldwide [12,14,17,18,22,27,28]. In agreement with these previous findings, the tet (O) gene was the predominant tet gene among our isolates. Resistance to macrolides is mainly due to the action of erythromycin ribosomal methylases encoded by erm genes or to macrolide efflux pumps encoded by mef genes [22]. In this study, ermB (85.7%) was the most frequently detected macrolide gene, replicating previous findings in S. suis isolates recovered from both pigs and humans [14,17,18,22,23,27,28]. This gene could contribute to lincosamide and streptogamin B resistance due to the detected clindamycin resistance in S. suis isolates. However, 14.3% of S. suis in our study did not harbor ermB, mef (A/E), or msrD genes, suggesting that, similarly to what has been reported in a previous study [12], other mechanisms such as 23S rRNA methyl transferase (cfr) or macrolide phosphotransferase (mph(C)) are responsible for the macrolide-resistant phenotype displayed by the isolates analyzed here [22].
Despite the longstanding worldwide usage of β-lactams in pigs, the majority of S. suis isolates remain susceptible to this class of antibiotics. In this investigation, all human S. suis isolates were susceptible to ceftriaxone and cefepime, and most were susceptible to penicillin. However, we detected penicillin non-susceptibility (intermediate resistance) in a relatively significant percentage (8.3%) of our clinical isolates. Although several reports have shown different resistance rates to penicillin (from 0.5 up to as high as 62%) among S. suis isolates from pigs [15,16,17,23,24,29], our data differ greatly from what has been described thus far in terms of penicillin non-susceptibility for other human S. suis isolates [16,18,19,20]; therefore, our results are alarming.
Our data show that isolates belonging to serotype 2 ST233 were the most common strain type associated with penicillin non-susceptibility. Serotype 14 (ST105) and serotype 24 (ST221 and ST234) were also found to be intermediately resistant to penicillin. Previous studies have reported penicillin non-susceptibility in serotypes 2, 7, and 21 (ST1097), as well as in non-serotypeable (ST776, ST1098) isolates [15,16,17,23,30]. The mechanisms of β-lactam resistance in S. suis are not fully known. Our data expand on previous findings that analyzed four pbp genes (pbp2x, pbp2b, pbp1a, and pbp2a) in S. suis strain R61, and discovered the presence of multiple amino acid substitutions throughout the entire sequence of the predicted translated protein sequences of these genes [31]. Substitutions were mostly high in the PBP2X and PBP2B proteins, which are the targets for resistance to β-lactam antibiotics, similar to previous reports [30,31]. Mutations in PBP proteins may affect enzyme catalysis, binding-site affinity, stability, or structural configuring changes that lead to resistance to the antibiotic. However, our S. suis serotype 14 (ST105), which was intermediately resistant to penicillin, did not present any substitutions all five PBP proteins, suggesting that other mechanisms are involved. Indeed, as shown in S. pneumoniae and S. gordonii, penicillin non-susceptibility may occur via the involvement of other factors, such as a putative glycosyltransferase (cpoA), a histidine protein kinase (ciaH), and/or a putative iron permease (spr1178) [32,33,34].
Our phylogenetic analysis of five PBP proteins showed that isolate clusters were concordant with STs or CCs. This seems to indicate that the diversity of the PBP proteins is associated with STs or CCs. Therefore, the diversity of five PBP proteins should be extensively compared and analyzed using isolates belonging to different STs or serotypes in the future. Similarly, an SNP-based phylogenetic tree based on whole-genome analysis of our 10 non-penicillin-susceptible isolates, and closely related strains, showed that isolate clustering followed grouping by STs or CCs. Based on the MLST database, the cluster of STs104/233/379 was closely related to ST907 (serotype 7) which was isolated from pigs in the United Kingdom. ST221/234 was related to ST908 (serotype 4 and 9), ST903 (non-serotypeable), ST858 (serotype 2), and ST859 (non-serotypeable), which all were isolated from pigs in the United Kingdom. It is interesting that our serotype 14-ST105 isolate was closely related to serotype 2-ST105 isolates from Vietnam, which may suggest capsular gene switching [35].
4. Materials and Methods
4.1. Bacterial Isolates
A total of 448 isolates of S. suis from human specimens and belonging to different collection dates, serotypes, sequence types (STs), and regions in Thailand were included in the current study (Table S2). Serotypes and STs of these selected isolates were already known [5,6,7,8,36,37]. Of 448 isolates, 392 were serotype 2 and 49 were serotype 14, whereas the rest were serotype 24 (N =3), serotype 5 (N = 2), serotype 4 (N = 1), and serotype 9 (N = 1). Details of these isolates and their origins are shown in Tables S1 and S2.
4.2. Antimicrobial Susceptibility Testing
The broth microdilution technique was used according to the standards defined in the Clinical and Laboratory Standard Institute (CLSI) guidelines 2020 (M100-30th edn; CLSI, 2020) to determine the minimum inhibitory concentrations (MICs) of penicillin (≤0.12 μg/mL = Susceptible; 0.25–2 μg/mL = Intermediate; ≥4 μg/mL = Resistance) and ceftriaxone (≤1 μg/mL = Susceptible; 2 μg/mL = Intermediate; ≥4 μg/mL = Resistance) [38]. Susceptibility to other antimicrobials, such as cefepime, azithromycin, erythromycin, tetracycline, clindamycin, levofloxacin, and chloramphenicol, were determined using a disk diffusion technique following the 2020 CLSI-M100 guidelines [38]. There are currently no breakpoints recommended for S. suis; therefore, those for viridans group streptococci were used, as defined in the 2020 CLSI-M100 guidelines [38]. Streptococcus pneumoniae strain ATCC 49619 was used for quality control purposes.
4.3. PCR Detection of Macrolide- and Tetracycline-Resistance Genes
Previously described PCR assays were used to detect three macrolide-resistance genes (ermB, mefA, and msrD) and 43 tetracycline-resistance genes (Table S3) [27,39].
4.4. Whole-Genome Sequencing (WGS)
The genomes of ten non-penicillin-susceptible S. suis isolates were further selected for WGS in order to investigate nucleotide variation in pbp genes. These isolates were chosen based on their different serotypes (2, 14, and 24) and STs, including four ST233 isolates, and one representative isolate each of ST25, ST104, ST105, ST221, ST234, and ST379. WGS was carried out on the Illumina platform at MicrobesNG (Birmingham, UK). Briefly, bacterial genomic DNA was extracted using ZymoBIOMICS DNA Kits (Zymo Research, Orange, CA, USA) and quantified in triplicates with the Quantit dsDNA HS assay in an Ependorff AF2200 plate reader. Genomic DNA libraries were prepared using a Nextera XT Library Prep Kit (Illumina, San Diego, CA, USA), following the manufacturer’s protocol. Pooled libraries were quantified using the Kapa Biosystems Library Quantification Kit for Illumina on a Roche light cycler 96 qPCR machine. Libraries were sequenced on an Illumina instrument using a 250 bp paired end protocol.
4.5. Bioinformatics Analysis
Illumina short-reads were adapter-trimmed using Trimmomatic 0.30 with a sliding window quality cutoff of Q15 [40]. De novo assemblies were performed using SPAdes version 3.7 (Columbia, SC, USA) [41], and contigs were annotated using Prokka 1.11 (Carlton, Australia) [42]. Antimicrobial-resistant genes were detected using ResFinder 4.1 (Center for Genomic Epidemiology, Lyngby, Denmark) [43] and the Comprehensive Antibiotic Resistance Database (CARD 2.0) (Hamilton, ON, Canada) [44]. Plasmid replicons were analyzed using PlasmidFinder 2.1 (Center for Genomic Epidemiology, Lyngby, Denmark) [45] and PLACNETw (Universidad de Cantabria & Instituto de Biomedicinay Biotecnología de Cantabria, Santander, Spain) [46]. Default parameters were used for all software programs unless otherwise specified.
To investigate penicillin non-susceptibility, we analyzed the predicted translated sequences of five pbp genes (pbp1a, pbp1b, pbp2a, pbp2b, and pbp2x) using local BLAST + and Clustal W. S. suis P1/7 (Genbank accession number NC_009648.1 (accessed on 21 June 2021)) was used as the penicillin-susceptible reference strain. A neighbor-joining phylogenetic tree was constructed using MEGA X with 1000 bootstrap replicates by applying the Dayhoff model [47]. The tree was visualized and annotated using Interactive Tree of Life (iTOL) v5 (Heidelberg, Germany) [48].
To search for the genetically closest relatives to the 10 selected penicillin-non-susceptible isolates, a modular single genome analysis following the core genome multilocus sequence typing (cgMLST) approach by BacWGSTdb 2.0 was used (Institute of Translational Medicine, Zhejiang University, Hangzhou, China) [49]. The genetically closest relatives were chosen for 5–10 strains based on small numbers of allelic differences with selection thresholds of 100 to 500, depending on the isolate under study. The genomic comparisons of 10 penicillin-non-susceptible isolates and the closest relatives selected from BacWGSTdb were conducted using a reference genome-based single-nucleotide polymorphism (SNP) strategy with CSI phylogeny (Center for Genomic Epidemiology, Lyngby, Denmark) [50]. Phylogenetic trees were built using MEGA X via the neighbor-joining method with 1000 bootstrap replicates by applying the Tamura three-parameter model [47]. The phylogenetic tree was visualized using the iTOL v5 [48]. S. suis S735, a type strain (accession no. CP003736 (accessed on 21 June 2021)), was used as the reference sequences for SNP analysis.
4.6. Accession Numbers
The genome sequences of the 10 non-penicillin-susceptible S. suis isolates were deposited in the NCBI GenBank under Bioproject accession number PRJNA691075 with the following strains: ID40747 (JAFEIV000000000), ID40085 (JAFEIX000000000), ID40484 (JAFEIY000000000), ID37647 (JAFEIZ000000000), ID30190 (JAFEIT000000000), ID32098 (JAFEIR000000000), ID32494 (JAFEIS000000000), ID27715 (JAFEIU000000000), ID41126 (JAFEIW000000000), and ID33329 (CP068708).
5. Conclusions
We have presented the characteristics of antimicrobial resistance of S. suis isolates recovered from humans. Resistance to tetracycline, macrolide, and clindamycin with tet (O) and ermB as the predominant resistance genes was commonly detected among isolates. Non-susceptibility to penicillin was shown in 8.3% of isolates, especially serotype 2-ST233 as the predominant group. In the “from farm to table” model, antimicrobial resistance is developed through the usage of antimicrobial agents on farms, and spread from animals to people via animal product consumption, direct contact with the animals, or through the environment. The “One Health” approach is a useful tool to combat AMR in bacteria by coordinating the human, animal, and environmental sectors. In addition, improvements in food hygiene standards and biosecurity measures at farms and in slaughtering procedures using hazard analysis and critical control point (HACCP) criteria are recommended, as well as better standards of hygiene in retail markets. Hence, restrictions on antimicrobial use, and the continuous control, monitoring, and surveillance of S. suis throughout the pork supply chain are top priority concerns and crucial for ensuring good public health outcomes.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/pathogens10091178/s1. Table S1. Antimicrobial resistance patterns and the resistance genes of S. suis by serotypes and sequence types, Table S2. Detail of Streptococcus suis used in this study, Table S3. PCR primers for the detection of tetracycline- and macrolide-resistant genes, Supplemental file 1. Alignment of the pbp1A sequence, Supplemental file 2. Alignment of the pbp1B sequence, Supplemental file 3. Alignment of the pbp2A sequence, Supplemental file 4. Alignment of the pbp2B sequence, Supplemental file 5. Alignment of the pbp2X sequence.
Author Contributions
Conceptualization, A.K., R.H., N.F. and M.G.; methodology, N.B., P.C. and P.B.; validation, A.K., R.H. and N.B.; formal analysis, N.B., P.C. and P.B.; resources, A.K.; data curation, A.K. and N.B.; writing-original draft preparation, A.K., N.F. and M.G.; writing-review and editing, A.K., N.F. and M.G.; supervision, A.K. and M.G.; funding acquisition, A.K. and N.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported in part by the Graduate Program Scholarship from The Graduate School, Kasetsart University, Thailand.
Institutional Review Board Statement
This study was reviewed and approved by the Ethics Committees of the Department of Medical Sciences, Ministry of Public Health, Thailand. Medical record reviews were conducted by the medical doctors under the protocol approved by the Ethics Committees. This study was conducted according to the principles of the Declaration of Helsinki.
Informed Consent Statement
Patient consent was waived due to the medical records were reviewed by attending physicians at the hospital using the clinical record form approved by the Ethics Committees and the study satisfied the conditions of the policy statement on ethical conduct for research involving humans.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.
Acknowledgments
We would like to thank MDPI English Editing Services for editing the draft of this manuscript.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Goyette-Desjardins, G.; Auger, J.P.; Xu, J.; Segura, M.; Gottschalk, M. Streptococcus suis, an important pig pathogen and emerging zoonotic agent—An update on the worldwide distribution based on serotyping and sequence typing. Emerg. Microbes. Infect. 2014, 3, e45. [Google Scholar] [CrossRef]
- Segura, M. Streptococcus suis research: Progress and challenges. Pathogens 2020, 9, 707. [Google Scholar] [CrossRef]
- Okura, M.; Osaki, M.; Nomoto, R.; Arai, S.; Osawa, R.; Sekizaki, T.; Takamatsu, D. Current Taxonomical situation of Streptococcus suis. Pathogens 2016, 24, E45. [Google Scholar] [CrossRef] [Green Version]
- Callejo, R.; Prieto, M.; Salamone, F.; Auger, J.P.; Goyette-Desjardins, G.; Gottschalk, M. Atypical Streptococcus suis in man, Argentina. Emerg. Infect. Dis. 2014, 20, 500–502. [Google Scholar] [CrossRef] [PubMed]
- Hatrongjit, R.; Kerdsin, A.; Gottschalk, M.; Takeuchi, D.; Hamada, S.; Oishi, K.; Akeda, Y. First human case report of sepsis due to infection with Streptococcus suis serotype 31 in Thailand. BMC Infect. Dis. 2015, 15, 392. [Google Scholar] [CrossRef]
- Kerdsin, A.; Oishi, K.; Sripakdee, S.; Boonkerd, N.; Polwichai, P.; Nakamura, S.; Uchida, R.; Sawanpanyalert, P.; Dejsirilert, S. Clonal dissemination of Streptococcus suis serotype 14 in Thailand. J. Med. Microbiol. 2009, 58, 1508–1513. [Google Scholar] [CrossRef] [PubMed]
- Kerdsin, A.; Dejsirilert, S.; Sawanpanyalert, P.; Boonnark, A.; Noithachang, W.; Sriyakum, D.; Simkum, S.; Chokngam, S.; Gottschalk, M.; Akeda, Y.; et al. Sepsis and spontaneous bacterial peritonitis in Thailand. Lancet 2011, 378, 960. [Google Scholar] [CrossRef]
- Kerdsin, A.; Hatrongjit, R.; Gottschalk, M.; Takeuchi, D.; Hamada, S.; Akeda, Y.; Oishi, K. Emergence of Streptococcus suis serotype 9 infection in humans. J. Microbiol. Immunol. Infect. 2017, 50, 545–546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nghia, H.D.; Hoa, N.T.; le Linh, D.; Campbell, J.; Diep, T.S.; Chau, N.V.; Mai, N.T.; Hien, T.T.; Spratt, B.; Farrar, J.; et al. Human case of Streptococcus suis serotype 16 infection. Emerg. Infect. Dis. 2008, 14, 155–157. [Google Scholar] [CrossRef] [Green Version]
- Palmieri, C.; Varaldo, P.E.; Facinelli, B. Streptococcus suis, an emerging drug-resistant animal and human pathogen. Front. Microbiol. 2011, 2, 235. [Google Scholar] [CrossRef] [Green Version]
- Segura, M.; Aragon, V.; Brockmeier, S.L.; Gebhart, C.; Greeff, A.; Kerdsin, A.; O’Dea, M.A.; Okura, M.; Saléry, M.; Schultsz, C.; et al. Update on Streptococcus suis research and prevention in the era of antimicrobial restriction: 4th International Workshop on S. suis. Pathogens 2020, 9, 374. [Google Scholar] [CrossRef]
- Gurung, M.; Tamang, M.D.; Moon, D.C.; Kim, S.R.; Jeong, J.H.; Jang, G.C.; Jung, S.C.; Park, Y.H.; Lim, S.K. Molecular basis of resistance to selected antimicrobial agents in the emerging zoonotic pathogen Streptococcus suis. J. Clin. Microbiol. 2015, 53, 2332–2336. [Google Scholar] [CrossRef] [Green Version]
- Hernandez-Garcia, J.; Wang, J.; Restif, O.; Holmes, M.A.; Mather, A.E.; Weinert, L.A.; Wileman, T.M.; Thomson, J.R.; Langford, P.R.; Wren, B.W.; et al. Patterns of antimicrobial resistance in Streptococcus suis isolates from pigs with or without streptococcal disease in England between 2009 and 2014. Vet Microbiol. 2017, 207, 117–124. [Google Scholar] [CrossRef]
- Ichikawa, T.; Oshima, M.; Yamagishi, J.; Muramatsu, C.; Asai, T. Changes in antimicrobial resistance phenotypes and genotypes in Streptococcus suis strains isolated from pigs in the Tokai area of Japan. J. Vet. Med. Sci. 2020, 82, 9–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matajira, C.E.C.; Moreno, L.Z.; Poor, A.P.; Gomes, V.T.M.; Dalmutt, A.C.; Parra, B.M.; Oliveira, C.H.; Barbosa, M.R.F.; Sato, M.I.Z.; Calderaro, F.F.; et al. Streptococcus suis in Brazil: Genotypic, virulence, and resistance profiling of strains isolated from pigs between 2001 and 2016. Pathogens 2020, 9, 31. [Google Scholar] [CrossRef] [Green Version]
- Yongkiettrakul, S.; Maneerat, K.; Arechanajan, B.; Malila, Y.; Srimanote, P.; Gottschalk, M.; Visessanguan, W. Antimicrobial susceptibility of Streptococcus suis isolated from diseased pigs, asymptomatic pigs, and human patients in Thailand. BMC Vet. Res. 2019, 15, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, C.; Zhang, P.; Wang, Y.; Fu, L.; Liu, L.; Xu, D.; Hou, Y.; Li, Y.; Fu, M.; Ding, S.; et al. Capsular serotypes, antimicrobial susceptibility, and the presence of transferable oxazolidinone resistance genes in Streptococcus suis isolated from healthy pigs in China. Vet. Microbiol. 2020, 247, 108750. [Google Scholar] [CrossRef] [PubMed]
- Bojarska, A.; Molska, E.; Janas, K.; Skoczyńska, A.; Stefaniuk, E.; Hryniewicz, W.; Sadowy, E. Streptococcus suis in invasive human infections in Poland: Clonality and determinants of virulence and antimicrobial resistance. Eur. J. Clin. Microbiol. Infect. Dis. 2016, 35, 917–925. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoa, N.T.; Chieu, T.T.; Nghia, H.D.; Mai, N.T.; Anh, P.H.; Wolbers, M.; Baker, S.; Campbell, J.I.; Chau, N.V.; Hien, T.T.; et al. The antimicrobial resistance patterns and associated determinants in Streptococcus suis isolated from humans in southern Vietnam, 1997–2008. BMC Infect. Dis. 2011, 11, 6. [Google Scholar] [CrossRef] [Green Version]
- Marie, J.; Morvan, H.; Berthelot-Hérault, F.; Sanders, P.; Kempf, I.; Gautier-Bouchardon, A.V.; Jouy, E.; Kobisch, M. Antimicrobial susceptibility of Streptococcus suis isolated from swine in France and from humans in different countries between 1996 and 2000. J. Antimicrob. Chemother. 2002, 50, 201–209. [Google Scholar] [CrossRef] [PubMed]
- Rayanakorn, A.; Ademi, Z.; Liew, D.; Lee, L.H. Burden of disease and productivity impact of Streptococcus suis infection in Thailand. PLoS Negl. Trop. Dis. 2021, 15, e0008985. [Google Scholar] [CrossRef] [PubMed]
- Dechêne-Tempier, M.; Marois-Créhan, C.; Libante, V.; Jouy, E.; Leblond-Bourget, N.; Payot, S. Update on the Mechanisms of Antibiotic Resistance and the Mobile Resistome in the Emerging Zoonotic Pathogen Streptococcus suis. Microorganisms 2021, 9, 1765. [Google Scholar] [CrossRef]
- Zhang, C.; Zhang, Z.; Song, L.; Fan, X.; Wen, F.; Xu, S.; Ning, Y. Antimicrobial resistance profile and genotypic characteristics of Streptococcus suis capsular type 2 isolated from clinical carrier sows and diseased pigs in China. Biomed. Res. Int. 2015, 2015, 284303. [Google Scholar] [CrossRef] [PubMed]
- van Hout, J.; Heuvelink, A.; Gonggrijp, M. Monitoring of antimicrobial susceptibility of Streptococcus suis in the Netherlands, 2013–2015. Vet. Microbiol. 2016, 194, 5–10. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.P.; Saegerman, C.; Douny, C.; Dinh, T.V.; Xuan, B.H.; Vu, B.D.; Hong, N.P.; Scippo, M.L. First survey on the use of antibiotics in pig and poultry production in the Red Delta region of Vietnam. Food Pub. Health 2013, 3, 247–256. [Google Scholar] [CrossRef]
- Pyörälä, S.; Baptiste, K.E.; Catry, B.; van Duijkeren, E.; Greko, C.; Moreno, M.A.; Pomba, M.C.; Rantala, M.; Ružauskas, M.; Sanders, P.; et al. Macrolides and lincosamides in cattle and pigs: Use and development of antimicrobial resistance. Vet. J. 2014, 200, 230–239. [Google Scholar] [CrossRef]
- Chen, L.; Song, Y.; Wei, Z.; He, H.; Zhang, A.; Jin, M. Antimicrobial susceptibility, tetracycline and erythromycin resistance genes, and multilocus sequence typing of Streptococcus suis isolates from diseased pigs in China. J. Vet. Med. Sci. 2013, 75, 583–587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yongkiettrakul, S.; Wongsurawat, T.; Jenjaroenpun, P.; Acheampong, D.A.; Srimanote, P.; Maneerat, K.; Visessanguan, W.; Nookaew, I. Genome sequences of antibiotic-resistant Streptococcus suis strains isolated from human patients and diseased and asymptomatic pigs in Thailand. Infect. Genet. Evol. 2021, 87, 104674. [Google Scholar] [CrossRef] [PubMed]
- Werinder, A.; Aspán, A.; Backhans, A.; Sjölund, M.; Guss, B.; Jacobson, M. Streptococcus suis in Swedish grower pigs: Occurrence, serotypes, and antimicrobial susceptibility. Acta Vet. Scand. 2020, 62, 36. [Google Scholar] [CrossRef]
- Niemann, L.; Eichhorn, I.; Müller, P.; Brauns, J.; Nathaus, R.; Schäkel, F.; Höltig, D.; Wendt, M.; Kadlec, K.; Schwarz, S. Draft genome sequences of three porcine Streptococcus suis isolates which differ in their susceptibility to penicillin. Microbiol. Resour. Announc. 2019, 8, e01711-18. [Google Scholar] [CrossRef] [Green Version]
- Hu, P.; Yang, M.; Zhang, A.; Wu, J.; Chen, B.; Hua, Y.; Yu, J.; Chen, H.; Xiao, J.; Jin, M. Comparative genomics study of multi-drug-resistance mechanisms in the antibiotic-resistant Streptococcus suis R61 strain. PLoS ONE 2011, 6, e24988. [Google Scholar] [CrossRef] [Green Version]
- Hakenbeck, R.; Grebe, T.; Zähner, D.; Stock, J.B. β-lactam resistance in Streptococcus pneumoniae: Penicillin-binding proteins and non-penicillin-binding proteins. Mol. Microbiol. 1999, 33, 673–678. [Google Scholar] [CrossRef]
- Haenni, M.; Moreillon, P. Mutations in penicillin-binding protein (PBP) genes and in non-PBP genes during selection of penicillin-resistant Streptococcus gordonii. Antimicrob. Agents Chemother. 2006, 50, 4053–4061. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fani, F.; Leprohon, P.; Légaré, D.; Ouellette, M. Whole genome sequencing of penicillin-resistant Streptococcus pneumoniae reveals mutations in penicillin-binding proteins and in a putative iron permease. Genome. Biol. 2011, 12, R115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okura, M.; Auger, J.P.; Shibahara, T.; Goyette-Desjardins, G.; Van Calsteren, M.R.; Maruyama, F.; Kawai, M.; Osaki, M.; Segura, M.; Gottschalk, M.; et al. Capsular polysaccharide switching in Streptococcus suis modulates host cell interactions and virulence. Sci. Rep. 2021, 11, 6513. [Google Scholar] [CrossRef] [PubMed]
- Kerdsin, A.; Dejsirilert, S.; Puangpatra, P.; Sripakdee, S.; Chumla, K.; Boonkerd, N.; Polwichai, P.; Tanimura, S.; Takeuchi, D.; Nakayama, T.; et al. Genotypic profile of Streptococcus suis serotype 2 and clinical features of infection in humans, Thailand. Emerg. Infect. Dis. 2011, 17, 836–842. [Google Scholar] [CrossRef] [PubMed]
- Kerdsin, A.; Akeda, Y.; Takeuchi, D.; Dejsirilert, S.; Gottschalk, M.; Oishi, K. Genotypic diversity of Streptococcus suis strains isolated from humans in Thailand. Eur. J. Clin. Microbiol. Infect. Dis. 2018, 37, 917–925. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing, 30th ed.; CLSI Document M100-S30; Clinical and Laboratory Standards Institute (CLSI): Wayne, PA, USA, 2020. [Google Scholar]
- Kang, Y.; Li, Q.; Yin, Z.; Shen, M.; Zhao, H.; Bai, Y.; Mei, L.; Hu, J. High diversity and abundance of cultivable tetracycline-resistant bacteria in soil following pig manure application. Sci. Rep. 2018, 8, 1489. [Google Scholar] [CrossRef] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [Green Version]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic. Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef] [PubMed]
- Carattoli, A.; Zankari, E.; García-Fernández, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Møller Aarestrup, F.; Hasman, H. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vielva, L.; de Toro, M.; Lanza, V.F.; de la Cruz, F. PLACNETw: A web-based tool for plasmid reconstruction from bacterial genomes. Bioinformatics 2017, 33, 3796–3798. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic. Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Feng, Y.; Zou, S.; Chen, H.; Yu, Y.; Ruan, Z. BacWGSTdb 2.0: A one-stop repository for bacterial whole-genome sequence typing and source tracking. Nucleic. Acids Res. 2021, 49, D644–D650. [Google Scholar] [CrossRef]
- Kaas, R.S.; Leekitcharoenphon, P.; Aarestrup, F.M.; Lund, O. Solving the problem of comparing whole bacterial genomes across different sequencing platforms. PLoS ONE 2014, 9, e104984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).