A Promising Tool in Serological Diagnosis: Current Research Progress of Antigenic Epitopes in Infectious Diseases
Abstract
1. Introduction
2. Definition and Classification
3. Epitope Mapping and Prediction
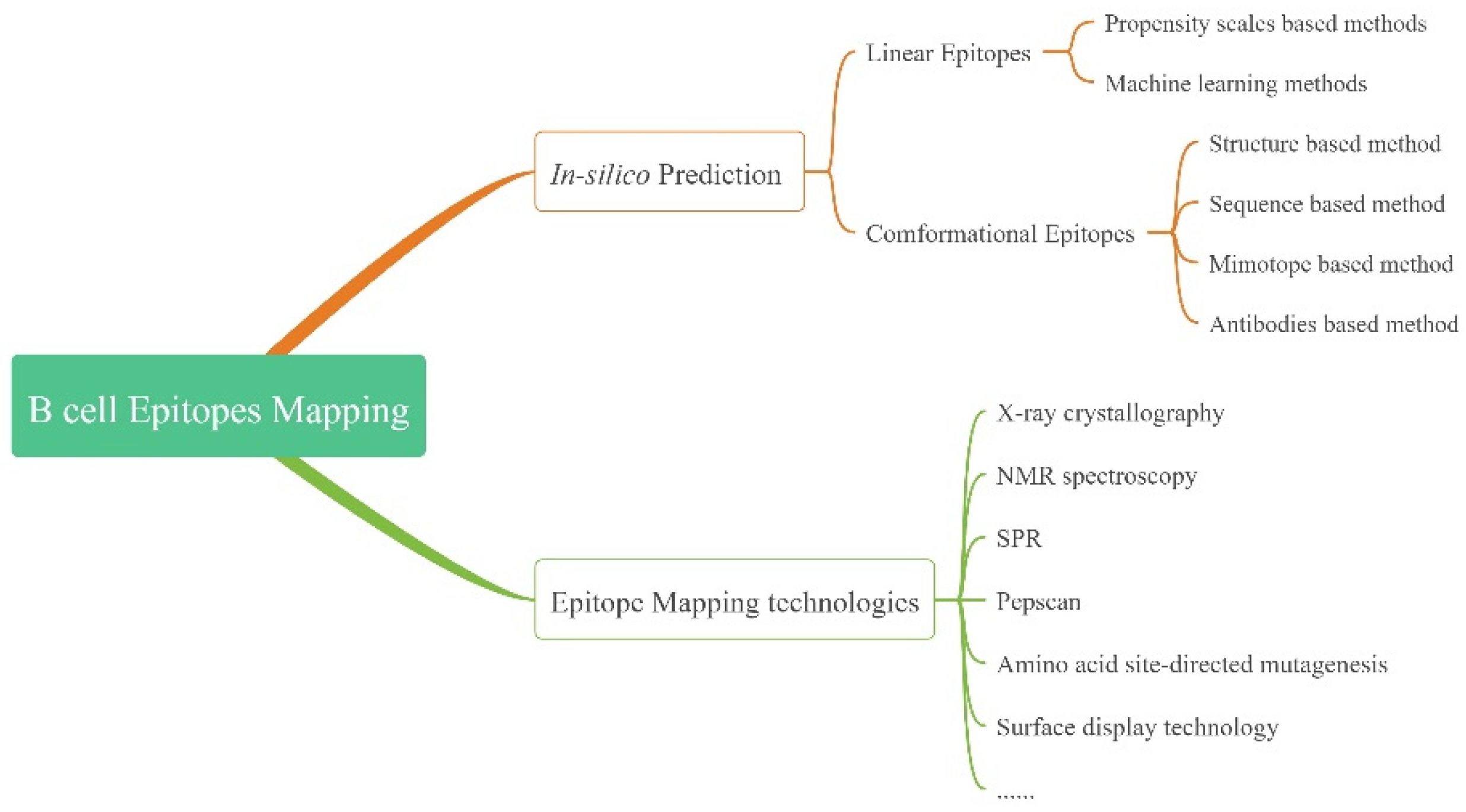
3.1. Prediction
3.1.1. LEs
3.1.2. CEs
3.2. Epitope Mapping Technologies
4. Applications in Diagnosis
4.1. Diagnosis in Bacterial Infection
4.2. Diagnosis in Viral Infection
4.2.1. SARS-CoV-2
4.2.2. Epstein–Barr Virus (EBV)
4.2.3. Dengue Virus (DENV)
4.2.4. Hepatitis Virus
4.2.5. Ebolavirus (EBOV)
4.2.6. Hantaviruses
4.3. Diagnosis in Parasitic Infections
4.4. Diagnosis in Fungal Infection
5. Discussion and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| PCR | Polymerase chain reaction | COVID-19 | Coronavirus disease 2019 |
| LEs/CEs | Linear/conformational epitopes | S | Spike |
| B/TCRs | B/T cell receptors | E | Envelope |
| MHC | Major histocompatibility complex | M | Membrane |
| Igs | immunoglobulins | N | Nucleocapsid |
| ML | Machine learning | ORF | Open reading frame |
| HMM | Hidden Markov model | EBV | Epstein–Barr virus |
| ROC | Receiver operating characteristic | NP | Nucleoprotein |
| ANN | Artificial neural network | NPC | Nasopharyngeal carcinoma |
| AUC | Area under the curve | LMP2 | Latent membrane protein 2 |
| ASEP | Antibody-specific epitope propensity | DENV | Dengue virus |
| FNN | Feed-forward neural network | NS1 | Non-structural protein 1 |
| RNN | Recurrent neural network | HBV/HCV | Hepatitis B/C virus |
| SVM | Support vector machine | HBeAg/HBcAg | Hepatitis B e/core antigen |
| RF | Random Forest | EBOV | Ebolavirus |
| NMR | Nuclear magnetic resonance | MBV | Marburgvirus |
| SPR | Surface plasmon resonance | RDTs | Rapid diagnostic tests |
| Pepscan | Peptide scanning | GP | Glycoprotein |
| ELISA | Enzyme-linked immunosorbent assay | HFRS | Hemorrhagic fever renal syndrome |
| WB | Western blotting | HCPS | Hantavirus cardiopulmonary syndrome |
| SARS-CoV-2 | Severe acute respiratory syndrome coronavirus 2 | SHNP | Seoul orthohantavirus nucleoprotein |
| TB | Tuberculosis | VL | Visceral leishmaniasis |
| RD | Regions of difference | VP | Viral protein |
| CFP-10 | Culture filtrate protein | SjSAP4 | S. japonicum saposin protein |
| ESAT-6 | Early secretory antigenic target | SjSP-13 | S. japonicum saposin-like protein |
| OMPs | outer membrane proteins | SmSPI | S. mansoni Serine protease inhibitor |
| MAT | microscopic agglutination test | BCLA | Brain Cyst Load-associated Antigen |
| SEA/SEB | Staphylococcal enterotoxin A/B | Hsp90 | Heat shock protein 90 |
| Opp | Oligopeptide permease | Sap: | Secreted aspartic protease |
| Osp | Outer surface protein | SjEV | S. japonicum Extracellular vesicles |
| PPV/NPV | Positive/negative predict value | ICT | Immunochromatographic test |
References
- Standing up to infectious disease. Nat. Microbiol. 2019, 4, 1. [CrossRef] [PubMed]
- Heiss, K.; Heidepriem, J.; Fischer, N.; Weber, L.K.; Dahlke, C.; Jaenisch, T.; Loeffler, F.F. Rapid Response to Pandemic Threats: Immunogenic Epitope Detection of Pandemic Pathogens for Diagnostics and Vaccine Development Using Peptide Microarrays. J. Proteome Res. 2020, 19, 4339–4354. [Google Scholar] [CrossRef] [PubMed]
- Hahn, A.; Podbielski, A.; Meyer, T.; Zautner, A.E.; Loderstadt, U.; Schwarz, N.G.; Kruger, A.; Cadar, D.; Frickmann, H. On detection thresholds-a review on diagnostic approaches in the infectious disease laboratory and the interpretation of their results. Acta Trop. 2020, 205, 105377. [Google Scholar] [CrossRef]
- Van Regenmortel, M.H. The concept and operational definition of protein epitopes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1989, 323, 451–466. [Google Scholar] [CrossRef] [PubMed]
- Kapingidza, A.B.; Kowal, K.; Chruszcz, M. Antigen-Antibody Complexes. Subcell. Biochem. 2020, 94, 465–497. [Google Scholar] [CrossRef] [PubMed]
- Fleri, W.; Paul, S.; Dhanda, S.K.; Mahajan, S.; Xu, X.; Peters, B.; Sette, A. The Immune Epitope Database and Analysis Resource in Epitope Discovery and Synthetic Vaccine Design. Front. Immunol. 2017, 8, 278. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Trincado, J.L.; Gomez-Perosanz, M.; Reche, P.A. Fundamentals and Methods for T- and B-Cell Epitope Prediction. J. Immunol. Res. 2017, 2017, 2680160. [Google Scholar] [CrossRef]
- Barlow, D.J.; Edwards, M.S.; Thornton, J.M. Continuous and discontinuous protein antigenic determinants. Nature 1986, 322, 747–748. [Google Scholar] [CrossRef]
- Chen, P.; Rayner, S.; Hu, K.H. Advances of bioinformatics tools applied in virus epitopes prediction. Virol. Sin. 2011, 26, 1–7. [Google Scholar] [CrossRef]
- Zinsli, L.V.; Stierlin, N.; Loessner, M.J.; Schmelcher, M. Deimmunization of protein therapeutics—Recent advances in experimental and computational epitope prediction and deletion. Comput. Struct. Biotechnol. J. 2021, 19, 315–329. [Google Scholar] [CrossRef]
- Sun, P.; Guo, S.; Sun, J.; Tan, L.; Lu, C.; Ma, Z. Advances in In-silico B-cell Epitope Prediction. Curr. Top. Med. Chem. 2019, 19, 105–115. [Google Scholar] [CrossRef] [PubMed]
- Ferdous, S.; Kelm, S.; Baker, T.S.; Shi, J.; Martin, A.C.R. B-cell epitopes: Discontinuity and conformational analysis. Mol. Immunol. 2019, 114, 643–650. [Google Scholar] [CrossRef]
- Hopp, T.P.; Woods, K.R. Prediction of protein antigenic determinants from amino acid sequences. Proc. Natl. Acad. Sci. USA 1981, 78, 3824–3828. [Google Scholar] [CrossRef] [PubMed]
- Alix, A.J. Predictive estimation of protein linear epitopes by using the program PEOPLE. Vaccine 1999, 18, 311–314. [Google Scholar] [CrossRef]
- Saha, S.; Raghava, G.P.S. BcePred: Prediction of Continuous B-Cell Epitopes in Antigenic Sequences Using Physico-chemical Properties. In Proceedings of the Artificial Immune Systems; Spring: Berlin/Heidelberg, Germany, 2004; pp. 197–204. [Google Scholar]
- Blythe, M.J.; Flower, D.R. Benchmarking B cell epitope prediction: Underperformance of existing methods. Protein Sci. 2005, 14, 246–248. [Google Scholar] [CrossRef] [PubMed]
- Larsen, J.E.; Lund, O.; Nielsen, M. Improved method for predicting linear B-cell epitopes. Immunome Res. 2006, 2, 2. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Saha, S.; Raghava, G.P. Prediction of continuous B-cell epitopes in an antigen using recurrent neural network. Proteins 2006, 65, 40–48. [Google Scholar] [CrossRef]
- Jespersen, M.C.; Peters, B.; Nielsen, M.; Marcatili, P. BepiPred-2.0: Improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Res. 2017, 45, W24–W29. [Google Scholar] [CrossRef]
- Kulkarni-Kale, U.; Bhosle, S.; Kolaskar, A.S. CEP: A conformational epitope prediction server. Nucleic Acids Res. 2005, 33, W168–W171. [Google Scholar] [CrossRef]
- Haste Andersen, P.; Nielsen, M.; Lund, O. Prediction of residues in discontinuous B-cell epitopes using protein 3D structures. Protein Sci. 2006, 15, 2558–2567. [Google Scholar] [CrossRef]
- Ponomarenko, J.; Bui, H.H.; Li, W.; Fusseder, N.; Bourne, P.E.; Sette, A.; Peters, B. ElliPro: A new structure-based tool for the prediction of antibody epitopes. BMC Bioinform. 2008, 9, 514. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Wu, D.; Xu, T.; Wang, X.; Xu, X.; Tao, L.; Li, Y.X.; Cao, Z.W. SEPPA: A computational server for spatial epitope prediction of protein antigens. Nucleic Acids Res. 2009, 37, W612–W616. [Google Scholar] [CrossRef] [PubMed]
- Sun, P.; Ju, H.; Liu, Z.; Ning, Q.; Zhang, J.; Zhao, X.; Huang, Y.; Ma, Z.; Li, Y. Bioinformatics resources and tools for conformational B-cell epitope prediction. Comput. Math. Methods Med. 2013, 2013, 943636. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Niu, Y.; Xiong, Y.; Ke, M. Prediction of conformational B-cell epitopes. Methods Mol. Biol. 2014, 1184, 185–196. [Google Scholar] [CrossRef]
- Wang, H.W.; Pai, T.W. Machine learning-based methods for prediction of linear B-cell epitopes. Methods Mol. Biol. 2014, 1184, 217–236. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.H.; Sun, P.P.; Lu, Y.; Guo, W.W.; Huang, Y.X.; Ma, Z.Q. MimoPro: A more efficient Web-based tool for epitope prediction using phage display libraries. BMC Bioinform. 2011, 12, 199. [Google Scholar] [CrossRef]
- Huang, J.; Gutteridge, A.; Honda, W.; Kanehisa, M. MIMOX: A web tool for phage display based epitope mapping. BMC Bioinform. 2006, 7, 451. [Google Scholar] [CrossRef]
- Soga, S.; Kuroda, D.; Shirai, H.; Kobori, M.; Hirayama, N. Use of amino acid composition to predict epitope residues of individual antibodies. Protein Eng. Des. Sel. 2010, 23, 441–448. [Google Scholar] [CrossRef]
- Krawczyk, K.; Liu, X.; Baker, T.; Shi, J.; Deane, C.M. Improving B-cell epitope prediction and its application to global antibody-antigen docking. Bioinformatics 2014, 30, 2288–2294. [Google Scholar] [CrossRef]
- Sela-Culang, I.; Ashkenazi, S.; Peters, B.; Ofran, Y. PEASE: Predicting B-cell epitopes utilizing antibody sequence. Bioinformatics 2015, 31, 1313–1315. [Google Scholar] [CrossRef]
- Sela-Culang, I.; Ofran, Y.; Peters, B. Antibody specific epitope prediction-emergence of a new paradigm. Curr. Opin. Virol. 2015, 11, 98–102. [Google Scholar] [CrossRef] [PubMed]
- Yao, B.; Zhang, L.; Liang, S.; Zhang, C. SVMTriP: A method to predict antigenic epitopes using support vector machine to integrate tri-peptide similarity and propensity. PLoS ONE 2012, 7, e45152. [Google Scholar] [CrossRef] [PubMed]
- Sweredoski, M.J.; Baldi, P. COBEpro: A novel system for predicting continuous B-cell epitopes. Protein Eng. Des. Sel. 2009, 22, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Niu, Y.; Xiong, Y.; Zhao, M.; Yu, R.; Liu, J. Computational prediction of conformational B-cell epitopes from antigen primary structures by ensemble learning. PLoS ONE 2012, 7, e43575. [Google Scholar] [CrossRef]
- Sela-Culang, I.; Benhnia, M.R.; Matho, M.H.; Kaever, T.; Maybeno, M.; Schlossman, A.; Nimrod, G.; Li, S.; Xiang, Y.; Zajonc, D.; et al. Using a combined computational-experimental approach to predict antibody-specific B cell epitopes. Structure 2014, 22, 646–657. [Google Scholar] [CrossRef]
- Toride King, M.; Brooks, C.L. Epitope Mapping of Antibody-Antigen Interactions with X-Ray Crystallography. Methods Mol. Biol. 2018, 1785, 13–27. [Google Scholar] [CrossRef]
- Varani, L.; Bankovich, A.J.; Liu, C.W.; Colf, L.A.; Jones, L.L.; Kranz, D.M.; Puglisi, J.D.; Garcia, K.C. Solution mapping of T cell receptor docking footprints on peptide-MHC. Proc. Natl. Acad. Sci. USA 2007, 104, 13080–13085. [Google Scholar] [CrossRef]
- Rosen, O.; Anglister, J. Epitope mapping of antibody-antigen complexes by nuclear magnetic resonance spectroscopy. Methods Mol. Biol. 2009, 524, 37–57. [Google Scholar] [CrossRef]
- Abbott, W.M.; Damschroder, M.M.; Lowe, D.C. Current approaches to fine mapping of antigen-antibody interactions. Immunology 2014, 142, 526–535. [Google Scholar] [CrossRef]
- Chavanieu, A.; Pugnière, M. Developments in SPR Fragment Screening. Expert Opin. Drug Discov. 2016, 11, 489–499. [Google Scholar] [CrossRef]
- Liu, C.; Liu, Q.; Hu, X. SPR phase detection for measuring the thickness of thin metal films. Opt. Express 2014, 22, 7574–7580. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.H.; Park, J.; Kang, S.; Kim, M. Surface plasmon resonance: A versatile technique for biosensor applications. Sensors 2015, 15, 10481–10510. [Google Scholar] [CrossRef] [PubMed]
- Shpacovitch, V.; Hergenröder, R. Surface Plasmon Resonance (SPR)-Based Biosensors as Instruments with High Versatility and Sensitivity. Sensors 2020, 20, 3010. [Google Scholar] [CrossRef]
- Qu, J.H.; Dillen, A.; Saeys, W.; Lammertyn, J.; Spasic, D. Advancements in SPR biosensing technology: An overview of recent trends in smart layers design, multiplexing concepts, continuous monitoring and in vivo sensing. Anal. Chim. Acta 2020, 1104, 10–27. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, J.; Chen, Y.; Wang, A.; Wei, Q.; Yang, S.; Feng, H.; Chai, S.; Liu, D.; Zhang, G. Identification of a dominant linear epitope on the VP2 capsid protein of porcine parvovirus and characterization of two monoclonal antibodies with neutralizing abilities. Int. J. Biol. Macromol. 2020, 163, 2013–2022. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Liu, H.; Li, J.; Liu, D.; Meng, R.; Zhang, Q.; Shaozhou, W.; Bai, X.; Zhang, T.; Liu, M.; et al. A Conserved Epitope Mapped with a Monoclonal Antibody against the VP3 Protein of Goose Parvovirus by Using Peptide Screening and Phage Display Approaches. PLoS ONE 2016, 11, e0147361. [Google Scholar] [CrossRef]
- Rojas, G.; Tundidor, Y.; Infante, Y.C. High throughput functional epitope mapping: Revisiting phage display platform to scan target antigen surface. mAbs 2014, 6, 1368–1376. [Google Scholar] [CrossRef]
- Leung, P.S.; Iwayama, T.; Coppel, R.L.; Gershwin, M.E. Site-directed mutagenesis of lysine within the immunodominant autoepitope of PDC-E2. Hepatology 1990, 12, 1321–1328. [Google Scholar] [CrossRef]
- Moreira, G.; Fuhner, V.; Hust, M. Epitope Mapping by Phage Display. Methods Mol. Biol. 2018, 1701, 497–518. [Google Scholar] [CrossRef]
- Liu, G.Y.; Mei, X.J.; Hu, M.J.; Yang, Y.; Liu, M.; Li, M.S.; Zhang, M.L.; Cao, M.J.; Liu, G.M. Analysis of the Allergenic Epitopes of Tropomyosin from Mud Crab Using Phage Display and Site-Directed Mutagenesis. J. Agric. Food Chem. 2018, 66, 9127–9137. [Google Scholar] [CrossRef]
- Chen, Y.Y.; Li, M.S.; Yun, X.; Xia, F.; Hu, M.J.; Jin, T.; Cao, M.J.; Lai, D.; Chen, G.; Liu, G.M. Site-Directed Mutations of Calcium-Binding Sites Contribute to Reducing the Immunoreactivity of the EF-Hand Sarcoplasmic Calcium-Binding Protein in Scylla paramamosain. J. Agric. Food Chem. 2021, 69, 428–436. [Google Scholar] [CrossRef] [PubMed]
- Park, M. Surface Display Technology for Biosensor Applications: A Review. Sensors 2020, 20, 2775. [Google Scholar] [CrossRef] [PubMed]
- Aghebati-Maleki, L.; Bakhshinejad, B.; Baradaran, B.; Motallebnezhad, M.; Aghebati-Maleki, A.; Nickho, H.; Yousefi, M.; Majidi, J. Phage display as a promising approach for vaccine development. J. Biomed. Sci. 2016, 23, 66. [Google Scholar] [CrossRef] [PubMed]
- Behr, M.; Wilson, M.; Gill, W. Comparative Cenomics of BCC Vaccines by Whole-Genome DNA Microarray. Science 1999, 284, 1520–1523. [Google Scholar] [CrossRef]
- Yang, H.; Chen, H.; Liu, Z.; Ma, H.; Qin, L.; Jin, R.; Zheng, R.; Feng, Y.; Cui, Z.; Wang, J.; et al. A novel B-cell epitope identified within Mycobacterium tuberculosis CFP10/ESAT-6 protein. PLoS ONE 2013, 8, e52848. [Google Scholar] [CrossRef]
- Goyal, B.; Kumar, K.; Gupta, D.; Agarwal, R.; Latawa, R.; Sheikh, J.A.; Verma, I. Utility of B-cell epitopes based peptides of RD1 and RD2 antigens for immunodiagnosis of pulmonary tuberculosis. Diagn. Microbiol. Infect. Dis. 2014, 78, 391–397. [Google Scholar] [CrossRef]
- Cheng, P.; Wang, L.; Gong, W. In silico Analysis of Peptide-Based Biomarkers for the Diagnosis and Prevention of Latent Tuberculosis Infection. Front. Microbiol. 2022, 13, 947852. [Google Scholar] [CrossRef]
- Zhao, H.; Wang, Y.; Zhao, L.; Dong, Z.; Mi, J.; Zhao, H.; Wang, J.; Zeng, J.; Wang, H.; Wang, L. Evaluation and verification of the characteristic peptides for detection of Staphylococcus aureus in food by targeted LC-MS/MS. Talanta 2021, 235, 122794. [Google Scholar] [CrossRef]
- Cheung, G.Y.C.; Bae, J.S.; Otto, M. Pathogenicity and virulence of Staphylococcus aureus. Virulence 2021, 12, 547–569. [Google Scholar] [CrossRef]
- Tam, K.; Torres, V.J. Staphylococcus aureus Secreted Toxins and Extracellular Enzymes. Microbiol. Spectr. 2019, 7. [Google Scholar] [CrossRef]
- Bhatti, A.R.; Micusan, V.V. Production and characterization of anti-peptide monoclonal antibodies with specificity for staphylococcal enterotoxins A and B. J. Microbiol. Methods 1999, 35, 143–149. [Google Scholar] [CrossRef]
- Pastells, C.; Acosta, G.; Pascual, N.; Albericio, F.; Royo, M.; Marco, M.P. An immunochemical strategy based on peptidoglycan synthetic peptide epitopes to diagnose Staphylococcus aureus infections. Anal. Chim. Acta 2015, 889, 203–211. [Google Scholar] [CrossRef] [PubMed]
- Schreier, S.; Doungchawee, G.; Chadsuthi, S.; Triampo, D.; Triampo, W. Leptospirosis: Current situation and trends of specific laboratory tests. Expert Rev. Clin. Immunol. 2013, 9, 263–280. [Google Scholar] [CrossRef] [PubMed]
- Kanagavel, M.; Shanmughapriya, S.; Anbarasu, K.; Natarajaseenivasan, K. B-cell-specific peptides of leptospira interrogans LigA for diagnosis of patients with acute leptospirosis. Clin. Vaccine Immunol. CVI 2014, 21, 354–359. [Google Scholar] [CrossRef] [PubMed]
- Anita, K.; Premlatha, M.M.; Kanagavel, M.; Akino Mercy, C.S.; Raja, V.; Shanmughapriya, S.; Natarajaseenivasan, K. Evaluation of combined B cell specific N-terminal immunogenic domains of LipL21 for diagnosis of leptospirosis. Int. J. Biol. Macromol. 2016, 91, 465–470. [Google Scholar] [CrossRef] [PubMed]
- Kanagavel, M.; Shanmughapriya, S.; Aishwarya, K.V.L.; Ponmurugan, K.; Murugan, K.; Al-Dhabi, N.A.; Natarajaseenivasan, K. Peptide specific monoclonal antibodies of Leptospiral LigA for acute diagnosis of leptospirosis. Sci. Rep. 2017, 7, 3250. [Google Scholar] [CrossRef]
- Hsu, S.H.; Yang, C.W. Insight into the Structure, Functions, and Dynamics of the Leptospira Outer Membrane Proteins with the Pathogenicity. Membranes 2022, 12, 300. [Google Scholar] [CrossRef]
- Aviat, F.; Rochereau-Roulet, S.; Branger, C.; Estavoyer, J.M.; Chatrenet, B.; Orsonneau, J.L.; Thorin, C.; Andre-Fontaine, G. Synthetic peptide issued from Hap1/LipL32 for new early serodiagnosis of human leptospirosis. Comp. Immunol. Microbiol. Infect. Dis. 2010, 33, 375–387. [Google Scholar] [CrossRef]
- Mukherjee, P.; Dutta, M.; Datta, P.; Dasgupta, A.; Pradhan, R.; Pradhan, M.; Kundu, M.; Basu, J.; Chakrabarti, P. The RD1-encoded antigen Rv3872 of Mycobacterium tuberculosis as a potential candidate for serodiagnosis of tuberculosis. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2007, 13, 146–152. [Google Scholar] [CrossRef]
- Tungtrakanpoung, R.; Pitaksajjakul, P.; Na-Ngarm, N.; Chaicumpa, W.; Ekpo, P.; Saengjaruk, P.; Froman, G.; Ramasoota, P. Mimotope of Leptospira from phage-displayed random peptide library is reactive with both monoclonal antibodies and patients’ sera. Vet. Microbiol. 2006, 115, 54–63. [Google Scholar] [CrossRef]
- Lin, X.; Chen, Y.; Yan, J. Recombinant multiepitope protein for diagnosis of leptospirosis. Clin. Vaccine Immunol. CVI 2008, 15, 1711–1714. [Google Scholar] [CrossRef] [PubMed]
- Frikha-Gargouri, O.; Gdoura, R.; Znazen, A.; Gargouri, B.; Gargouri, J.; Rebai, A.; Hammami, A. Evaluation of an in silico predicted specific and immunogenic antigen from the OmcB protein for the serodiagnosis of Chlamydia trachomatis infections. BMC Microbiol. 2008, 8, 217. [Google Scholar] [CrossRef] [PubMed]
- Rahman, K.S.; Darville, T.; Russell, A.N.; O’Connell, C.M.; Wiesenfeld, H.C.; Hillier, S.L.; Lee, D.E.; Kaltenboeck, B. Comprehensive Molecular Serology of Human Chlamydia trachomatis Infections by Peptide Enzyme-Linked Immunosorbent Assays. mSphere 2018, 3, e00253-18. [Google Scholar] [CrossRef]
- Rahman, K.S.; Darville, T.; Wiesenfeld, H.C.; Hillier, S.L.; Kaltenboeck, B. Mixed Chlamydia trachomatis Peptide Antigens Provide a Specific and Sensitive Single-Well Colorimetric Enzyme-Linked Immunosorbent Assay for Detection of Human Anti-C. trachomatis Antibodies. mSphere 2018, 3, e00484-18. [Google Scholar] [CrossRef]
- Rahman, K.S.; Kaltenboeck, B. Multi-peptide ELISAs overcome cross-reactivity and inadequate sensitivity of conventional Chlamydia pneumoniae serology. Sci. Rep. 2019, 9, 15078. [Google Scholar] [CrossRef] [PubMed]
- Signorino, G.; Arnaboldi, P.M.; Petzke, M.M.; Dattwyler, R.J. Identification of OppA2 linear epitopes as serodiagnostic markers for Lyme disease. Clin. Vaccine Immunol. CVI 2014, 21, 704–711. [Google Scholar] [CrossRef]
- Pulzova, L.; Flachbartova, Z.; Bencurova, E.; Potocnakova, L.; Comor, L.; Schreterova, E.; Bhide, M. Identification of B-cell epitopes of Borrelia burgdorferi outer surface protein C by screening a phage-displayed gene fragment library. Microbiol. Immunol. 2016, 60, 669–677. [Google Scholar] [CrossRef]
- Schreterova, E.; Bhide, M.; Potocnakova, L.; Borszekova Pulzova, L. Design, construction and evaluation of multi-epitope antigens for diagnosis of Lyme disease. Ann. Agric. Environ. Med. AAEM 2017, 24, 696–701. [Google Scholar] [CrossRef]
- Toumanios, C.; Prisco, L.; Dattwyler, R.J.; Arnaboldi, P.M. Linear B Cell Epitopes Derived from the Multifunctional Surface Lipoprotein BBK32 as Targets for the Serodiagnosis of Lyme Disease. mSphere 2019, 4, e00111-19. [Google Scholar] [CrossRef]
- Reifert, J.; Kamath, K.; Bozekowski, J.; Lis, E.; Horn, E.J.; Granger, D.; Theel, E.S.; Shon, J.; Sawyer, J.R.; Daugherty, P.S. Serum Epitope Repertoire Analysis Enables Early Detection of Lyme Disease with Improved Sensitivity in an Expandable Multiplex Format. J. Clin. Microbiol. 2021, 59, e01836-20. [Google Scholar] [CrossRef]
- Tokarz, R.; Tagliafierro, T.; Caciula, A.; Mishra, N.; Thakkar, R.; Chauhan, L.V.; Sameroff, S.; Delaney, S.; Wormser, G.P.; Marques, A.; et al. Identification of immunoreactive linear epitopes of Borrelia miyamotoi. Ticks Tick-Borne Dis. 2020, 11, 101314. [Google Scholar] [CrossRef] [PubMed]
- Yin, D.; Li, L.; Song, X.; Li, H.; Wang, J.; Ju, W.; Qu, X.; Song, D.; Liu, Y.; Meng, X.; et al. A novel multi-epitope recombined protein for diagnosis of human brucellosis. BMC Infect. Dis. 2016, 16, 219. [Google Scholar] [CrossRef] [PubMed]
- Yin, D.; Bai, Q.; Li, L.; Xu, K.; Zhang, J. Study on immunogenicity and antigenicity of a novel brucella multiepitope recombined protein. Biochem. Biophys. Res. Commun. 2021, 540, 37–41. [Google Scholar] [CrossRef] [PubMed]
- Dehghani, S.; Sabzehei, F.; Taromchi, A.H.; Mobaien, A.R.; Arsang-Jang, S. Hybrid recombinant Omp 22, 25, and 31 immunodominant epitopes can be used for serodiagnosis of brucellosis. J. Immunol. Methods 2021, 497, 113123. [Google Scholar] [CrossRef] [PubMed]
- Yin, D.; Bai, Q.; Wu, X.; Li, H.; Shao, J.; Sun, M.; Jiang, H.; Zhang, J. Paper-based ELISA diagnosis technology for human brucellosis based on a multiepitope fusion protein. PLoS Negl. Trop. Dis. 2021, 15, e0009695. [Google Scholar] [CrossRef]
- Wang, C.; Horby, P.W.; Hayden, F.G.; Gao, G.F. A novel coronavirus outbreak of global health concern. Lancet 2020, 395, 470–473. [Google Scholar] [CrossRef]
- Poh, C.M.; Carissimo, G.; Wang, B.; Amrun, S.N.; Lee, C.Y.-P.; Chee, R.S.-L.; Fong, S.-W.; Yeo, N.K.-W.; Lee, W.-H.; Torres-Ruesta, A.; et al. Two linear epitopes on the SARS-CoV-2 spike protein that elicit neutralising antibodies in COVID-19 patients. Nat. Commun. 2020, 11, 2806. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, Z.; Tian, S.; Li, B.; Feng, T.; He, J.; Jiang, M.; Tang, X.; Mei, S.; Li, H.; et al. A newly identified linear epitope on non-RBD region of SARS-CoV-2 spike protein improves the serological detection rate of COVID-19 patients. BMC Microbiol. 2021, 21, 194. [Google Scholar] [CrossRef]
- Cai, X.F.; Chen, J.; Li Hu, J.; Long, Q.X.; Deng, H.J.; Liu, P.; Fan, K.; Liao, P.; Liu, B.Z.; Wu, G.C.; et al. A Peptide-Based Magnetic Chemiluminescence Enzyme Immunoassay for Serological Diagnosis of Coronavirus Disease 2019. J. Infect. Dis. 2020, 222, 189–193. [Google Scholar] [CrossRef]
- Guo, J.Y.; Liu, I.J.; Lin, H.T.; Wang, M.J.; Chang, Y.L.; Lin, S.C.; Liao, M.Y.; Hsu, W.C.; Lin, Y.L.; Liao, J.C.; et al. Identification of COVID-19 B-cell epitopes with phage-displayed peptide library. J. Biomed. Sci. 2021, 28, 43. [Google Scholar] [CrossRef]
- Kesarwani, V.; Gupta, R.; Vetukuri, R.R.; Kushwaha, S.K.; Gandhi, S. Identification of Unique Peptides for SARS-CoV-2 Diagnostics and Vaccine Development by an In Silico Proteomics Approach. Front. Immunol. 2021, 12, 725240. [Google Scholar] [CrossRef] [PubMed]
- Phan, I.Q.; Subramanian, S.; Kim, D.; Murphy, M.; Pettie, D.; Carter, L.; Anishchenko, I.; Barrett, L.K.; Craig, J.; Tillery, L.; et al. In silico detection of SARS-CoV-2 specific B-cell epitopes and validation in ELISA for serological diagnosis of COVID-19. Sci. Rep. 2021, 11, 4290. [Google Scholar] [CrossRef] [PubMed]
- Can, H.; Köseoğlu, A.E.; Erkunt Alak, S.; Güvendi, M.; Döşkaya, M.; Karakavuk, M.; Gürüz, A.Y.; Ün, C. In silico discovery of antigenic proteins and epitopes of SARS-CoV-2 for the development of a vaccine or a diagnostic approach for COVID-19. Sci. Rep. 2020, 10, 22387. [Google Scholar] [CrossRef] [PubMed]
- Javadi Mamaghani, A.; Arab-Mazar, Z.; Heidarzadeh, S.; Ranjbar, M.M.; Molazadeh, S.; Rashidi, S.; Niazpour, F.; Naghi Vishteh, M.; Bashiri, H.; Bozorgomid, A.; et al. In-silico design of a multi-epitope for developing sero-diagnosis detection of SARS-CoV-2 using spike glycoprotein and nucleocapsid antigens. Netw. Model. Anal. Health Inform. Bioinform. 2021, 10, 61. [Google Scholar] [CrossRef]
- Amrun, S.N.; Lee, C.Y.; Lee, B.; Fong, S.W.; Young, B.E.; Chee, R.S.; Yeo, N.K.; Torres-Ruesta, A.; Carissimo, G.; Poh, C.M.; et al. Linear B-cell epitopes in the spike and nucleocapsid proteins as markers of SARS-CoV-2 exposure and disease severity. EBioMedicine 2020, 58, 102911. [Google Scholar] [CrossRef]
- Wang, X.; Lam, J.Y.; Chen, L.; Au, S.W.; To, K.K.W.; Yuen, K.Y.; Kok, K.H. Mining of linear B cell epitopes of SARS-CoV-2 ORF8 protein from COVID-19 patients. Emerg. Microbes Infect. 2021, 10, 1016–1023. [Google Scholar] [CrossRef]
- Cai, Y.; Song, Y.; Cen, D.; Zhang, C.; Mao, S.; Ye, X.; Xiong, Y.; Jiang, P.; Chen, J.; Xue, X.; et al. Novel ELISA for serodiagnosis of nasopharyngeal carcinoma based on a B cell epitope of Epstein-Barr virus latent membrane protein 2. Oncol. Lett. 2018, 16, 4372–4378. [Google Scholar] [CrossRef]
- Dunmire, S.K.; Verghese, P.S.; Balfour, H.H., Jr. Primary Epstein-Barr virus infection. J. Clin. Virol. 2018, 102, 84–92. [Google Scholar] [CrossRef]
- Nowalk, A.; Green, M. Epstein-Barr Virus. Microbiol. Spectr. 2016, 4, 127–134. [Google Scholar] [CrossRef]
- Casey, J.L.; Coley, A.M.; Street, G.; Parisi, K.; Devine, P.L.; Foley, M. Peptide mimotopes selected from a random peptide library for diagnosis of Epstein-Barr virus infection. J. Clin. Microbiol. 2006, 44, 764–771. [Google Scholar] [CrossRef]
- Lin, X.; Chen, S.; Xue, X.; Lu, L.; Zhu, S.; Li, W.; Chen, X.; Zhong, X.; Jiang, P.; Sename, T.S.; et al. Chimerically fused antigen rich of overlapped epitopes from latent membrane protein 2 (LMP2) of Epstein-Barr virus as a potential vaccine and diagnostic agent. Cell. Mol. Immunol. 2016, 13, 492–501. [Google Scholar] [CrossRef] [PubMed]
- Xue, X.; Zhu, S.; Li, W.; Chen, J.; Ou, Q.; Zheng, M.; Gong, W.; Zhang, L. Identification and characterization of novel B-cell epitopes within EBV latent membrane protein 2 (LMP2). Viral Immunol. 2011, 24, 227–236. [Google Scholar] [CrossRef] [PubMed]
- Muller, D.A.; Depelsenaire, A.C.; Young, P.R. Clinical and Laboratory Diagnosis of Dengue Virus Infection. J. Infect. Dis. 2017, 215, S89–S95. [Google Scholar] [CrossRef] [PubMed]
- Olagnier, D.; Amatore, D.; Castiello, L.; Ferrari, M.; Palermo, E.; Diamond, M.S.; Palamara, A.T.; Hiscott, J. Dengue Virus Immunopathogenesis: Lessons Applicable to the Emergence of Zika Virus. J. Mol. Biol. 2016, 428, 3429–3448. [Google Scholar] [CrossRef]
- Maldaner, F.R.; Aragao, F.J.; dos Santos, F.B.; Franco, O.L.; da Rocha Queiroz Lima, M.; de Oliveira Resende, R.; Vasques, R.M.; Nagata, T. Dengue virus tetra-epitope peptide expressed in lettuce chloroplasts for potential use in dengue diagnosis. Appl. Microbiol. Biotechnol. 2013, 97, 5721–5729. [Google Scholar] [CrossRef]
- Zhu, T.; He, J.; Chen, W.; Ho, H.P.; Kong, S.K.; Wang, C.; Long, J.; Fong-Chuen Loo, J.; Gu, D. Development of peptide-based chemiluminescence enzyme immunoassay (CLEIA) for diagnosis of dengue virus infection in human. Anal. Biochem. 2018, 556, 112–118. [Google Scholar] [CrossRef]
- Nagar, P.K.; Savargaonkar, D.; Anvikar, A.R. Detection of Dengue Virus-Specific IgM and IgG Antibodies through Peptide Sequences of Envelope and NS1 Proteins for Serological Identification. J. Immunol. Res. 2020, 2020, 1820325. [Google Scholar] [CrossRef]
- Bergamaschi, G.; Fassi, E.M.A.; Romanato, A.; D’Annessa, I.; Odinolfi, M.T.; Brambilla, D.; Damin, F.; Chiari, M.; Gori, A.; Colombo, G.; et al. Computational Analysis of Dengue Virus Envelope Protein (E) Reveals an Epitope with Flavivirus Immunodiagnostic Potential in Peptide Microarrays. Int. J. Mol. Sci. 2019, 20, 1921. [Google Scholar] [CrossRef]
- Nguyen, N.M.; Duong, B.T.; Azam, M.; Phuong, T.T.; Park, H.; Thuy, P.T.B.; Yeo, S.J. Diagnostic Performance of Dengue Virus Envelope Domain III in Acute Dengue Infection. Int. J. Mol. Sci. 2019, 20, 3464. [Google Scholar] [CrossRef]
- Zomosa-Signoret, V.C.; Morales-Gonzalez, K.R.; Estrada-Rodriguez, A.E.; Rivas-Estilla, A.M.; Deveze-Garcia, M.C.; Galaviz-Aguilar, E.; Vidaltamayo, R. Alanine Substitution Inactivates Cross-Reacting Epitopes in Dengue Virus Recombinant Envelope Proteins. Viruses 2020, 12, 208. [Google Scholar] [CrossRef]
- Wu, H.C.; Huang, Y.L.; Chao, T.T.; Jan, J.T.; Huang, J.L.; Chiang, H.Y.; King, C.C.; Shaio, M.F. Identification of B-cell epitope of dengue virus type 1 and its application in diagnosis of patients. J. Clin. Microbiol. 2001, 39, 977–982. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.C.; Jung, M.Y.; Chiu, C.Y.; Chao, T.T.; Lai, S.C.; Jan, J.T.; Shaio, M.F. Identification of a dengue virus type 2 (DEN-2) serotype-specific B-cell epitope and detection of DEN-2-immunized animal serum samples using an epitope-based peptide antigen. J. Gen. Virol. 2003, 84, 2771–2779. [Google Scholar] [CrossRef] [PubMed]
- Versiani, A.F.; Rocha, R.P.; Mendes, T.A.O.; Pereira, G.C.; Coelho Dos Reis, J.G.A.; Bartholomeu, D.C.; da Fonseca, F.G. Identification of B-Cell Epitopes with Potential to Serologicaly Discrimnate Dengue from Zika Infections. Viruses 2019, 11, 1079. [Google Scholar] [CrossRef] [PubMed]
- de Souza, M.Q.; Galdino, A.S.; dos Santos, J.C.; Soares, M.V.; de Nóbrega, Y.C.; Alvares Ada, C.; de Freitas, S.M.; Torres, F.A.; Felipe, M.S. A recombinant multiepitope protein for hepatitis B diagnosis. BioMed Res. Int. 2013, 2013, 148317. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.J.; Chen, Z.M.; Wei, M.; Liu, J.Q.; Li, Z.L.; Shi, T.S.; Nian, S.; Fu, R.; Wu, Y.T.; Zhang, Y.L.; et al. Specific determination of hepatitis B e antigen by antibodies targeting precore unique epitope facilitates clinical diagnosis and drug evaluation against hepatitis B virus infection. Emerg. Microbes Infect. 2021, 10, 37–50. [Google Scholar] [CrossRef] [PubMed]
- Dipti, C.A.; Jain, S.K.; Navin, K. A novel recombinant multiepitope protein as a hepatitis C diagnostic intermediate of high sensitivity and specificity. Protein Expr. Purif. 2006, 47, 319–328. [Google Scholar] [CrossRef]
- He, J.; Xiu, B.; Wang, G.; Chen, K.; Feng, X.; Song, X.; Zhu, C.; Yang, X.; Bai, G.; Ling, S.; et al. Construction, expression, purification and biotin labeling of a single recombinant multi-epitope antigen for double-antigen sandwich ELISA to detect hepatitis C virus antibody. Protein Pept. Lett. 2011, 18, 839–847. [Google Scholar] [CrossRef]
- Hua, R.; Jiang, X.; Qi, L.; Guan, S.; Kuai, Z.; Qiao, Y.; Xu, Y.; Gong, X.; Shi, Y.; Kong, W.; et al. Screening HCV genotype-specific epitope peptides based on conserved sequence analysis and B cell epitope prediction in HCV E2 region. Immunol. Res. 2018, 66, 67–73. [Google Scholar] [CrossRef]
- Quiroga, J.A.; Castillo, I.; Llorente, S.; Bartolomé, J.; Barril, G.; Carreño, V. Identification of serologically silent occult hepatitis C virus infection by detecting immunoglobulin G antibody to a dominant HCV core peptide epitope. J. Hepatol. 2009, 50, 256–263. [Google Scholar] [CrossRef]
- Changula, K.; Yoshida, R.; Noyori, O.; Marzi, A.; Miyamoto, H.; Ishijima, M.; Yokoyama, A.; Kajihara, M.; Feldmann, H.; Mweene, A.S.; et al. Mapping of conserved and species-specific antibody epitopes on the Ebola virus nucleoprotein. Virus Res. 2013, 176, 83–90. [Google Scholar] [CrossRef]
- Becquart, P.; Mahlakoiv, T.; Nkoghe, D.; Leroy, E.M. Identification of continuous human B-cell epitopes in the VP35, VP40, nucleoprotein and glycoprotein of Ebola virus. PLoS ONE 2014, 9, e96360. [Google Scholar] [CrossRef] [PubMed]
- Babirye, P.; Musubika, C.; Kirimunda, S.; Downing, R.; Lutwama, J.J.; Mbidde, E.K.; Weyer, J.; Paweska, J.T.; Joloba, M.L.; Wayengera, M. Identity and validity of conserved B cell epitopes of filovirus glycoprotein: Towards rapid diagnostic testing for Ebola and possibly Marburg virus disease. BMC Infect. Dis. 2018, 18, 498. [Google Scholar] [CrossRef]
- Conte, F.P.; Tinoco, B.C.; Santos Chaves, T.; Oliveira, R.C.; Figueira Mansur, J.; Mohana-Borges, R.; Lemos, E.R.S.; Neves, P.; Rodrigues-da-Silva, R.N. Identification and validation of specific B-cell epitopes of hantaviruses associated to hemorrhagic fever and renal syndrome. PLoS Negl. Trop. Dis. 2019, 13, e0007915. [Google Scholar] [CrossRef]
- Kalaiselvan, S.; Sankar, S.; Ramamurthy, M.; Ghosh, A.R.; Nandagopal, B.; Sridharan, G. Prediction of B Cell Epitopes Among Hantavirus Strains Causing Hemorragic Fever with Renal Syndrome. J. Cell. Biochem. 2017, 118, 1182–1188. [Google Scholar] [CrossRef] [PubMed]
- Kalaiselvan, S.; Sankar, S.; Ramamurthy, M.; Ghosh, A.R.; Nandagopal, B.; Sridharan, G. Prediction of Pan-Specific B-Cell Epitopes from Nucleocapsid Protein of Hantaviruses Causing Hantavirus Cardiopulmonary Syndrome. J. Cell. Biochem. 2017, 118, 2320–2324. [Google Scholar] [CrossRef] [PubMed]
- Siman-Tov, D.D.; Zemel, R.; Tur Kaspa, R.; Gershoni, J.M. The use of epitope arrays in immunodiagnosis of infectious disease: Hepatitis C virus, a case study. Anal. Biochem. 2013, 432, 63–70. [Google Scholar] [CrossRef]
- Hajissa, K.; Zakaria, R.; Suppian, R.; Mohamed, Z. An evaluation of a recombinant multiepitope based antigen for detection of Toxoplasma gondii specific antibodies. BMC Infect. Dis. 2017, 17, 807. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, G.; Ou, J.; Yin, H.; Zhang, D. Analyzing and identifying novel B cell epitopes within Toxoplasma gondii GRA4. Parasites Vectors 2014, 7, 474. [Google Scholar] [CrossRef]
- Javadi Mamaghani, A.; Fathollahi, A.; Spotin, A.; Ranjbar, M.M.; Barati, M.; Aghamolaie, S.; Karimi, M.; Taghipour, N.; Ashrafi, M.; Tabaei, S.J.S. Candidate antigenic epitopes for vaccination and diagnosis strategies of Toxoplasma gondii infection: A review. Microb. Pathog. 2019, 137, 103788. [Google Scholar] [CrossRef]
- Akhoundi, M.; Downing, T.; Votypka, J.; Kuhls, K.; Lukes, J.; Cannet, A.; Ravel, C.; Marty, P.; Delaunay, P.; Kasbari, M.; et al. Leishmania infections: Molecular targets and diagnosis. Mol. Asp. Med. 2017, 57, 1–29. [Google Scholar] [CrossRef]
- Song, Y.; Zhao, Y.; Pan, K.; Shen, B.; Fang, R.; Hu, M.; Zhao, J.; Zhou, Y. Characterization and evaluation of a recombinant multiepitope peptide antigen MAG in the serological diagnosis of Toxoplasma gondii infection in pigs. Parasites Vectors 2021, 14, 408. [Google Scholar] [CrossRef] [PubMed]
- Denny, P.W.; Kalesh, K. How can proteomics overhaul our understanding of Leishmania biology? Expert Rev. Proteom. 2020, 17, 789–792. [Google Scholar] [CrossRef] [PubMed]
- van Griensven, J.; Diro, E. Visceral leishmaniasis. Infect. Dis. Clin. N. Am. 2012, 26, 309–322. [Google Scholar] [CrossRef] [PubMed]
- Machado, A.S.; Ramos, F.F.; Oliveira-da-Silva, J.A.; Santos, T.T.O.; Ludolf, F.; Tavares, G.S.V.; Costa, L.E.; Lage, D.P.; Steiner, B.T.; Chaves, A.T.; et al. A Leishmania infantum hypothetical protein evaluated as a recombinant protein and specific B-cell epitope for the serodiagnosis and prognosis of visceral leishmaniasis. Acta Trop. 2020, 203, 105318. [Google Scholar] [CrossRef]
- Mendes, T.M.; Roma, E.H.; Costal-Oliveira, F.; Dhom-Lemos, L.C.; Toledo-Machado, C.M.; Bruna-Romero, O.; Bartholomeu, D.C.; Fujiwara, R.T.; Chavez-Olortegui, C. Epitope mapping of recombinant Leishmania donovani virulence factor A2 (recLdVFA2) and canine leishmaniasis diagnosis using a derived synthetic bi-epitope. PLoS Negl. Trop. Dis. 2017, 11, e0005562. [Google Scholar] [CrossRef]
- Faria, A.R.; de Castro Veloso, L.; Coura-Vital, W.; Reis, A.B.; Damasceno, L.M.; Gazzinelli, R.T.; Andrade, H.M. Novel recombinant multiepitope proteins for the diagnosis of asymptomatic leishmania infantum-infected dogs. PLoS Negl. Trop. Dis. 2015, 9, e3429. [Google Scholar] [CrossRef]
- Menezes-Souza, D.; Mendes, T.A.; Nagem, R.A.; Santos, T.T.; Silva, A.L.; Santoro, M.M.; de Carvalho, S.F.; Coelho, E.A.; Bartholomeu, D.C.; Fujiwara, R.T. Mapping B-cell epitopes for the peroxidoxin of Leishmania (Viannia) braziliensis and its potential for the clinical diagnosis of tegumentary and visceral leishmaniasis. PLoS ONE 2014, 9, e99216. [Google Scholar] [CrossRef]
- McManus, D.P.; Dunne, D.W.; Sacko, M.; Utzinger, J.; Vennervald, B.J.; Zhou, X.N. Schistosomiasis. Nat. Rev. Dis. Prim. 2018, 4, 13. [Google Scholar] [CrossRef]
- Chen, Y.; Giri, B.R.; Li, X.; He, X.; Jing, Z.; Cheng, G. Preliminary evaluation of the diagnostic potential of Schistosoma japonicum extracellular vesicle proteins for Schistosomiasis japonica. Acta Trop. 2020, 201, 105184. [Google Scholar] [CrossRef]
- Mu, Y.; Gordon, C.A.; Olveda, R.M.; Ross, A.G.; Olveda, D.U.; Marsh, J.M.; McManus, D.P.; Cai, P. Identification of a linear B-cell epitope on the Schistosoma japonicum saposin protein, SjSAP4: Potential as a component of a multi-epitope diagnostic assay. PLoS Negl. Trop. Dis. 2022, 16, e0010619. [Google Scholar] [CrossRef]
- Galvani, N.C.; Machado, A.S.; Lage, D.P.; Martins, V.T.; de Oliveira, D.; Freitas, C.S.; Vale, D.L.; Fernandes, B.B.; Oliveira-da-Silva, J.A.; Reis, T.A.R.; et al. Sensitive and specific serodiagnosis of tegumentary leishmaniasis using a new chimeric protein based on specific B-cell epitopes of Leishmania antigenic proteins. Microb. Pathog. 2022, 162, 105341. [Google Scholar] [CrossRef] [PubMed]
- Jamal, F.; Dikhit, M.R.; Singh, M.K.; Shivam, P.; Kumari, S.; Pushpanjali, S.; Dubey, A.K.; Kumar, P.; Narayan, S.; Gupta, A.K.; et al. Identification of B-cell Epitope of Leishmania donovani and its application in diagnosis of visceral leishmaniasis. J. Biomol. Struct. Dyn. 2017, 35, 3569–3580. [Google Scholar] [CrossRef] [PubMed]
- Soto, M.; Requena, J.M.; Quijada, L.; Alonso, C. Multicomponent chimeric antigen for serodiagnosis of canine visceral leishmaniasis. J. Clin. Microbiol. 1998, 36, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Boarino, A.; Scalone, A.; Gradoni, L.; Ferroglio, E.; Vitale, F.; Zanatta, R.; Giuffrida, M.G.; Rosati, S. Development of recombinant chimeric antigen expressing immunodominant B epitopes of Leishmania infantum for serodiagnosis of visceral leishmaniasis. Clin. Diagn. Lab. Immunol. 2005, 12, 647–653. [Google Scholar] [CrossRef]
- Faria, A.R.; Costa, M.M.; Giusta, M.S.; Grimaldi, G., Jr.; Penido, M.L.; Gazzinelli, R.T.; Andrade, H.M. High-throughput analysis of synthetic peptides for the immunodiagnosis of canine visceral leishmaniasis. PLoS Negl. Trop. Dis. 2011, 5, e1310. [Google Scholar] [CrossRef]
- Costa, L.E.; Salles, B.C.S.; Santos, T.T.O.; Ramos, F.F.; Lima, M.P.; Lima, M.I.S.; Portela, A.S.B.; Chavez-Fumagalli, M.A.; Duarte, M.C.; Menezes-Souza, D.; et al. Antigenicity of phage clones and their synthetic peptides for the serodiagnosis of canine and human visceral leishmaniasis. Microb. Pathog. 2017, 110, 14–22. [Google Scholar] [CrossRef]
- Carvalho, A.; Mendes, T.A.O.; Coelho, E.A.F.; Duarte, M.C.; Menezes-Souza, D. New antigens for the serological diagnosis of human visceral leishmaniasis identified by immunogenomic screening. PLoS ONE 2018, 13, e0209599. [Google Scholar] [CrossRef]
- Bremer Hinckel, B.C.; Marlais, T.; Airs, S.; Bhattacharyya, T.; Imamura, H.; Dujardin, J.C.; El-Safi, S.; Singh, O.P.; Sundar, S.; Falconar, A.K.; et al. Refining wet lab experiments with in silico searches: A rational quest for diagnostic peptides in visceral leishmaniasis. PLoS Negl. Trop. Dis. 2019, 13, e0007353. [Google Scholar] [CrossRef]
- Ramos, F.F.; Tavares, G.S.V.; Ludolf, F.; Machado, A.S.; Santos, T.T.O.; Goncalves, I.A.P.; Dias, A.C.S.; Alves, P.T.; Fraga, V.G.; Bandeira, R.S.; et al. Diagnostic application of sensitive and specific phage-exposed epitopes for visceral leishmaniasis and human immunodeficiency virus coinfection. Parasitology 2021, 148, 1706–1714. [Google Scholar] [CrossRef]
- Maksimov, P.; Zerweck, J.; Maksimov, A.; Hotop, A.; Gross, U.; Pleyer, U.; Spekker, K.; Daubener, W.; Werdermann, S.; Niederstrasser, O.; et al. Peptide microarray analysis of in silico-predicted epitopes for serological diagnosis of Toxoplasma gondii infection in humans. Clin. Vaccine Immunol. 2012, 19, 865–874. [Google Scholar] [CrossRef]
- Dai, J.; Jiang, M.; Wang, Y.; Qu, L.; Gong, R.; Si, J. Evaluation of a recombinant multiepitope peptide for serodiagnosis of Toxoplasma gondii infection. Clin. Vaccine Immunol. 2012, 19, 338–342. [Google Scholar] [CrossRef] [PubMed]
- Dard, C.; Swale, C.; Brenier-Pinchart, M.P.; Farhat, D.C.; Bellini, V.; Robert, M.G.; Cannella, D.; Pelloux, H.; Tardieux, I.; Hakimi, M.A. A brain cyst load-associated antigen is a Toxoplasma gondii biomarker for serodetection of persistent parasites and chronic infection. BMC Biol. 2021, 19, 25. [Google Scholar] [CrossRef] [PubMed]
- Mickum, M.L.; Prasanphanich, N.S.; Song, X.; Dorabawila, N.; Mandalasi, M.; Lasanajak, Y.; Luyai, A.; Secor, W.E.; Wilkins, P.P.; Van Die, I.; et al. Identification of Antigenic Glycans from Schistosoma mansoni by Using a Shotgun Egg Glycan Microarray. Infect. Immun. 2016, 84, 1371–1386. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, G.B.F.; Resende, D.M.; Siqueira, L.M.V.; Lopes, M.D.; Lopes, D.O.; Coelho, P.M.Z.; Teixeira-Carvalho, A.; Ruiz, J.C.; Fonseca, C.T. Selecting targets for the diagnosis of Schistosoma mansoni infection: An integrative approach using multi-omic and immunoinformatics data. PLoS ONE 2017, 12, e0182299. [Google Scholar] [CrossRef]
- De Benedetti, S.; Di Pisa, F.; Fassi, E.M.A.; Cretich, M.; Musico, A.; Frigerio, R.; Mussida, A.; Bombaci, M.; Grifantini, R.; Colombo, G.; et al. Structure, Immunoreactivity, and In Silico Epitope Determination of SmSPI S. mansoni Serpin for Immunodiagnostic Application. Vaccines 2021, 9, 322. [Google Scholar] [CrossRef]
- Ma, L.; Zhao, W.; Hou, X.; Liu, M.; Li, Y.; Shen, L.; Xu, X. Identification of linear epitopes in SjSP-13 of Schistosoma japonicum using a GST-peptide fusion protein microplate array. Parasites Vectors 2019, 12, 507. [Google Scholar] [CrossRef]
- Yao, M.X.; Sun, X.D.; Gao, Y.H.; Cheng, Z.B.; Deng, W.W.; Zhang, J.J.; Wang, H. Multi-epitope chimeric antigen used as a serological marker to estimate Plasmodium falciparum transmission intensity in the border area of China-Myanmar. Infect. Dis. Poverty 2016, 5, 98. [Google Scholar] [CrossRef][Green Version]
- Strickland, A.B.; Shi, M. Mechanisms of fungal dissemination. Cell. Mol. Life Sci. CMLS 2021, 78, 3219–3238. [Google Scholar] [CrossRef]
- Brown, G.D.; Denning, D.W.; Gow, N.A.; Levitz, S.M.; Netea, M.G.; White, T.C. Hidden killers: Human fungal infections. Sci. Transl. Med. 2012, 4, 165rv113. [Google Scholar] [CrossRef]
- von Lilienfeld-Toal, M.; Wagener, J.; Einsele, H.; Cornely, O.A.; Kurzai, O. Invasive Fungal Infection. Dtsch. Arztebl. Int. 2019, 116, 271–278. [Google Scholar] [CrossRef]
- Moreira, A.L.E.; Oliveira, M.A.P.; Silva, L.O.S.; Inácio, M.M.; Bailão, A.M.; Parente-Rocha, J.A.; Cruz-Leite, V.R.M.; Paccez, J.D.; de Almeida Soares, C.M.; Weber, S.S.; et al. Immunoproteomic Approach of Extracellular Antigens From Paracoccidioides Species Reveals Exclusive B-Cell Epitopes. Front. Microbiol. 2019, 10, 2968. [Google Scholar] [CrossRef] [PubMed]
- Almeida, M.A.; Almeida-Paes, R.; Guimarães, A.J.; Valente, R.H.; Soares, C.M.A.; Zancopé-Oliveira, R.M. Immunoproteomics Reveals Pathogen’s Antigens Involved in Homo sapiens-Histoplasma capsulatum Interaction and Specific Linear B-Cell Epitopes in Histoplasmosis. Front. Cell. Infect. Microbiol. 2020, 10, 591121. [Google Scholar] [CrossRef] [PubMed]
- Martins, L.M.; de Andrade, H.M.; Vainstein, M.H.; Wanke, B.; Schrank, A.; Balaguez, C.B.; dos Santos, P.R.; Santi, L.; Pires Sda, F.; da Silva, A.S.; et al. Immunoproteomics and immunoinformatics analysis of Cryptococcus gattii: Novel candidate antigens for diagnosis. Future Microbiol. 2013, 8, 549–563. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Su, Q.P.; Wang, G.Y.; Wen, D.Z.; Zhang, Y.H.; Bao, H.Z.; Wang, L. Production of hybrid phage displaying secreted aspartyl proteinase epitope of Candida albicans and its application for the diagnosis of disseminated candidiasis. Mycoses 2007, 50, 165–171. [Google Scholar] [CrossRef]
- Quanping, S.; Yanyan, H.; Yicun, W.; Zhigang, J.; Yuling, G.; Li, W. The use of hybrid phage displaying antigen epitope and recombinant protein in the diagnosis of systemic Candida albicans infection in rabbits and cancer patients. Diagn. Microbiol. Infect. Dis. 2010, 68, 382–389. [Google Scholar] [CrossRef]
- Caldini, C.P.; Xander, P.; Kioshima, É.S.; Bachi, A.L.; de Camargo, Z.P.; Mariano, M.; Lopes, J.D. Synthetic peptides mimic gp75 from Paracoccidioides brasiliensis in the diagnosis of paracoccidioidomycosis. Mycopathologia 2012, 174, 1–10. [Google Scholar] [CrossRef]
- Portes, L.D.S.; Kioshima, E.S.; de Camargo, Z.P.; Batista, W.L.; Xander, P. Subtractive phage display selection for screening and identification of peptide sequences with potential use in serodiagnosis of paracoccidioidomycosis caused by Paracoccidioides brasiliensis. Lett. Appl. Microbiol. 2017, 65, 346–353. [Google Scholar] [CrossRef]
- de Serpa Brandão, R.M.; Soares Martins, L.M.; de Andrade, H.M.; Faria, A.R.; Soares Leal, M.J.; da Silva, A.S.; Wanke, B.; dos Santos Lazéra, M.; Vainstein, M.H.; Mendes, R.P.; et al. Immunoreactivity of synthetic peptides derived from proteins of Cryptococcus gattii. Future Microbiol. 2014, 9, 871–878. [Google Scholar] [CrossRef]
- Vahedi, F.; Ghasemi, Y.; Atapour, A.; Zomorodian, K.; Ranjbar, M.; Monabati, A.; Nezafat, N.; Savardashtaki, A. B-Cell Epitope Mapping from Eight Antigens of Candida albicans to Design a Novel Diagnostic Kit: An Immunoinformatics Approach. Int. J. Pept. Res. Ther. 2022, 28, 110. [Google Scholar] [CrossRef]
- de Serpa Brandão, R.M.S.; Faria, A.R.; de Andrade, H.M.; Soares Martins, L.M.; da Silva, A.S.; do Monte, S.J.H. Novel recombinant multiepitope proteins for the detection of anti-Cryptococcus antibodies. Future Microbiol. 2018, 13, 429–436. [Google Scholar] [CrossRef]
- Tomás, A.L.; Cardoso, F.; Esteves, F.; Matos, O. Serological diagnosis of pneumocystosis: Production of a synthetic recombinant antigen for immunodetection of Pneumocystis jirovecii. Sci. Rep. 2016, 6, 36287. [Google Scholar] [CrossRef] [PubMed]
- Tomás, A.L.; Cardoso, F.; de Sousa, B.; Matos, O. Detection of anti-Pneumocystis jirovecii antibodies in human serum using a recombinant synthetic multi-epitope kexin-based antigen. Eur. J. Clin. Microbiol. Infect. Dis. Off. Publ. Eur. Soc. Clin. Microbiol. 2020, 39, 2205–2209. [Google Scholar] [CrossRef] [PubMed]
- Inácio, M.M.; Cruz-Leite, V.R.M.; Moreira, A.L.E.; Mattos, K.; Paccez, J.D.; Ruiz, O.H.; Venturini, J.; de Souza Carvalho Melhem, M.; Paniago, A.M.M.; de Almeida Soares, C.M.; et al. Challenges in Serologic Diagnostics of Neglected Human Systemic Mycoses: An Overview on Characterization of New Targets. Pathogens 2022, 11, 569. [Google Scholar] [CrossRef] [PubMed]
- Potocnakova, L.; Bhide, M.; Pulzova, L.B. An Introduction to B-Cell Epitope Mapping and In Silico Epitope Prediction. J. Immunol. Res. 2016, 2016, 6760830. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.R.; Zhu, X.Z.; Liu, D.; Liu, L.L.; Tong, M.L.; Yang, T.C. Are nontreponemal tests suitable for monitoring syphilis treatment efficacy? Evidence from rabbit infection models. Clin. Microbiol. Infect. 2020, 26, 240–246. [Google Scholar] [CrossRef]
- Luo, Y.; Xie, Y.; Xiao, Y. Laboratory Diagnostic Tools for Syphilis: Current Status and Future Prospects. Front. Cell. Infect. Microbiol. 2020, 10, 574806. [Google Scholar] [CrossRef]
| Methods | Year | Description | Availability | |
|---|---|---|---|---|
| Linear Epitopes | ||||
| Propensity scales-based | Hopp and Woods [13] | 1981 | Hopp–Woods hydrophilicity. | Not stated |
| PEOPLE [14] | 1999 | Antigenic index AG, which includes secondary structure, hydrophilicity, surface accessibility and flexibility. | Not stated | |
| BcePred [15] | 2004 | Combination of residue properties hydrophilicity, flexibility, polarity and surface solvent accessibility. | http://crdd.osdd.net/raghava/bcepred/index.html (accessed on 1 September 2022) | |
| ML-based | BepiPred [17] | 2006 | Combine an HMM with an amino acid propensity scale. | Not currently available online |
| ABCpred [18] | 2006 | Based on standard feed-forward (FNN) and recurrent neural network (RNN). | http://www.imtech.res.in/raghava/abcpred/ (accessed on 1 September 2022) | |
| SVMTriP [33] | 2012 | Use support vector machine (SVM) to combine the tripeptide similarity and propensity scores. | http://sysbio.unl.edu/SVMTriP/ (accessed on 1 September 2022) | |
| BepiPred 2.0 [19] | 2017 | Random forest (RF) algorithm trained on epitopes derived from antibody–antigen complex structures. | https://services.healthtech.dtu.dk/service.php?BepiPred-2.0 (accessed on 1 September 2022) | |
| Conformational Epitopes | ||||
| Sequence-based | COBEpro [34] | 2009 | A two-step method, which first uses a SVM model to assign an epitopic propensity score to fragments within the given peptide sequence and then calculates an epitopic propensity score for each residue based on the scores in the first stage. | http://scratch.proteomics.ics.uci.edu (accessed on 1 September 2022) |
| Bprediction [35] | 2012 | Adopt ensemble learning approach to incorporate various sequence-derived features, and develop an ensemble model. | Not currently available online | |
| Structure-based | CEP [20] | 2005 | Use accessibility of residues and spatial distance cutoff to predict epitopes of protein antigens with known structures | Not currently available online |
| DiscoTope [21] | 2006 | Use solvent accessibility, amino acid statistics and spatial information. | Not currently available online (DiscoTope2.0: https://services.healthtech.dtu.dk/service.php?DiscoTope-2.0) (accessed on 1 September 2022) | |
| ElliPro [22] | 2008 | Implement Thornton’s method, and together with a residue clustering algorithm, the MODELLER program and the Jmol viewer for predicting CEs. | http://tools.immuneepitope.org/ellipro/ (accessed on 1 September 2022) | |
| SEPPA [23] | 2009 | Employ the concept of “unit patch of residue triangle” to describe the local spatial context of protein surface and “clustering coefficient” to describe the spatial compactness of surface residues. Then, the two features are combined to predict epitopes. | Not currently available online (SEPPA3.0: http://www.badd-cao.net/seppa3/index.html) (accessed on 1 September 2022) | |
| Mimotope-based | MIMOX [28] | 2006 | Map a single mimotope or a consensus sequence of a set of mimotopes onto the corresponding antigen structure and search for all of the clusters of residues that could represent the native epitope. | Not currently available online |
| MimoPro [27] | 2011 | Operate a searching algorithm on a series of overlapping patches on the surface of a protein. These patches are then transformed to a number of graphs using an adaptable distance threshold regulated by an appropriate compactness factor. | Not currently available online | |
| PepMapper [24] | 2012 | An ensemble approach to incorporate two servers: Pep-3D-Search and MimoPro. | Not currently available online | |
| Antibody-based | Shinji Soga [29] | 2010 | Predict epitopes for individual antibodies by narrowing down candidate epitope residues using the Propose ASEP index proposed in this method. | Not stated |
| EpiPred [30] | 2014 | Combine conformational matching of the antibody–antigen structures and a specific antibody–antigen score. | http://www.stats.ox.ac.uk/research/proteins/resources (accessed on 1 September 2022) | |
| PEASE [36] | 2014 | Use antibody–antigen contact preferences, as well as other properties computed from the antibody sequence and antigen structure or sequence. | Not currently available online | |
| # | Epitope Mapping Technologies | Advantages | Disadvantages |
|---|---|---|---|
| 1 | X-ray crystallography | Gold standard method that provides detailed information about the epitope and paratope. | Laborious, time-consuming and complicated for eutectics. |
| 2 | NMR spectroscopy | Providing a dynamic picture of the antibody–antigen complex in solution. | Restricted to small proteins and peptides, time-consuming and complicated. |
| 3 | SPR | Highly sensitive, requires no additional biomarkers and can be dynamically tracked. | Low sensitivity for small molecule detection and high environmental requirements for the SPR sensor. |
| 4 | Pepscan | Low-cost and rapid. | Unable to provide complete epitope information. |
| 5 | Amino acid site-directed mutagenesis | Simple and quick to screen several hundreds or thousands of proteins. | Difficult to identify whether the mutation has disrupted the folding of the protein or is genuinely a key interacting residue. |
| 6 | Surface display technology | High-throughput screening, highly stable and easy to operate. | Different display systems have their own shortcomings; for example, the insertion of exogenous proteins in the phage display system may affect the assembly of phage, or the insertion of certain protein sequences into the bacterial surface system may lead to low protein secretion. |
| Pathogens | Year | Associated Proteins/Peptides | Main Outcome | Ref. | ||
|---|---|---|---|---|---|---|
| Sensitivity | Specificity | Others | ||||
| Mycobacterium tuberculosis | 2007 | Rv3872 | P8 + P9: for pulmonary TB: 94%, for extra-pulmonary TB: 90% | Both 100% | / | [70] |
| 2013 | ESAT-6 and CFP-10 | / | / | E5 peptide and CFP10/ESAT-6 protein obtain similar results. | [56] | |
| 2014 | ESAT-6, CFP-10, CFP-21 and MPT-64 | Combined peptides: for smear-positive pulmonary TB: 83.3%, for smear-negative pulmonary TB: 62.5%, for sarcoidosis: 4.16% | 100% | / | [57] | |
| 2022 | Rv1981c, Rv2659c, Rv3879c | / | / | The polypeptide molecule can induce robust immune responses, and may be a new diagnostic biomarker for latent TB infection | [58] | |
| Staphylococcus aureus | 1998 | SEA and SEB | / | / | The mAbs for epitopes were able to quantitate the native SEA or SEB at nanogram levels. | [62] |
| 2015 | PSau5 and PSau7 | / | / | A competitive ELISA based on epitopes can detect S. aureus down to 104 CFU mL−1 | [63] | |
| Leptospira | 2006 | Phage mimotopes | / | / | Mimotopes reacting with both mAbs and patients’ sera have potential for further use as diagnostic reagent. | [71] |
| 2008 | OmpL1, LipL21, and LipL32 | / | / | The multiepitope protein recognized IgG and IgM in all the sera that were MAT positive. | [72] | |
| 2010 | Hap1/LipL32 | / | IgM: 89% IgG: 100% | The peptide is an earlier serological diagnosis of human leptospirosis than MAT. | [69] | |
| 2014 | LigA | Epitopes 1 and 2: 97.9% | Epitopes 1 and 2: 99.1% | / | [65] | |
| 2016 | LipL21(r-I-LipL21) | 92.59% | 92.86% | / | [66] | |
| 2017 | LK90543 and LK901110 of LigA | 77~89% | 93~96% | The results of mAb-based dot blot ELISA; The mAbs are targeted for epitopes. | [67] | |
| Chlamydia trachomatis | 2008 | OmcB | 23.9% | 94.3% | / | [73] |
| 2018 | 11 peptides from 8 proteins | 11 peptides: 91.8% 5 optimal peptides: 86.5% | Both 98% | / | [74] | |
| 2018 | 12 peptides from different proteins | Ctr Mix1:94% | Ctr Mix1: 98% | / | [75] | |
| Chlamydia pneumoniae | 2019 | Mixed peptides from different proteins | CpnMixF12: 91% | CpnMixF12: 95% | / | [76] |
| Borrelia burgdorferi | 2014 | OppA2 | OppA (191-225): 44.2% | OppA (191-225): 95.5% | / | [77] |
| 2016 | OspC | / | / | Six OspC epitopes capable of distinguishing between Lyme disease patient and healthy control sera. | [78] | |
| 2017 | OspA and OspC | A/C-2 and A/C-7.1: 80.17% and 91.37% | A/C-2 and A/C-7.1: 52.83% and 73.58% | / | [79] | |
| 2019 | BBK32 | BBK32(51–80): 33.3% | BBK32(51–80): 94.7% | / | [80] | |
| 2021 | Epitope motifs | 77% | 99% | Results of diagnosing early Lyme disease | [81] | |
| Borrelia miyamotoi | 2020 | Several peptides identified by peptide array | / | / | The panel of linear peptides may have greater potential for differential diagnosis. | [82] |
| brucella | 2016 | OMP16, 2b, 31, and BP26 (periplasmic protein) | 88.89% | 85.54% | / | [83] |
| 2021 | OMPs | 96.49% (95% CI, 87.89 to 99.57) | 94.44% (95% CI, 81.34 to 99.32 | The results are obtained at optimum cut off value 0.6195 | [84] | |
| 2021 | Recombinant protein (OMP 22, 25, and 31) | 84.37% | 83.78% | / | [85] | |
| 2021 | Multiepitope protein (BP26, OMP31, 16, 2b and 25) | 92.38% | 98.35% | PPV: 98.26% NPV:91.67%. | [86] | |
| Pathogens | Year | Associated Proteins/Peptides | Main Outcome | Ref. | ||
|---|---|---|---|---|---|---|
| Sensitivity | Specificity | Others | ||||
| SARS-CoV-2 | 2020 | S protein (S14P5 and S21P2) | / | / | Two epitopes are strongly recognized by sera from COVID-19 convalescent patients. One is specific to SARS-CoV-2, the other region, which could potentially function as a pan-SARS target. | [88] |
| 2020 | S14P5, S20P2, S21P2 and N4P5 | N4P5: >96% | N4P5: 100% | The magnitude of IgG responses to S14P5, S21P2 and N4P5 were strongly associated with disease severity | [96] | |
| 2020 | orf1a/b, S, and N proteins | / | / | Positive rate IgG:71.4% IgM: 57.2% | [90] | |
| 2020 | 27 proteins | / | / | Nucleocapsid protein and a highly antigenic GGDGKMKD epitope were identified as ideal antigens to be used in the development of serodiagnostic assays. | [94] | |
| 2021 | S protein | / | / | S14P5 + S21P2 + P104: Positive reaction rate for all patients and asymptomatic infections: 92.7% and 86.7% | [89] | |
| 2021 | N and S protein | / | / | Several B-cell epitopes having potential diagnostic performance have been identified | [91] | |
| 2021 | S region | / | / | Selected four peptides for SARS-CoV-2 diagnosis in silico. | [92] | |
| 2021 | ORF8 protein | / | / | Peptides 1, 2, 8 and 15 were recognized in ≥75% COVID-19 patients. | [97] | |
| Epstein–Barr virus | 2006 | Peptides F1, A3, gp125, and A2 | F1: 88%, A3:85% gp125: 71% A2: 54% gp125 + F1: 99% | 100% | / | [101] |
| 2011 | LMP2 | / | / | Positive rates in the NPC group: Epitope1/2/3: 90.82% / 62.56% / 69.39% | [103] | |
| 2016 | LMP2 | 52.84% | 95.40% | / | [102] | |
| 2018 | LMP2 | 91.91% | 93.14% | / | [98] | |
| Dengue virus | 2001 | NS1 of DENV-1 | 95% | 100% | / | [112] |
| 2003 | NS1 of DENV-2 | / | / | This mAb and its epitope-based peptide antigen will be useful for serologic diagnosis of DENV-2 infection. | [113] | |
| 2013 | E protein of DENV 1 to 4 serotypes | 71.7% | 100% | / | [106] | |
| 2018 | E protein of DENV-1 | E1: 82.5% E7: 79.2% E1 + E7: 85.0% | E1: 94.6% E7: 92.9% E1+ E7:96.4% | / | [107] | |
| 2019 | NS4B protein | IgG: 87.88% IgM: 79.17% | IgG: 93.55% IgM: 82.61% | / | [114] | |
| 2019 | E protein | E01: 100% | E01: 75% | / | [109] | |
| 2020 | E and NS1proteins | 73.33–96.66% | 82.14–100% | / | [108] | |
| 2020 | E protein | / | / | Substituting key residues for alanine within linear epitopes on the surface of the DENV E protein abolishes the contribution of some cross-reacting epitopes to its antigenicity. | [111] | |
| Hepatitis virus | 2013 | HBcAg recombinant hepatitis B core multiepitope antigen | / | / | Performance of recombinant antigen responds as well as the commercial antigen. | [115] |
| 2021 | N-terminal residues of HBeAg | / | 100% | A novel HBeAg immunoassay using high-affinity mAbs recognizing unique N-terminal epitope on precore protein, eliminating the confounding signal from the secreted HBcAg. | [116] | |
| 2006 | Recombinant multiepitope protein (core 1b, core 3g, NS3, NS4 I, NS 4 II, and NS5.) | 99.8% | 100% | Sensitivity and specificity can be comparable with commercially available anti-HCV EIA | [117] | |
| 2009 | Core protein | / | / | 40.7% patients with occult HCV infection showed IgG anti-HCV core reactivity. | [120] | |
| 2011 | Core, NS3, NS4 and NS5 proteins | / | / | With good sensitivity and specificity | [118] | |
| 2013 | Epitope arrays | / | / | Combinatorial epitopes proved to be effective for the discrimination between positive and negative sera as well as serotyping of HCV. | [127] | |
| 2018 | E2 region | / | / | HC-13 has a high degree Of specificity in E2 region among the genotypes. | [119] | |
| Ebolavirus | 2013 | NP | / | / | / | [121] |
| 2014 | GP, NP, and VP40 and VP35 | / | / | The B-cell epitopes identified may represent important tools for developing new antibody-based detection methods. | [122] | |
| 2018 | GP | / | / | These conserved B cell epitopes of GP1, 2 and their derivative antibodies are targets presently for development of RDTs for Ebolavirus disease. | [123] | |
| Hantaviruses | 2017 | NP | / | / | The reported pan-specific epitopes can be developed for test detecting antibodies to hantaviruses causing HCPS | [126] |
| 2017 | NP | / | / | It predicted a conserved 20-mer peptide used for development of geographic region-specific immunoassays | [125] | |
| 2019 | NP | / | / | SHNP(G72-D110) and SHNP(P251-D264) epitopes are promising targets to development of highly specific tools to HFRS orthohantavirus diagnosis. | [124] | |
| Pathogens | Year | Associated Proteins/Peptides | Main Outcome | Ref. | ||
|---|---|---|---|---|---|---|
| Sensitivity | Specificity | Others | ||||
| Leishmania | 2015 | Multiepitope proteins PQ10 and PQ20 | PQ10:88.8% PQ20:84.9% | PQ-10: 80% PQ-20: 65% | PQ10 was able to detect 80% of asymptomatic infected dogs. | [137] |
| 2022 | Chimeric protein ChimLeish (LiHyT, LiHyD, LiHyV, and LiHyP) | 100% | 100% | For tegumentary leishmaniasis diagnosis. | [142] | |
| Leishmania donovani | 2017 | P1P2 (P1:RFFVQGDGIGQHSLQEALERR and P2:RRVAVLVLLDRL) | P1P2: 90% ICT strip: 100% | P1P2:100% ICT strip: 95.2% | Colloidal gold conjugated anti-P1P2 antibody ICT strip anti-P1P2 antibody | [143] |
| 2017 | recLdVFA2 | 98% | 99% | For canine VL | [136] | |
| Leishmania infantum | 1998 | LiP2a, LiP2b, LiP0 and H2A | 79–93% | 96–100% | For canine VL | [144] |
| 2005 | K9, K26, and K39 | Human/canine: 82%/96% | Both 99% | / | [145] | |
| 2011 | Peptides: PSLc1- PSLc10 | PSLc10: 88.70% | PSLc8 and Mix10: 95.00% | Best results of peptides in serum samples from symptomatic and asymptomatic dogs | [146] | |
| 2017 | B10 Peptide/Phage and C01Peptide/Phage | 90.5%/100% and 91.5%/92.3% | 89.9%/98.1% and 85.5%/98.1% | / | [147] | |
| 2018 | 6 peptides of 3 proteins (Protein ID: LinJ.30.2730, LinJ.32.0280, LinJ.27.0980) | Peptide-6, Mix I, II, III and IV: 100.00% (CI 95%: 94.87–100.00%) | Mix IV: 100.00%, (CI 95%: 97.36–100.00), Peptide-6 and Mix III: 99.28%, (CI 95%: 96.03–99.98) | Mix IV have the ability to identify VL cases and simultaneously to discriminate infections caused by Trypanosoma cruzi parasite with high accuracy (100.00%) | [148] | |
| 2019 | Peptide EpQ11 | 79–84% | / | None of the sera from nonendemic healthy control patients were positive. | [149] | |
| 2020 | Synthetic peptide PeptC of protein LiHyC | 100% | 100% | / | [135] | |
| 2021 | Peptides: Pep2, Pep3 and Pep4 | 100% | 100% | Have potential to diagnose VL and VL/HIV coinfection. | [150] | |
| Toxoplasma gondii | 2012 | GRA1, GRA2, GRA4, SAG1, NTPase1, NTPase2 and MIC3 | 69% | 84% | Overall sensitivity is 69%. The assay has different diagnostic sensitivity in different types of patients, as can be seen in the references | [151] |
| 2012 | SAG1, SAG2, SAG3, GRA5, GRA6, and P35 | IgG: 94.4% IgM: 96.9% | IgG and IgM: 100% | / | [152] | |
| 2017 | SAG1, GRA2 and GRA7 | 85.43% | 81.25% | / | [128] | |
| 2021 | SAG1, GRA1, ROP2, GRA4, and MIC3 | 79.1% | 88.6% | IgG ELISA | [132] | |
| 2021 | BCLA | / | / | Peptides significantly increased the test sensitivity. | [153] | |
| Schistosoma mansoni | 2016 | FLDNF | / | / | / | [154] |
| 2017 | 7 proteins | 96.15% | 100% | The ELISA performance is achieved by using peptide 5, which could discriminate between individuals living in an endemic area that were actively infected from those that were not. | [155] | |
| 2021 | SmSPI | / | / | Three predicted immunoreactive epitopes of SmSPI are potential biomarkers for serodiagnostic Schistosomiasis. | [156] | |
| Schistosoma japonicum | 2019 | SjSP-13 | 76.7% (95% CI: 68.8–84.5%) | 100% | Two adjacent peptides (p7 and p8) | [157] |
| 2020 | SjEV proteins | / | / | Combined epitope protein demonstrated a modest sensitivity for detection of S. japonica | [140] | |
| 2022 | SjSAP4 and SjSP-13 | 84.0% | 100%. | A dual epitope-ELISA (SjSAP4-Peptide + SjSP-13V2-Peptide-ELISA) | [141] | |
| Plasmodium falciparum | 2016 | M.RCAg-1 (11 epitopes) | / | / | M.RCAg-1 was well-recognized by the naturally acquired anti-malaria antibodies and can be used as a tool for assessing malaria transmission intensity in the border area of China-Myanmar. | [158] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, J.; Chen, J.; Peng, Y.; Xie, Y.; Xiao, Y. A Promising Tool in Serological Diagnosis: Current Research Progress of Antigenic Epitopes in Infectious Diseases. Pathogens 2022, 11, 1095. https://doi.org/10.3390/pathogens11101095
Zhou J, Chen J, Peng Y, Xie Y, Xiao Y. A Promising Tool in Serological Diagnosis: Current Research Progress of Antigenic Epitopes in Infectious Diseases. Pathogens. 2022; 11(10):1095. https://doi.org/10.3390/pathogens11101095
Chicago/Turabian StyleZhou, Jiahuan, Jiayi Chen, Yunchi Peng, Yafeng Xie, and Yongjian Xiao. 2022. "A Promising Tool in Serological Diagnosis: Current Research Progress of Antigenic Epitopes in Infectious Diseases" Pathogens 11, no. 10: 1095. https://doi.org/10.3390/pathogens11101095
APA StyleZhou, J., Chen, J., Peng, Y., Xie, Y., & Xiao, Y. (2022). A Promising Tool in Serological Diagnosis: Current Research Progress of Antigenic Epitopes in Infectious Diseases. Pathogens, 11(10), 1095. https://doi.org/10.3390/pathogens11101095






