Multidistrict Host–Pathogen Interaction during COVID-19 and the Development Post-Infection Chronic Inflammation
Abstract
:1. COVID-19: From Infection to Host Antiviral Response
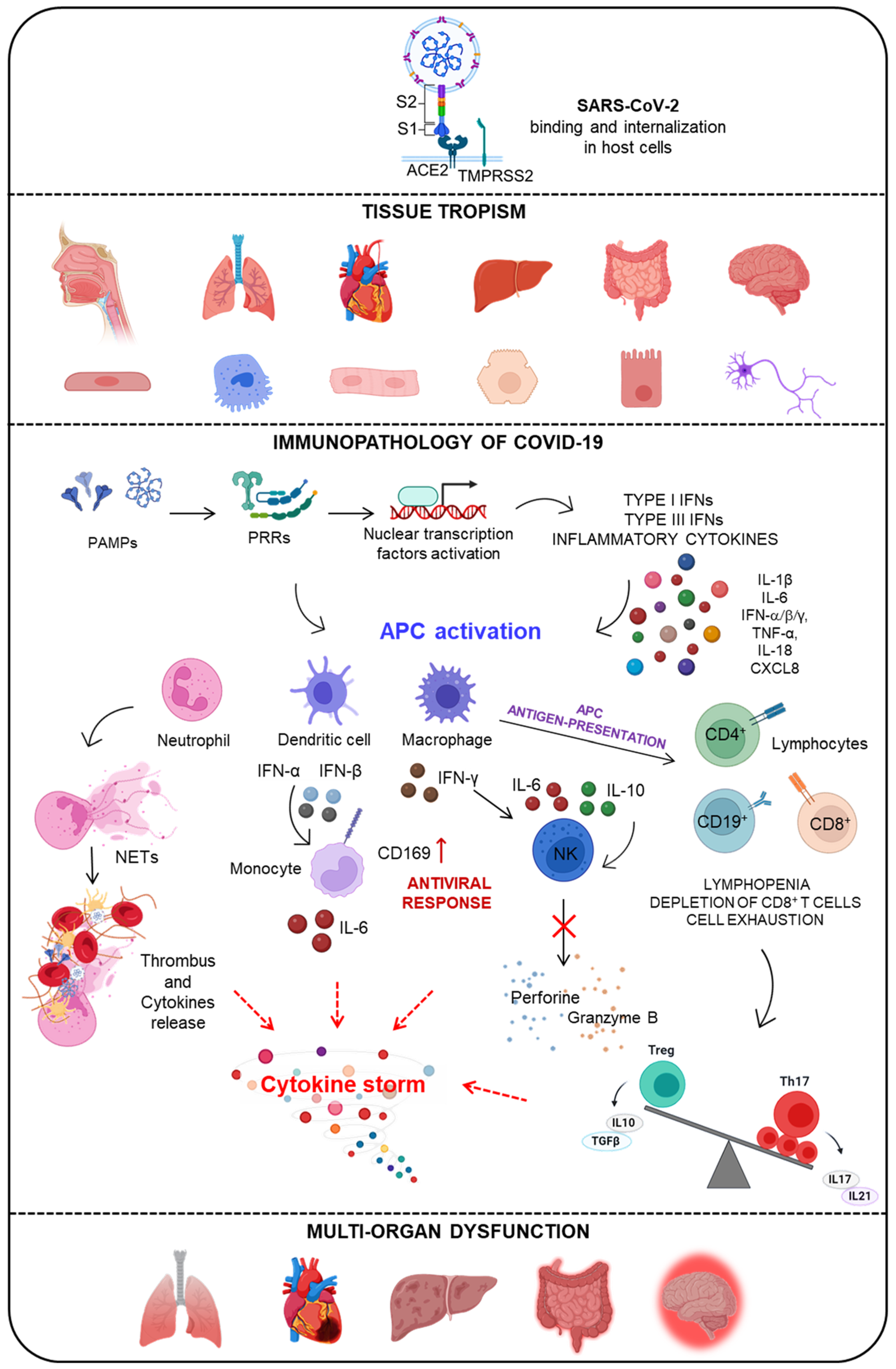
1.1. SARS-CoV-2 Infection and COVID-19 Disease
1.2. Multi-Systemic Alteration Due to SARS-CoV-2 Infection
1.3. Immunopathology of COVID-19 Disease
1.3.1. The Activation of the Innate Immune Response by Viral-Associated Molecular Patterns
1.3.2. The SARS-CoV-2-Induced Activation of the IFN System
1.3.3. Adaptive Immune Dysregulation and Hyper Inflammation in COVID-19
2. The Role of Microbiota in Health and Disease
2.1. The Different Districts of the Mucosal Immune System
2.2. Microbiota Alteration during Respiratory Virus Infections
2.3. The Contribution of Microbiota to COVID-19 Disease
Microbiota Alterations in Oral Cavity and Nasopharynx during COVID-19
2.4. Lung Microbiota during COVID-19
2.5. Changes in Intestinal Microbiota in COVID-19 Patients
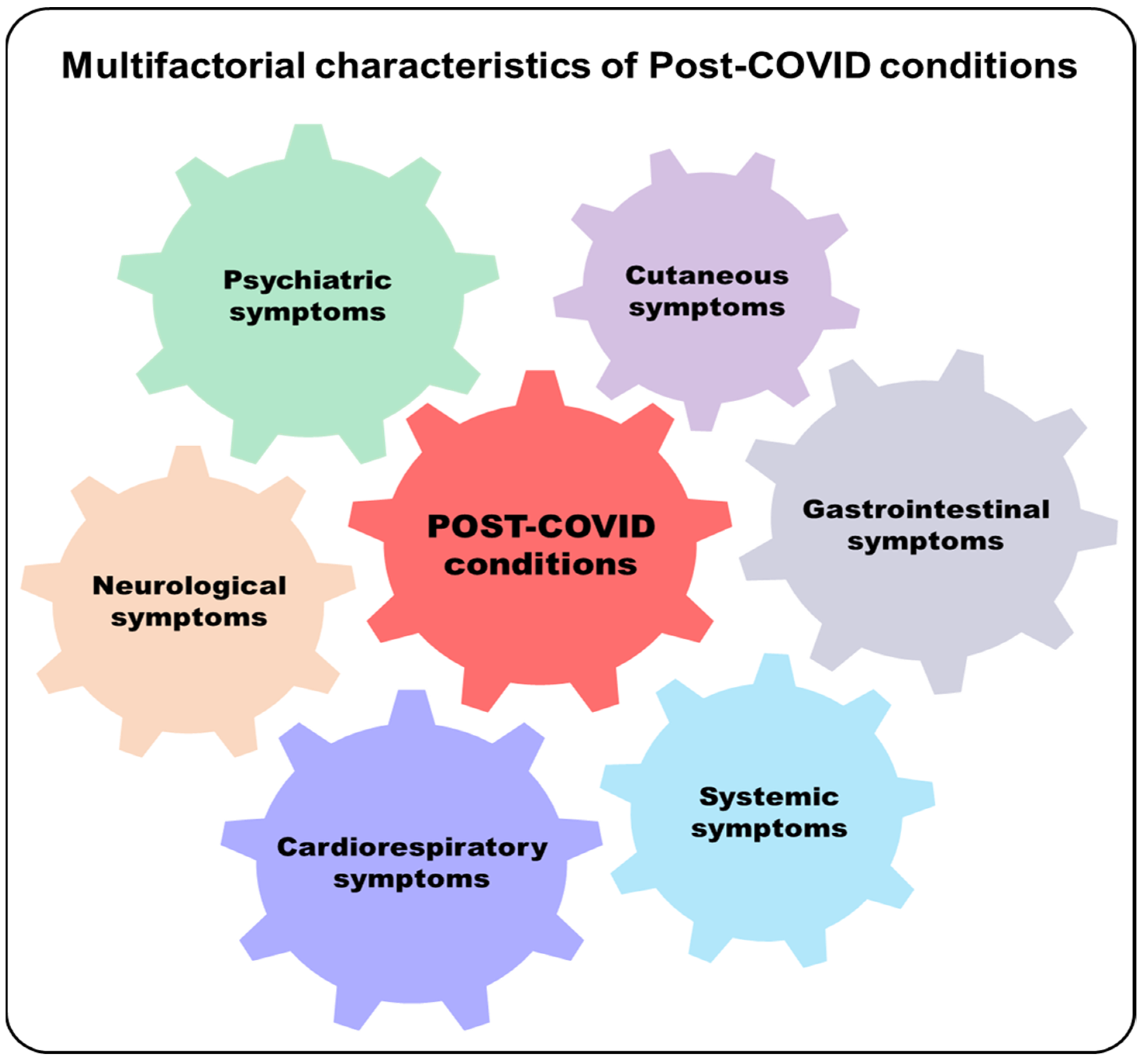
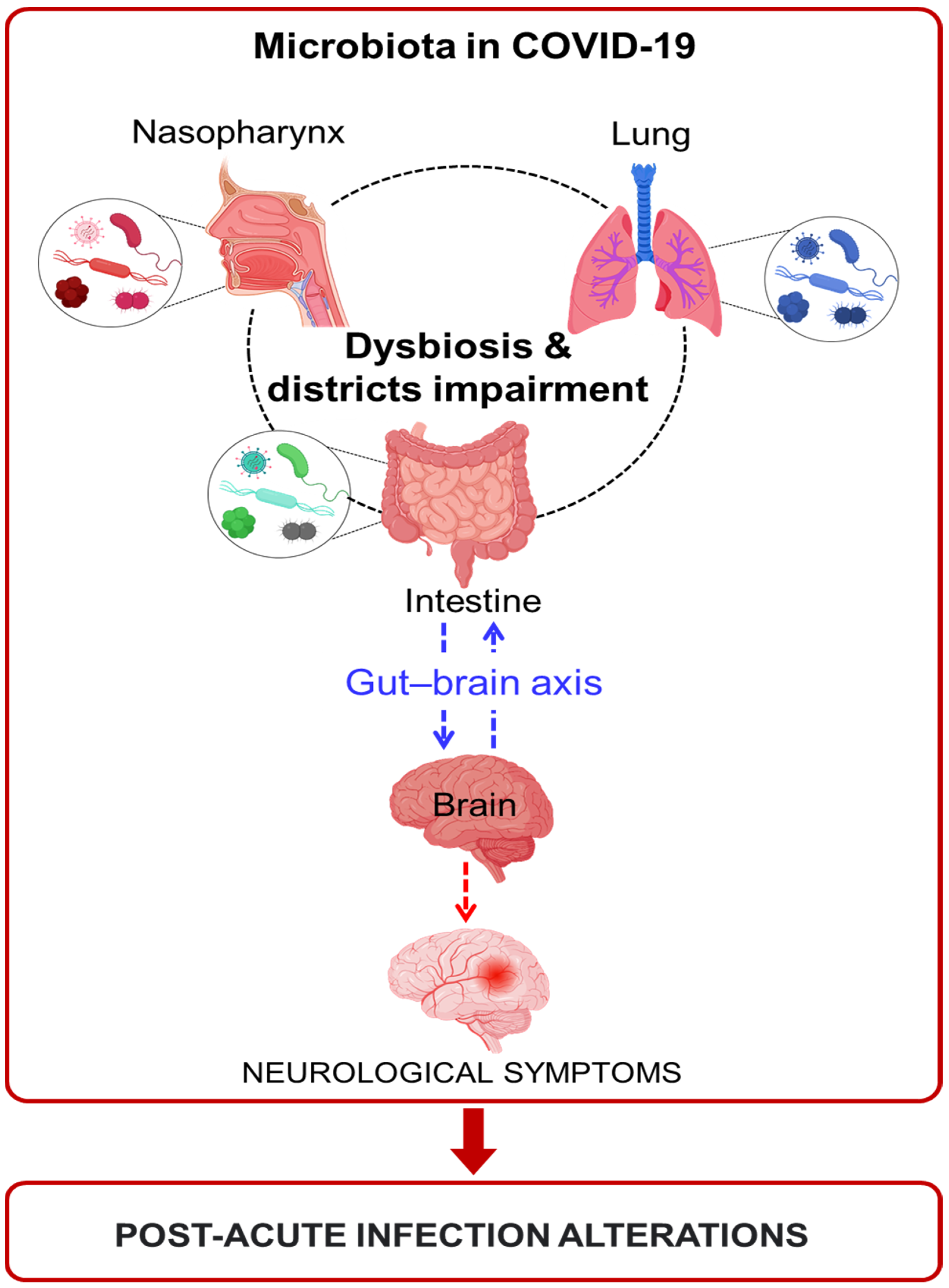
3. Post-Acute COVID-19 Syndrome
Potential Involvement of Microbiota in Post-Acute COVID-19 Syndrome
4. Discussion and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Li, Q.; Guan, X.; Wu, P.; Wang, X.; Zhou, L.; Tong, Y.; Ren, R.; Leung, K.S.M.; Lau, E.H.Y.; Wong, J.Y.; et al. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia. N. Engl. J. Med. 2020, 382, 1199–1207. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.F.; Kok, K.H.; Zhu, Z.; Chu, H.; To, K.K.; Yuan, S.; Yuen, K.Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020, 9, 221–236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.; Zhang, Y.; Wu, L.; Niu, S.; Song, C.; Zhang, Z.; Lu, G.; Qiao, C.; Hu, Y.; Yuen, K.Y.; et al. Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2. Cell 2020, 181, 894–904. [Google Scholar] [CrossRef] [PubMed]
- Tang, T.; Bidon, M.; Jaimes, J.A.; Whittaker, G.R.; Daniel, S. Coronavirus membrane fusion mechanism offers a potential target for antiviral development. Antivir. Res. 2020, 178, 104792. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280. [Google Scholar] [CrossRef]
- Masters, P. The Molecular Biology of Coronaviruses. Adv. Virus Res. 2006, 66, 193–292. [Google Scholar]
- Umakanthan, S.; Sahu, P.; Ranade, A.V.; Bukelo, M.M.; Rao, J.S.; Abrahao-Machado, L.F.; Dahal, S.; Kumar, H.; Kv, D. Origin, transmission, diagnosis and management of coronavirus disease 2019 (COVID-19). Postgrad Med. J. 2020, 96, 1142753–1142758. [Google Scholar]
- Jiang, F.; Deng, L.; Zhang, L.; Cai, Y.; Cheung, C.W.; Xia, Z. Review of the clinical charateristics of coronavirus disease 2019 (COVID-19). J. Gen. Intern. Med. 2020, 35, 1545–1549. [Google Scholar] [CrossRef] [Green Version]
- Wang, T.; Du, Z.; Zhu, F.; Cao, Z.; An, Y.; Gao, Y.; Jiang, B. Comorbidities and multi-Organ injuries in the treatment of COVID-19. Lancet 2020, 395, e52. [Google Scholar] [CrossRef]
- Dos Santos, A.C.M.; Dos Santos, B.R.C.; Dos Santos, B.B.; De Moura, E.L.; Ferreira, J.M.; Dos Santos, L.K.C.; Oliveira, S.P.; Dias, R.B.F.; Pereira E Silva, A.C.; De Farias, K.F.; et al. Genetic polymorphisms as multi-Biomarkers in severe acute respiratory syndrome (SARS) by coronavirus infection: A systematic review of candidate gene association studies. Infect. Genet. Evol. 2021, 93, 104846. [Google Scholar] [CrossRef]
- Monticelli, M.; Mele, B.H.; Andreotti, G.; Cubellis, M.V.; Riccio, G. Why does SARS-CoV-2 hit in different ways? Host genetic factors can influence the acquisition or the course of COVID-19. Eur. J. Med. Genet. 2021, 64, 104227. [Google Scholar] [CrossRef] [PubMed]
- Di Maria, E.; Latini, A.; Borgiani, P.; Novelli, G. Genetic variants of the human host influencing the coronavirus-associated phenotypes (SARS, MERS and COVID-19): Rapid systematic review and field synopsis. Hum Genom. 2020, 14, 30. [Google Scholar] [CrossRef] [PubMed]
- Posadas-Sánchez, R.; Fragoso, J.M.; Sánchez-Muñoz, F.; Rojas-Velasco, G.; Ramírez-Bello, J.; López-Reyes, A.; Martínez-Gómez, L.E.; Sierra-Fernández, C.; Rodríguez-Reyna, T.; Regino-Zamarripa, N.E.; et al. Association of the Transmembrane Serine Protease-2 (TMPRSS2) Polymorphisms with COVID-19. Viruses 2022, 14, 1976. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Bastard, P.; Liu, Z.; Le Pen, J.; Moncada-Velez, M.; Chen, J.; Ogishi, M.; Sabli, I.K.D.; Hodeib, S.; Korol, C.; et al. Inborn errors of type I IFN immunity in patients with life-Threatening COVID-19. Science 2020, 370, eabd4570. [Google Scholar] [CrossRef] [PubMed]
- Manry, J.; Bastard, P.; Gervais, A.; Le Voyer, T.; Rosain, J.; Philippot, Q.; Michailidis, E.; Hoffmann, H.H.; Eto, S.; Garcia-Prat, M.; et al. The risk of COVID-19 death is much greater and age dependent with type I IFN autoantibodies. Proc. Natl. Acad. Sci. USA 2022, 119, e2200413119. [Google Scholar] [CrossRef] [PubMed]
- Bastard, P.; Vazquez, S.; Liu, J.; Laurie, M.T.; Wang, C.Y.; Gervais, A.; Le Voyer, T.; Bizien, L.; Zamecnik, C.; Philippot, Q.; et al. Vaccine breakthrough hypoxemic COVID-19 pneumonia in patients with auto-Abs neutralizing type I IFNs. Sci. Immunol. 2022, eabp8966. [Google Scholar] [CrossRef]
- Stenmark, K.R.; Frid, M.G.; Gerasimovskaya, E.; Zhang, H.; McCarthy, M.K.; Thurman, J.M.; Morrison, T.E. Mechanisms of SARS-CoV-2-induced lung vascular disease: Potential role of complement. Pulm. Circ. 2021, 11, 20458940211015799. [Google Scholar] [CrossRef]
- Wang, D.; Hu, B.; Hu, C.; Zhu, F.; Liu, X.; Zhang, J.; Wang, B.; Xiang, H.; Cheng, Z.; Xiong, Y.; et al. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus–Infected Pneumonia in Wuhan, China. JAMA 2020, 323, 1061–1069. [Google Scholar] [CrossRef]
- Chai, X.Q.; Hu, L.F.; Zhang, Y.; Lu, Z.; Ke, A.; Zhou, J.; Shi, G.; Fang, N.; Fan, J.; Cai, J.; et al. Specific ACE2 expression in cholangiocytes may cause liver damage after COVID-19 infection. bioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Li, M.Y.; Li, L.; Zhang, Y.; Wang, X.S. Expression of the SARS-CoV-2 cell receptor gene ACE2 in a wide variety of human tissues. Infect. Dis. Poverty 2020, 9, 45. [Google Scholar] [CrossRef]
- Wu, J.; Liu, J.; Li, S.; Peng, Z.; Xiao, Z.; Wang, X.; Yan, R.; Luo, J. Detection and analysis of nucleic acid in various biological samples of COVID-19 patients. Travel Med. Infect. Dis. 2020, 37, 101673. [Google Scholar] [CrossRef]
- Helms, J.; Kremer, S.; Merdji, H.; Clere-Jehl, R.; Schenck, M.; Kummerlen, C.; Collange, O.; Boulay, C.; Fafi-Kremer, S.; Ohana, M.; et al. Neurologic Features in Severe SARS-CoV-2 Infection. N. Engl. J. Med. 2020, 382, 2268–2270. [Google Scholar] [CrossRef] [PubMed]
- Wijeratne, T.; Crewther, S. Post-COVID 19 Neurological Syndrome (PCNS); a novel syndrome with challenges for the global neurology community. J. Neurol. Sci. 2020, 419, 117179. [Google Scholar] [CrossRef] [PubMed]
- Hikmet, F.; Méar, L.; Edvinsson, Å.; Micke, P.; Uhlén, M.; Lindskog, C. The protein expression profile of ACE2 in human tissues. Mol. SystBiol. 2020, 16, e9610. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Li, Y.; Liu, Q.; Yao, Q.; Wang, X.; Zhang, H.; Chen, R.; Ren, L.; Min, J.; Deng, F.; et al. SARS-CoV-2 cell tropism and multiorgan infection. Cell Discov. 2021, 7, 17. [Google Scholar] [CrossRef]
- Bräuninger, H.; Stoffers, B.; Fitzek, A.D.E.; Meißner, K.; Aleshcheva, G.; Schweizer, M.; Weimann, J.; Rotter, B.; Warnke, S.; Edler, C. Cardiac SARS-CoV-2 infection is associated with pro-inflammatory transcriptomic alterations within the heart. Cardiovasc. Res. 2022, 118, 542–555. [Google Scholar] [CrossRef]
- Wanner, N.; Andrieux, G.; Badia-I-Mompel, P.; Edler, C.; Pfefferle, S.; Lindenmeyer, M.T.; Schmidt-Lauber, C.; Czogalla, J.; Wong, M.N.; Okabayashi, Y.; et al. Molecular consequences of SARS-CoV-2 liver tropism. Nat. Metab. 2022, 4, 310–319. [Google Scholar] [CrossRef]
- Schaller, M.A.; Sharma, Y.; Dupee, Z.; Nguyen, D.; Urueña, J.; Smolchek, R.; Loeb, J.C.; Machuca, T.N.; Lednicky, J.A.; Odde, D.J.; et al. Ex vivo SARS-CoV-2 infection of human lung reveals heterogeneous host defense and therapeutic responses. JCI Insight 2021, 6, e148003. [Google Scholar] [CrossRef]
- Lehmann, M.; Allers, K.; Heldt, C.; Meinhardt, J.; Schmidt, F.; Rodriguez-Sillke, Y.; Kunkel, D.; Schumann, M.; Böttcher, C.; Stahl-Hennig, C.; et al. Human small intestinal infection by SARS-CoV-2 is characterized by a mucosal infiltration with activated CD8+ T cells. Mucosal Immunol. 2021, 14, 1381–1392. [Google Scholar] [CrossRef]
- De Melo, G.D.; Lazarini, F.; Levallois, S.; Hautefort, C.; Michel, V.; Larrous, F.; Verillaud, B.; Aparicio, C.; Wagner, S.; Gheusi, G.; et al. COVID-19-related anosmia is associated with viral persistence and inflammation in human olfactory epithelium and brain infection in hamsters. Sci. Transl. Med. 2021, 13, eabf8396. [Google Scholar] [CrossRef]
- Meinhardt, J.; Radke, J.; Dittmayer, C.; Franz, J.; Thomas, C.; Mothes, R.; Laue, M.; Schneider, J.; Brünink, S.; Greuel, S. Olfactory transmucosal SARS-CoV-2 invasion as a port of central nervous system entry in individuals with COVID-19. Nat. Neurosci. 2021, 24, 168–175. [Google Scholar] [CrossRef]
- Takeda, K.; Akira, S. Toll-Like receptors. Curr. Protoc. Immunol. 2015, 109, 14.12.1–14.12.10. [Google Scholar] [CrossRef] [PubMed]
- Kumar, H.; Kawai, T.; Akira, S. Pathogen recognition by the innate immune system. Int. Rev. Immunol. 2011, 30, 16–34. [Google Scholar] [CrossRef] [PubMed]
- Ricci, D.; Etna, M.P.; Rizzo, F.; Sandini, S.; Severa, M.; Coccia, E.M. Innate Immune Response to SARS-CoV-2 Infection: From Cells to Soluble Mediators. Int. J. Mol. Sci. 2021, 22, 7017. [Google Scholar] [CrossRef]
- Salvi, V.; Nguyen, H.O.; Sozio, F.; Schioppa, T.; Gaudenzi, C.; Laffranchi, M.; Scapini, P.; Passari, M.; Barbazza, I.; Tiberio, L.; et al. SARS-CoV-2-associated ssRNAs activate inflammation and immunity via TLR7/8. JCI Insight 2021, 6, e150542. [Google Scholar] [CrossRef] [PubMed]
- Zheng, M.; Karki, R.; Williams, E.P.; Yang, D.; Fitzpatrick, E.; Vogel, P.; Jonsson, C.B.; Kanneganti, T.D. TLR2 senses the SARS-CoV-2 envelope protein to produce inflammatory cytokines. Nat. Immunol. 2021, 22, 829–838. [Google Scholar] [CrossRef] [PubMed]
- Sung, P.S.; Yang, S.P.; Peng, Y.C.; Sun, C.P.; Tao, M.H.; Hsieh, S.L. CLEC5A and TLR2 are critical in SARS-CoV-2-induced NET formation and lung inflammation. J. Biomed. Sci. 2022, 29, 52. [Google Scholar] [CrossRef] [PubMed]
- Kell, A.M.; Gale, M., Jr. RIG-I in RNA virus recognition. Virology 2015, 479, 110–121. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.L.; Ye, H.Q.; Liu, S.Q.; Deng, C.L.; Li, X.D.; Shi, P.Y.; Zhang, B. West Nile Virus NS1 Antagonizes Interferon Beta Production by Targeting RIG-I and MDA5. J. Virol. 2017, 91, e02396-16. [Google Scholar] [CrossRef] [Green Version]
- Lopes-Pacheco, M.; Silva, P.L.; Cruz, F.F.; Battaglini, D.; Robba, C.; Pelosi, P.; Morales, M.M.; Caruso Neves, C.; Rocco, P.R.M. Pathogenesis of Multiple Organ Injury in COVID-19 and Potential Therapeutic Strategies. Front. Physiol. 2021, 12, 593223. [Google Scholar] [CrossRef]
- Abdelmoaty, M.M.; Yeapuri, P.; Machhi, J.; Olson, K.E.; Shahjin, F.; Kumar, V.; Zhou, Y.; Liang, J.; Pandey, K.; Acharya, A.; et al. Defining the Innate Immune Responses for SARS-CoV-2-Human Macrophage Interactions. Front. Immunol. 2021, 12, 741502. [Google Scholar] [CrossRef]
- Severa, M.; Diotti, R.A.; Etna, M.P.; Rizzo, F.; Fiore, S.; Ricci, D.; Iannetta, M.; Sinigaglia, A.; Lodi, A.; Mancini, N.; et al. Differential plasmacytoid dendritic cell phenotype and type I Interferon response in asymptomatic and severe COVID-19 infection. PLoS Pathog. 2021, 17, e1009878. [Google Scholar] [CrossRef] [PubMed]
- Kumaran Satyanarayanan, S.; El Kebir, D.; Soboh, S.; Butenko, S.; Sekheri, M.; Saadi, J.; Peled, N.; Assi, S.; Othman, A.; Schif-Zuck, S.; et al. IFN-β is a macrophage-derived effector cytokine facilitating the resolution of bacterial inflammation. Nat. Commun. 2019, 10, 3471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marks, Z.R.C.; Campbell, N.; deWeerd, N.A.; Lim, S.S.; Gearing, L.J.; Bourke, N.M.; Hertzog, P.J. Properties and functions of the novel type I interferon epsilon. Semin. Immunol. 2019, 43, 101328. [Google Scholar] [CrossRef] [PubMed]
- Woodby, B.L.; Songock, W.K.; Scott, M.L.; Raikhy, G.; Bodily, J.M. Induction of Interferon Kappa in Human Papillomavirus 16 Infection by Transforming Growth Factor Beta-Induced Promoter Demethylation. J. Virol. 2018, 92, e01714-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, S.F.; Zhao, F.R.; Shao, J.J.; Xie, Y.L.; Chang, H.Y.; Zhang, Y.G. Interferon-Omega: Current status in clinical applications. Int. Immunopharmacol. 2017, 52, 253–260. [Google Scholar] [CrossRef]
- Kak, G.; Raza, M.; Tiwari, B.K. Interferon-gamma (IFN-γ): Exploring its implications in infectious diseases. Biomol. Concepts 2018, 9, 64–79. [Google Scholar] [CrossRef]
- Lozhkov, A.A.; Klotchenko, S.A.; Ramsay, E.S.; Moshkoff, H.D.; Moshkoff, D.A.; Vasin, A.V.; Salvato, M.S. The Key Roles of Interferon Lambda in Human Molecular Defense against Respiratory Viral Infections. Pathogens 2020, 9, 989. [Google Scholar] [CrossRef]
- Min, Y.Q.; Huang, M.; Sun, X.; Deng, F.; Wang, H.; Ning, Y.J. Immune evasion of SARS-CoV-2 from interferon antiviral system. Comput. Struct. Biotechnol. J. 2021, 19, 4217–4225. [Google Scholar] [CrossRef]
- Yang, E.; Li, M.M.H. All About the RNA: Interferon-Stimulated Genes That Interfere With Viral RNA Processes. Front. Immunol. 2020, 11, 605024. [Google Scholar] [CrossRef]
- Setaro, A.C.; Gaglia, M.M. All hands on deck: SARS-CoV-2 proteins that block early anti-viral interferon responses. Curr. Res. Virol. Sci. 2021, 2, 100015. [Google Scholar] [CrossRef]
- Bastard, P.; Rosen, L.B.; Zhang, Q.; Michailidis, E.; Hoffmann, H.H.; Zhang, Y.; Dorgham, K.; Philippot, Q.; Rosain, J.; Béziat, V.; et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science 2020, 370, eabd4585. [Google Scholar] [CrossRef] [PubMed]
- Cobat, A. Genotype count for all coding variants in type I IFN genes investigated in 659 life-threatening COVID-19 patients and 534 asymptomatic/mild infected controls. Dryad 2020. [Google Scholar] [CrossRef]
- Elena, R.S.; Maria, A.M.; Marta, N.; Somaye, J.; Stefano, R.; Sergio, U.; Salvatore, R.; Pietro, M.; Leonardo, A.S. Increased Presence of Antibodies against Type I Interferons and Human Endogenous Retrovirus W in Intensive Care Unit COVID-19 Patients. Microbiol. Spectr. 2022, 10, e0128022. [Google Scholar]
- Puryear, W.B.; Akiyama, H.; Geer, S.D.; Ramirez, N.P.; Yu, X.; Reinhard, B.M.; Gummuluru, S. Interferon-Inducible mechanism of dendritic cell-mediated HIV-1 dissemination is dependent on Siglec-1/CD169. PLoS Pathog. 2013, 9, e1003291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bourgoin, P.; Biéchelé, G.; AitBelkacem, I.; Morange, P.E.; Malergue, F. Role of the interferons in CD64 and CD169 expressions in whole blood: Relevance in the balance between viral-or bacterial-Oriented immune responses. Immun. Inflamm. Dis. 2020, 8, 106–123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pino, M.; Erkizia, I.; Benet, S.; Erikson, E.; Fernández-Figueras, M.T.; Guerrero, D.; Izquierdo-Useros, N.; Dalmau, J.; Ouchi, D.; Rausell, A.; et al. HIV-1 immune activation induces Siglec-1 expression and enhances viral trans-Infection in blood and tissue myeloid cells. Retrovirology 2015, 12, 37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bourgoin, P.; Soliveres, T.; Barbaresi, A.; Loundou, A.; Belkacem, I.A.; Arnoux, I.; Bernot, D.; Loosveld, M.; Morange, P.E.; Michelet, P.; et al. CD169 and CD64 could help differentiate bacterial from COVID-19 or other viral infections in the Emergency Department. Cytom. A 2021, 99, 435–445. [Google Scholar] [CrossRef]
- Doehn, J.M.; Tabeling, C.; Biesen, R.; Saccomanno, J.; Madlung, E.; Pappe, E.; Gabriel, F.; Kurth, F.; Meisel, C.; Corman, V.M.; et al. CD169/SIGLEC1 is expressed on circulating monocytes in COVID-19 and expression levels are associated with disease severity. Infection 2021, 49, 757–762. [Google Scholar] [CrossRef]
- Minutolo, A.; Petrone, V.; Fanelli, M.; Iannetta, M.; Giudice, M.; Ait Belkacem, I.; Zordan, M.; Vitale, P.; Rasi, G.; Sinibaldi-Vallebona, P.; et al. High CD169 Monocyte/Lymphocyte Ratio Reflects Immunophenotype Disruption and Oxygen Need in COVID-19 Patients. Pathogens 2021, 10, 1639. [Google Scholar] [CrossRef]
- Sadeghi, A.; Tahmasebi, S.; Mahmood, A.; Kuznetsova, M.; Valizadeh, H.; Taghizadieh, A.; Nazemiyeh, M.; Aghebati-Maleki, L.; Jadidi-Niaragh, F.; Abbaspour-Aghdam, S.; et al. Th17 and Treg cells function in SARS-CoV-2 patients compared with healthy controls. J. Cell Physiol. 2021, 236, 2829–2839. [Google Scholar] [CrossRef]
- Tavakolpour, S.; Rakhshandehroo, T.; Wei, E.X.; Rashidian, M. Lymphopenia during the COVID-19 infection: What it shows and what can be learned. Immunol. Lett. 2020, 225, 31–32. [Google Scholar] [CrossRef] [PubMed]
- Signore, A.; Lauri, C.; Colandrea, M.; Di Girolamo, M.; Chiodo, E.; Grana, C.M.; Campagna, G.; Aceti, A. Lymphopenia in patients affected by SARS-CoV-2 infection is caused by margination of lymphocytes in large bowel: An [18F]FDG PET/CT study. Eur. J. Nucl. Med. Mol. Imaging 2022, 49, 3419–3429. [Google Scholar] [CrossRef] [PubMed]
- Jarjour, N.N.; Masopust, D.; Jameson, S.C. T Cell Memory: Understanding COVID-19. Immunity 2021, 54, 14–18. [Google Scholar] [CrossRef] [PubMed]
- De Biasi, S.; Meschiari, M.; Gibellini, L.; Bellinazzi, C.; Borella, R.; Fidanza, L.; Gozzi, L.; Iannone, A.; Lo Tartaro, D.; Mattioli, M.; et al. Marked T cell activation, senescence, exhaustion and skewing towards TH17 in patients with COVID-19 pneumonia. Nat. Commun. 2020, 11, 3434. [Google Scholar] [CrossRef]
- Westmeier, J.; Paniskaki, K.; Karaköse, Z.; Werner, T.; Sutter, K.; Dolff, S.; Overbeck, M.; Limmer, A.; Liu, J.; Zheng, X.; et al. Impaired Cytotoxic CD8+ T Cell Response in Elderly COVID-19 Patients. MBio 2020, 11, e02243-20. [Google Scholar] [CrossRef]
- Bellesi, S.; Metafuni, E.; Hohaus, S.; Maiolo, E.; Marchionni, F.; D’Innocenzo, S.; La Sorda, M.; Ferraironi, M.; Ramundo, F.; Fantoni, M.; et al. Increased CD95 (Fas) and PD-1 expression in peripheral blood T lymphocytes in COVID-19 patients. Br. J. Haematol. 2020, 191, 207–211. [Google Scholar] [CrossRef]
- Diao, B.; Wang, C.; Tan, Y.; Chen, X.; Liu, Y.; Ning, L.; Chen, L.; Li, M.; Liu, Y.; Wang, G.; et al. Reduction and Functional Exhaustion of T Cells in Patients With Coronavirus Disease 2019 (COVID-19). Front. Immunol. 2020, 11, 827. [Google Scholar] [CrossRef]
- Bobcakova, A.; Petriskova, J.; Vysehradsky, R.; Kocan, I.; Kapustova, L.; Barnova, M.; Diamant, Z.; Jesenak, M. Immune Profile in Patients With COVID-19: Lymphocytes Exhaustion Markers in Relationship to Clinical Outcome. Front. Cell Infect. Microbiol. 2021, 11, 646688. [Google Scholar] [CrossRef]
- Ahn, E.; Araki, K.; Hashimoto, M.; Li, W.; Riley, J.L.; Cheung, J.; Sharpe, A.H.; Freeman, G.J.; Irving, B.A.; Ahmed, R. Role of PD-1 during effector CD8 T cell differentiation. Proc. Natl. Acad. Sci. USA 2018, 115, 4749–4754. [Google Scholar] [CrossRef] [Green Version]
- Phetsouphanh, C.; Darley, D.R.; Wilson, D.B.; Howe, A.; Munier, C.M.L.; Patel, S.K.; Juno, J.A.; Burrell, L.M.; Kent, S.J.; Dore, G.J.; et al. Immunological dysfunction persists for 8 months following initial mild-to-moderate SARS-CoV-2 infection. Nat. Immunol. 2022, 23, 210–216. [Google Scholar] [CrossRef]
- Huang, Q.; Wu, X.; Zheng, X.; Luo, S.; Xu, S.; Weng, J. Targeting inflammation and cytokine storm in COVID-19. Pharmacol. Res. 2020, 159, 105051. [Google Scholar] [CrossRef] [PubMed]
- Zuo, Y.; Yalavarthi, S.; Shi, H.; Gockman, K.; Zuo, M.; Madison, J.A.; Blair, C.; Weber, A.; Barnes, B.J.; Egeblad, M.; et al. Neutrophil extracellular traps in COVID-19. JCI Insight 2020, 5, e138999. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Xie, J.; Zhao, L.; Fei, X.; Zhang, H.; Tan, Y.; Nie, X.; Zhou, L.; Liu, Z.; Ren, Y.; et al. Alveolar macrophage dysfunction and cytokine storm in the pathogenesis of two severe COVID-19 patients. EbioMedicine 2020, 57, 102833. [Google Scholar] [CrossRef] [PubMed]
- Cifaldi, L.; Prencipe, G.; Caiello, I.; Bracaglia, C.; Locatelli, F.; De Benedetti, F.; Strippoli, R. Inhibition of natural killer cell cytotoxicity by interleukin-6: Implications for the pathogenesis of macrophage activation syndrome. Arthritis Rheumatol. 2015, 67, 3037–3046. [Google Scholar] [CrossRef]
- Zhou, F.; Yu, T.; Du, R.; Fan, G.; Liu, Y.; Liu, Z.; Xiang, J.; Wang, Y.; Song, B.; Gu, X.; et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: A retrospective cohort study. Lancet 2020, 395, 1054–1062. [Google Scholar] [CrossRef]
- Simon, J.C.; Marchesi, J.R.; Mougel, C.; Selosse, M.A. Host-microbiota interactions: From holobiont theory to analysis. Microbiome 2019, 7, 5. [Google Scholar] [CrossRef] [Green Version]
- Ruan, W.; Engevik, M.A.; Spinler, J.K.; Versalovic, J. Healthy Human Gastrointestinal Microbiome: Composition and Function After a Decade of Exploration. Dig. Dis. Sci. 2020, 65, 695–705. [Google Scholar] [CrossRef] [Green Version]
- Dekaboruah, E.; Suryavanshi, M.V.; Chettri, D.; Verma, A.K. Human microbiome: An academic update on human body site specific surveillance and its possible role. Arch. Microbiol. 2020, 202, 2147–2167. [Google Scholar] [CrossRef]
- Malard, F.; Dore, J.; Gaugler, B.; Mohty, M. Introduction to host microbiome symbiosis in health and disease. Mucosal Immunol. 2021, 14, 547–554. [Google Scholar] [CrossRef]
- Rowland, I.; Gibson, G.; Heinken, A.; Scott, K.; Swann, J.; Thiele, I.; Tuohy, K. Gut microbiota functions: Metabolism of nutrients and other food components. Eur. J. Nutr. 2018, 57, 1–24. [Google Scholar] [CrossRef] [Green Version]
- Brandtzaeg, P. Function of mucosa-associated lymphoid tissue in antibody formation. Immunol. Investig. 2010, 39, 303–355. [Google Scholar] [CrossRef] [PubMed]
- Belkaid, Y.; Hand, T.W. Role of the microbiota in immunity and inflammation. Cell 2014, 157, 121–141. [Google Scholar] [CrossRef] [PubMed]
- Mörbe, U.M.; Jørgensen, P.B.; Fenton, T.M.; von Burg, N.; Riis, L.B.; Spencer, J.; Agace, W.W. Human gut-associated lymphoid tissues (GALT); diversity, structure, and function. Mucosal Immunol. 2021, 14, 793–802. [Google Scholar] [CrossRef] [PubMed]
- Randall, T.D. Bronchus-associated lymphoid tissue (BALT) structure and function. Adv. Immunol. 2010, 107, 187–241. [Google Scholar] [PubMed]
- Kogame, T.; Kabashima, K.; Egawa, G. Putative Immunological Functions of Inducible Skin-Associated Lymphoid Tissue in the Context of Mucosa-Associated Lymphoid Tissue. Front. Immunol. 2021, 12, 733484. [Google Scholar] [CrossRef]
- Bankvall, M.; Jontell, M.; Wold, A.; Östman, S. Tissue-Specific Differences in Immune Cell Subsets Located in the Naso-oropharyngeal-associated Lymphoid Tissues. Scand J. Immunol. 2018, 87, 15–27. [Google Scholar] [CrossRef] [Green Version]
- Mastropasqua, R.; Agnifili, L.; Fasanella, V.; Nubile, M.; Gnama, A.A.; Falconio, G.; Perri, P.; Di Staso, S.; Mariotti, C. The Conjunctiva-Associated LymphoidTissue in ChronicOcular Surface Diseases. Microsc. Microanal. 2017, 23, 697–707. [Google Scholar] [CrossRef]
- Brandtzaeg, P. Mucosal immunity: Induction, dissemination, and effector functions. Scand J. Immunol. 2009, 70, 505–515. [Google Scholar] [CrossRef]
- Clevers, H. The intestinal crypt, a prototype stem cell compartment. Cell 2013, 154, 274–284. [Google Scholar] [CrossRef] [Green Version]
- Audi, A.; AlIbrahim, M.; Kaddoura, M.; Hijazi, G.; Yassine, H.M.; Zarake, T.H. Seasonality of Respiratory Viral Infections: Will COVID-19 Follow Suit? Front. Public Health 2020, 8, 567184. [Google Scholar] [CrossRef]
- Kirtipal, N.; Bharadwaj, S.; Kang, S.G. From SARS to SARS-CoV-2, insights on structure, pathogenicity and immunity aspects of pandemic human coronaviruses. Infect. Genet. Evol. 2020, 85, 104502. [Google Scholar] [CrossRef] [PubMed]
- Chunxi, L.; Haiyue, L.; Yanxia, L.; Jianbing, P.; Jin, S. The Gut Microbiota and Respiratory Diseases: New Evidence. J. Immunol. Res. 2020, 2340670. [Google Scholar] [CrossRef] [PubMed]
- Gu, S.; Chen, Y.; Wu, Z.; Chen, Y.; Gao, H.; Lv, L.; Guo, F.; Zhang, X.; Luo, R.; Huang, C.; et al. Alterations of the Gut Microbiota in Patients with Coronavirus Disease 2019 or H1N1 Influenza. Clin. Infect. Dis. 2020, 71, 2669–2678. [Google Scholar] [CrossRef] [PubMed]
- Qin, N.; Zheng, B.; Yao, J.; Guo, L.; Zuo, J.; Wu, L.; Zhou, J.; Liu, L.; Guo, J.; Ni, S.; et al. Influence of H7N9 virus infection and associated treatment on human gut microbiota. Sci. Rep. 2015, 5, 14771. [Google Scholar] [CrossRef] [Green Version]
- Sencio, V.; Machado, M.G.; Trottein, F. The lung-Gut axis during viral respiratory infections: The impact of gut dysbiosis on secondary disease outcomes. Mucosal Immunol. 2021, 14, 296–304. [Google Scholar] [CrossRef]
- Caselli, E.; Fabbri, C.; D’Accolti, M.; Soffritti, I.; Bassi, C.; Mazzacane, S.; Franchi, M. Defining the oral microbiome by whole-genome sequencing and resistome analysis: The complexity of the healthy picture. BMC Microbiol. 2020, 20, 120. [Google Scholar] [CrossRef]
- Johnson, T.J.; Nishida, R.T.; Sonpar, A.P.; Lin, Y.J.; Watson, K.A.; Smith, S.W.; Conly, J.M.; Evans, D.H.; Olfert, J.S. Viral load of SARS-CoV-2 in droplets and bioaerosols directly captured during breathing, speaking and coughing. Sci. Rep. 2022, 12, 3484. [Google Scholar] [CrossRef]
- De Maio, F.; Posteraro, B.; Ponziani, F.R.; Cattani, P.; Gasbarrini, A.; Sanguinetti, M. Nasopharyngeal Microbiota Profiling of SARS-CoV-2 Infected Patients. Biol. Proced. Online 2020, 22, 18. [Google Scholar] [CrossRef]
- Budding, E.; Sieswerda, B.B.; Wintermans, M.P.; Bos, M. An age dependent pharyngeal microbiota signature associated with SARS-CoV-2 infection. Prepr. Lancet 2020. [Google Scholar] [CrossRef]
- Liu, J.; Liu, S.; Zhang, Z.; Lee, X.; Wu, W.; Huang, Z.; Lei, Z.; Xu, W.; Chen, D.; Wu, X.; et al. Association between the nasopharyngeal microbiome and metabolome in patients with COVID-19. Synth. Syst. Biotechnol. 2021, 6, 135–143. [Google Scholar] [CrossRef]
- Beck, J.M.; Young, V.B.; Huffnagle, G.B. The microbiome of Lung. Iransl. Res. 2012, 160, 258–266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, J.; Li, X.; Gao, Y.; Zhou, J.; Wang, S.; Huang, B.; Wu, J.; Cao, Q.; Chen, Y.; Wang, Z.; et al. The lung tissue microbiota features of 20 deceased patients with COVID-19. J. Infect. 2020, 81, e64–e67. [Google Scholar] [CrossRef] [PubMed]
- Spagnolello, O.; Pinacchio, C.; Santinelli, L.; Vassalini, P.; Innocenti, G.P.; De Girolamo, G.; Fabris, S.; Giovanetti, M.; Angeletti, S.; Russo, A.; et al. Targeting microbiome: An alternative strategy for fighting SARS-CoV-2 infection. Chemotherapy 2021, 66, 24–32. [Google Scholar] [CrossRef]
- Doi, Y.; Murray, G.L.; Peleg, A.Y. Acinetobacter baumannii: Evolution of antimicrobial resistance-treatment options. Semin. Respir. Crit. Care Med. 2015, 36, 85–98. [Google Scholar] [PubMed] [Green Version]
- Alegre-González, D.; Herrera, S.; Bernal, J.; Soriano, A.; Bodro, M. Disseminated Cryptococcus neoformans infection associated to COVID-19. Med. Mycol. Case Rep. 2021, 34, 35–37. [Google Scholar] [CrossRef] [PubMed]
- Jandhyala, S.M.; Talukdar, R.; Subramanyam, C.; Vuyyuru, H.; Sasikala, M.; Nageshwar Reddy, D. Role of the normal gut microbiota. World J. Gastroenterol. 2015, 21, 8787–8803. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, A.; Harty, S.; Moeller, A.H.; Klein, S.L.; Erdman, S.E.; Friston, K.J.; Carmody, R.N. The gut microbiome as a biomarker of differential susceptibility to SARS-CoV-2. Trends Mol. Med. 2021, 27, 1115–1134. [Google Scholar] [CrossRef]
- De Maio, F.; Ianiro, G.; Coppola, G.; Santopaolo, F.; Abbate, V.; Bianco, D.M.; Del Zompo, F.; De Matteis, G.; Leo, M.; Nesci, A.; et al. Improved gut microbiota features after the resolution of SARS-CoV-2 infection. Gut. Pathog. 2021, 13, 62. [Google Scholar] [CrossRef]
- Zuo, T.; Zhang, F.; Lui, G.C.Y.; Yeoh, Y.K.; Li, A.Y.L.; Zhan, H.; Wan, Y.; Chung, A.C.K.; Cheung, C.P.; Chen, N.; et al. Alterations in gut microbiota of patients with COVID-19 during time of hospitalization. Gastrenterology 2020, 159, 944–955. [Google Scholar] [CrossRef]
- Rocchi, G.; Giovanetti, M.; Benedetti, F.; Borsetti, A.; Ceccarelli, G.; Zella, D.; Altomare, A.; Ciccozzi, M.; Guarino, M.P.L. Gut Microbiota and COVID-19: Potential Implications for Disease Severity. Pathogens 2022, 11, 1050. [Google Scholar] [CrossRef]
- Zuo, T.; Zhan, H.; Zhang, F.; Liu, Q.; Tso, E.Y.K.; Lui, G.C.Y.; Chen, N.; Li, A.; Lu, W.; Chan, P.K.S. Alterations in fecal fungal microbiome of patients with COVID-19 during time of hospitalization until discharge. Gastrenterology 2020, 159, 1302–1310. [Google Scholar] [CrossRef] [PubMed]
- Romani, L.; Del Chierico, F.; Macari, G.; Pane, S.; Ristori, M.V.; Guarrasi, V.; Gardini, S.; Pascucci, G.R.; Cotugno, N.; Perno, C.F.; et al. CACTUS Study Team. The Relationship Between Pediatric Gut Microbiota and SARS-CoV-2 Infection. Front. Cell Infect. Microbiol. 2022, 12, 908492. [Google Scholar] [CrossRef] [PubMed]
- CDC Post-COVID Conditions: Information for Healthcare Providers. Available online: https://www.cdc.gov/coronavirus/2019-ncov/hcp/clinical-care/post-covid-conditions.html (accessed on 22 September 2022).
- Chevinsky, J.R.; Tao, G.; Lavery, A.M.; Kukielka, E.A.; Click, E.S.; Malec, D.; Kompaniyets, L.; Bruce, B.B.; Yusuf, H.; Goodman, A.B. Late Conditions Diagnosed 1–4 Months Following an Initial Coronavirus Disease 2019 (COVID-19) Encounter: A Matched-Cohort Study Using Inpatient and Outpatient Administrative Data-United States, 1 March–30 June 2020. Clin. Infect. Dis. 2021, 73, S5–S16. [Google Scholar] [CrossRef] [PubMed]
- O’Laughlin, K.N.; Thompson, M.; Hota, B.; Gottlieb, M.; Plumb, I.D.; Chang, A.M.; Wisk, L.E.; Hall, A.J.; Wang, R.C.; Spatz, E.S.; et al. Study protocol for the Innovative Support for Patients with SARS-CoV-2 Infections Registry (INSPIRE): A longitudinal study of the medium and long-term sequelae of SARS-CoV-2 infection. PLoS ONE 2022, 17, e0264260. [Google Scholar] [CrossRef]
- Soriano, J.B.; Murthy, S.; Marshall, J.C.; Relan, P.; Diaz, J.V.; WHO Clinical Case Definition Working Group on Post-COVID-19 Condition. A clinical case definition of post-COVID-19 condition by a Delphi consensus. Lancet Infect. Dis. 2022, 22, e102–e107. [Google Scholar] [CrossRef]
- Carfì, A.; Bernabei, R.; Landi, F.; Gemelli Against COVID-19 Post-Acute Care Study Group. Persistent Symptoms in Patients After Acute COVID-19. JAMA 2020, 324, 603–605. [Google Scholar] [CrossRef]
- Peluso, M.J.; Deeks, S.G. Early clues regarding the pathogenesis of long-COVID. Trends Immunol. 2022, 43, 268–270. [Google Scholar] [CrossRef]
- WHO. Updates 3 & 4 in Relation to COVID-19 Coding in ICD-10. Available online: https://www.who.int/publications/m/item/updates-3-4-in-relation-to-covid-19-coding-in-icd-10 (accessed on 1 January 2021).
- Holms, R.D. Long COVID (PASC) Is Maintained by a Self-Sustaining Pro-Inflammatory TLR4/RAGE-Loop of S100A8/A9 > TLR4/RAGE Signalling, Inducing Chronic Expression of IL-1b, IL-6 and TNFa: Anti-Inflammatory Ezrin Peptides as Potential Therapy. Immuno 2022, 2, 512–533. [Google Scholar] [CrossRef]
- Queiroz, M.A.F.; Neves, P.F.M.D.; Lima, S.S.; Lopes, J.D.C.; Torres, M.K.D.S.; Vallinoto, I.M.V.C.; Bichara, C.D.A.; Dos Santos, E.F.; de Brito, M.T.F.M.; da Silva, A.L.S.; et al. Cytokine Profiles Associated With Acute COVID-19 and Long COVID-19 Syndrome. Front. Cell Infect. Microbiol. 2022, 12, 922422. [Google Scholar] [CrossRef]
- Ryan, F.J.; Hope, C.M.; Masavuli, M.G.; Lynn, M.A.; Mekonnen, Z.A.; Yeow, A.E.L.; Garcia-Valtanen, P.; Al-Delfi, Z.; Gummow, J.; Ferguson, C.; et al. Long-term perturbation of the peripheral immune system months after SARS-CoV-2 infection. BMC Med. 2022, 20, 26. [Google Scholar] [CrossRef]
- Tran, V.T.; Riveros, C.; Clepier, B.; Desvarieux, M.; Collet, C.; Yordanov, Y.; Ravaud, P. Development and Validation of the Long Coronavirus Disease (COVID) Symptom and Impact Tools: A Set of Patient-Reported Instruments Constructed From Patients’ Lived Experience. Clin. Infect. Dis. 2022, 74, 278–287. [Google Scholar] [CrossRef] [PubMed]
- Förster, M.; Weyers, V.; Küry, P.; Barnett, M.; Hartung, H.P.; Kremer, D. Neurological manifestations of severe acute respiratory syndrome coronavirus 2-a controversy ‘gone viral’. Brain Commun. 2020, 2, fcaa149. [Google Scholar] [CrossRef] [PubMed]
- Boldrini, M.; Canoll, P.D.; Klein, R.S. How COVID-19 Affects the Brain. JAMA Psychiatry 2021, 78, 682–683. [Google Scholar] [CrossRef] [PubMed]
- Nalbandian, A.; Sehgal, K.; Gupta, A.; Madhavan, M.V.; McGroder, C.; Stevens, J.S.; Cook, J.R.; Nordvig, A.S.; Shalev, D.; Sehrawat, T.S.; et al. Post-acute COVID-19 syndrome. Nat. Med. 2021, 27, 601–615. [Google Scholar] [CrossRef]
- Nath, A.; Smith, B. Neurological Complications of COVID-19: From Bridesmaid to Bride. Arq. Neuro-Psiquiatr. 2020, 78, 459–460. [Google Scholar] [CrossRef]
- Taquet, M.; Geddes, J.R.; Husain, M.; Luciano, S.; Harrison, P.J. 6-month neurological and psychiatric outcomes in 236 379 survivors of COVID-19: A retrospective cohort study using electronic health records. Lancet Psychiatry 2021, 8, 416–427. [Google Scholar] [CrossRef]
- Mao, L.; Jin, H.; Wang, M.; Hu, Y.; Chen, S.; He, Q.; Chang, J.; Hong, C.; Zhou, Y.; Wang, D.; et al. Neurologic Manifestations of Hospitalized Patients With Coronavirus Disease 2019 in Wuhan, China. JAMA Neurol. 2020, 77, 683–690. [Google Scholar] [CrossRef] [Green Version]
- Fortini, A.; Torrigiani, A.; Sbaragli, S.; Lo Forte, A.; Crociani, A.; Cecchini, P.; Innocenti Bruni, G.; Faraone, A. COVID-19: Persistence of symptoms and lung alterations after 3–6 months from hospital discharge. Infection 2021, 49, 1007–1015. [Google Scholar] [CrossRef]
- Liu, Q.; Mak, J.W.Y.; Su, Q.; Yeoh, Y.K.; Lui, G.C.; Ng, S.S.S.; Zhang, F.; Li, A.Y.L.; Lu, W.; Hui, D.S.; et al. Gut microbiota dynamics in a prospective cohort of patients with post-acute COVID-19 syndrome. Gut 2022, 71, 544–552. [Google Scholar] [CrossRef]
- Sohn, K.M.; Lee, S.G.; Kim, H.J.; Cheon, S.; Jeong, H.; Lee, J.; Kim, I.S.; Silwal, P.; Kim, Y.J.; Paik, S.; et al. COVID-19 Patients Upregulate Toll-like Receptor 4-mediated Inflammatory Signaling That Mimics Bacterial Sepsis. J. Med. Sci. 2020, 35, e343. [Google Scholar] [CrossRef]
- Pujani, M.; Raychaudhuri, S.; Verma, N.; Kaur, H.; Agarwal, S.; Singh, M.; Jain, M.; Chandoke, R.K.; Singh, K.; Sidam, D.; et al. Association of Hematologic biomarkers and their combinations with disease severity and mortality in COVID-19—An Indian perspective. Am. J. Blood Res. 2021, 11, 180–190. [Google Scholar] [PubMed]
- Mohamed Khosroshahi, L.; Rezaei, N. Dysregulation of the immune response in coronavirus disease 2019. Cell Biol. Int. 2021, 45, 702–707. [Google Scholar] [CrossRef] [PubMed]
- Rebendenne, A.; Valadão, A.L.C.; Tauziet, M.; Maarifi, G.; Bonaventure, B.; McKellar, J.; Planès, R.; Nisole, S.; Arnaud-Arnould, M.; Moncorgé, O.; et al. SARS-CoV-2 triggers an MDA-5-dependent interferon response which is unable to control replication in lung epithelial cells. J. Virol. 2021, 95, e02415-20. [Google Scholar] [CrossRef] [PubMed]
- Hadjadj, J.; Yatim, N.; Barnabei, L.; Corneau, A.; Boussier, J.; Smith, N.; Péré, H.; Charbit, B.; Bondet, V.; Chenevier-Gobeaux, C.; et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science 2020, 369, 718–724. [Google Scholar] [CrossRef]
- Chen, P.; Tang, Y.; He, W.; Yang, R.; Lan, Z.; Chen, R.; Zhang, P. Potential Pathophysiological Mechanisms Underlying Multiple Organ Dysfunction in Cytokine Release Syndrome. Mediat. Inflamm. 2022, 2022, 7137900. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.H.; Zhou, H.W.; Wu, W.K.; Fu, T.; Yan, M.; He, Z.; Sun, S.W.; Ji, Z.H.; Shao, Z.J. Alterations in the Composition of Intestinal DNA Virome in Patients With COVID-19. Front. Cell Infect. Microbiol. 2021, 11, 790422. [Google Scholar] [CrossRef]
- Cao, J.; Wang, C.; Zhang, Y.; Lei, G.; Xu, K.; Zhao, N.; Lu, J.; Meng, F.; Yu, L.; Yan, J.; et al. Integrated gut virome and bacteriome dynamics in COVID-19 patients. Gut Microbes 2021, 13, 1887722. [Google Scholar] [CrossRef]
- Bai, G.H.; Lin, S.C.; Hsu, Y.H.; Chen, S.Y. The Human Virome: Viral Metagenomics, Relations with Human Diseases, and Therapeutic Applications. Viruses 2022, 14, 278. [Google Scholar] [CrossRef]
- Frank, J.A.; Feschotte, C. Co-Option of endogenous viral sequences for host cell function. Curr. Opin. Virol. 2017, 25, 81–89. [Google Scholar] [CrossRef]
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [Google Scholar]
- Wang, W.; Jovel, J.; Halloran, B.; Wine, E.; Patterson, J.; Ford, G.; O’Keefe, S.; Meng, B.; Song, D.; Zhang, Y.; et al. Metagenomic Analysis of Microbiome in Colon Tissue from Subjects with Inflammatory Bowel Diseases Reveals Interplay of Viruses and Bacteria. Inflamm. Bowel. Dis. 2015, 21, 1419–1427. [Google Scholar] [CrossRef] [PubMed]
- Balestrieri, E.; Minutolo, A.; Petrone, V.; Fanelli, M.; Iannetta, M.; Zordan, M.; Vitale, P.; Charvet, B.; Horvat, B. Evidence of the pathogenic HERV-W envelope expression in T lymphocytes in association with the respiratory outcome of COVID-19 patients. EBioMedicine 2021, 66, 103341. [Google Scholar] [CrossRef] [PubMed]
- Kitsou, K.; Kotanidou, A.; Paraskevis, D.; Karamitros, T.; Katzourakis, A.; Tedder, R.; Hurst, T.; Sapounas, S.; Kotsinas, A.; Gorgoulis, V.; et al. Upregulation of Human Endogenous Retroviruses in Bronchoalveolar Lavage Fluid of COVID-19 Patients. Microbiol. Spectr. 2021, 9, e0126021. [Google Scholar] [CrossRef] [PubMed]
- Temerozo, J.R.; Fintelman-Rodrigues, N.; Dos Santos, M.C.; Hottz, E.D.; Sacramento, C.Q.; de Paula Dias da Silva, A.; Mandacaru, S.C.; Dos Santos Moraes, E.C.; Trugilho, M.R.O.; Gesto, J.S.M.; et al. Human endogenous retrovirus K in the respiratory tract is associated with COVID-19 physiopathology. Microbiome 2022, 10, 65. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Wen, X.L. Gut microbiota and inflammatory bowel disease: The current status and perspectives. World J. Clin. Cases. 2021, 9, 321. [Google Scholar] [CrossRef]
- Dopkins, N.; O’Mara, M.M.; Singh, B.; Marston, J.L.; Bendall, M.L.; Nixon, D.F. How human endogenous retroviruses interact with the microbiota in health and disease. Trends Microbiol. 2022, 30, 812–815. [Google Scholar] [CrossRef]
- Küry, P.; Nath, A.; Créange, A. Human endogenous retroviruses in neurological diseases. Trends Mol. Med. 2018, 24, 379–394. [Google Scholar] [CrossRef] [Green Version]
- Matteucci, C.; Balestrieri, E.; Argaw-Denboba, A.; Sinibaldi-Vallebona, P. Human endogenous retrovirusesrole in cancer cell stemness. Semin. Cancer Biol. 2018, 53, 17–30. [Google Scholar] [CrossRef]
- Zubair, A.S.; McAlpine, L.S.; Gardin, T.; Farhadian, S.; Kuruvilla, D.E.; Spudich, S. Neuropathogenesis and Neurologic Manifestations of the Coronaviruses in the Age of Coronavirus Disease 2019: A Review. JAMA Neurol. 2020, 77, 1018–1027. [Google Scholar] [CrossRef]
- Cryan, J.F.; O’Riordan, K.J.; Cowan, C.S.M.; Sandhu, K.V.; Bastiaanssen, T.F.S.; Boehme, M.; Codagnone, M.G.; Cussotto, S.; Fulling, C.; Golubeva, A.V.; et al. The Microbiota-Gut-Brain Axis. Physiol. Rev. 2019, 99, 1877–2013. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fanelli, M.; Petrone, V.; Buonifacio, M.; Delibato, E.; Balestrieri, E.; Grelli, S.; Minutolo, A.; Matteucci, C. Multidistrict Host–Pathogen Interaction during COVID-19 and the Development Post-Infection Chronic Inflammation. Pathogens 2022, 11, 1198. https://doi.org/10.3390/pathogens11101198
Fanelli M, Petrone V, Buonifacio M, Delibato E, Balestrieri E, Grelli S, Minutolo A, Matteucci C. Multidistrict Host–Pathogen Interaction during COVID-19 and the Development Post-Infection Chronic Inflammation. Pathogens. 2022; 11(10):1198. https://doi.org/10.3390/pathogens11101198
Chicago/Turabian StyleFanelli, Marialaura, Vita Petrone, Margherita Buonifacio, Elisabetta Delibato, Emanuela Balestrieri, Sandro Grelli, Antonella Minutolo, and Claudia Matteucci. 2022. "Multidistrict Host–Pathogen Interaction during COVID-19 and the Development Post-Infection Chronic Inflammation" Pathogens 11, no. 10: 1198. https://doi.org/10.3390/pathogens11101198
APA StyleFanelli, M., Petrone, V., Buonifacio, M., Delibato, E., Balestrieri, E., Grelli, S., Minutolo, A., & Matteucci, C. (2022). Multidistrict Host–Pathogen Interaction during COVID-19 and the Development Post-Infection Chronic Inflammation. Pathogens, 11(10), 1198. https://doi.org/10.3390/pathogens11101198






